Abstract
Functional genomics requires structural and functional studies of a large number of proteins. While the production of proteins through over-expression in cultured cells is a relatively routine procedure, the subsequent protein purification from the cell lysate often represents a significant challenge. The most direct way of protein purification from a cell lysate is affinity purification using an affinity probe to the target protein. It is extremely difficult to develop antibodies, classical affinity probes, for a protein in the cell lysate; their development requires a pure protein. Thus, isolating the protein from the cell lysate requires antibodies, while developing antibodies requires a pure protein. Here we resolve this loop problem. We introduce AptaPIC, Aptamer-facilitated Protein Isolation from Cells, a technology that integrates (i) the development of aptamers for a protein in cell lysate and (ii) the utilization of the developed aptamers for protein isolation from the cell lysate. Using MutS protein as a target, we demonstrate that this technology is applicable to the target protein being at an expression level as low as 0.8% of the total protein in the lysate. AptaPIC has the potential to considerably speed up the purification of proteins and, thus, accelerate their structural and functional studies.
INTRODUCTION
As we entered the post-genomic era, the focus has shifted from the analysis of genetic sequences to structural and functional studies of the proteins they encode. The fast progress in such studies requires the production of a large number of proteins in a pure form. Recombinant proteins are typically produced by their over-expression in cultured cells followed by affinity purification from the cell lysate (1). While protein expression in cells is a well developed and relatively generic procedure, the affinity purification of the protein from the cell lysate remains a lengthy and labor-intensive procedure, which limits the production of new proteins.
Usually for the purpose of affinity purification, a protein is genetically fused with a generic affinity tag and the resultant fusion protein (the protein plus the tag) is isolated from the cell lysate using an affinity probe for the tag (2). It is never clear a priori whether the tag will affect the protein. The exogenous nature of the tag may introduce unpredictable changes such as: changes in protein conformation that can lead to alteration of the protein's biological activity, decreased protein yield and toxicity (3). Moreover, it is often difficult to efficiently cleave the tag without fragmenting the target protein. Using the tag would be unnecessary if an affinity probe, such as an antibody, was available for the protein itself. However, antibodies are usually developed for a purified protein; development of antibodies for a protein in cell lysate is so laborious and time consuming that this possible approach did not find a wide application, if any (4).
Aptamers are synthetic oligonucleic acid molecules that bind their targets with high affinity and specificity (5–7), and can substitute antibodies in numerous applications (8–11). DNA aptamers are affinity ligands selected in vitro by different affinity methods from large combinatorial libraries of random DNA sequences (naive libraries) (12). Briefly, a naive DNA library is mixed with a target protein. DNA bound to the target is separated from unbound DNA and amplified by PCR to produce an aptamer-enriched DNA library. The procedure is repeated several times (rounds) starting with the enriched library obtained in the previous round. When affinity of the enriched library to the target stops improving with additional rounds of enrichment, the selection is stopped and an enriched DNA library with the best affinity to the target is considered as a pool of aptamers. The above-described procedure for selection of DNA aptamers is often called systematic evolution of ligands by exponential enrichment (SELEX). Here we use terms ‘aptamers’ and ‘DNA aptamers’ interchangeably. We also use interchangeably terms ‘a round of aptamer selection’ and ‘a round of library enrichment’. Aptamers have been previously selected for a variety of targets ranging from metal ions (13), to whole cells (14), and have proven to be dependable even in applications where antibodies have failed (such as detection of a protein in an ultra-wide concentration range or identification of cell-specific biomarkers) (15–17).
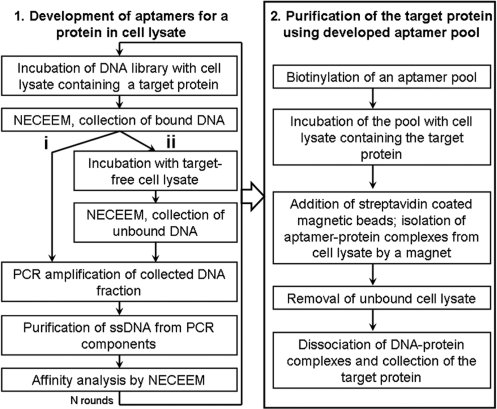
If aptamers are selected for a protein target in a cell lysate, they can be immediately used as affinity probes for purification of the target protein from the cell lysate. This is especially important for the purification of new proteins for which established purification procedures are not available. To the best of our knowledge, aptamers have never been developed from a naive DNA library for a specific full-sizes protein in a complex mixture, such as cell lysate, without any prior purification of the protein. Here, we introduce a new integrated procedure for: (i) the development of DNA aptamers for a protein target in cell lysate and (ii) application of the developed aptamer pool for purification of the target protein from the cell lysate. We name this technique AptaPIC—Aptamer facilitated Protein Isolation from Cells (Figure 1). In the first part of AptaPIC, an aptamer pool is selected for a target protein that is present in a cell lysate without any prior purification of the target protein. In the second part, the developed aptamer pool, containing a very large number of individual aptamers, is used as an affinity probe for purification of the target protein from cell lysate (there is no need to sequence individual aptamers).
Figure 1.
Aptamer-facilitated protein isolation from cells (AptaPIC). (i) denotes aptamer selection by consecutive rounds of positive selection. (ii) denotes aptamer selection with alternating positive and negative selection.
In this proof-of-principle work, we used T. aquaticus MutS, a recombinant prokaryotic protein expressed in Escherichia coli (1), as a target for aptamer selection from the E. coli cell lysate. While one may think that a protein that has DNA binding as its biological function may be more susceptible to aptamer development than a protein that does not have such a function, we argue that this assumption does not have sufficient evidence. In fact, the literature data suggests that DNA/RNA-binding proteins are no better targets for aptamer development than non-DNA/RNA-binding proteins (see Supplementary Data).
A recombinant protein expressed in E. coli may constitute up to 40–50% of the total bacterial protein (18). Depending on the nature of the protein none, some, or most of the recombinant protein may be present in insoluble aggregates—inclusion bodies. T. aquaticus MutS, the target of our selection, was previously recombinantly expressed in E. coli at a high level; the vast majority of which was shown to be in the soluble fraction (1). To provide a more challenging scenario of low-level expression, we demonstrated that library enrichment for binding MutS could be achieved with MutS being only at a level of 0.8% of the total protein in the cell lysate. To have this low and controlled level of MutS in the cell lysate, we added T. aquaticus MutS, recombinantly expressed in E. coli and purified from the E. coli cell lysate, to the MutS-free E. coli cell lysate. Adding MutS into the E. coli cell lysate rather than expressing it, is not expected to have an adverse effect because the protein was originally expressed in E. coli. Moreover, it provided us with the flexibility required for the study of aptamer selection for MutS present in the cell lysate at different levels.
Aptamers have been previously developed for complex targets such as mixtures of proteins and intact cells (17,19–21), but never before for a target in a whole-cell lysate. Cell lysate contains three groups of proteins that can interfere with aptamer selection. The first group is proteases and nucleases that can degrade the target protein and aptamers, respectively. The second group is DNA-binding proteins that can ruin the selection by nonspecific binding to DNA and contaminating the obtained pools with non-aptamers. The third group is highly abundant non-target proteins, which could become predominant targets in aptamer selection. The first two groups of proteins do not interfere with aptamer selection when using intact cells as targets. Their presence in cell lysate, however, makes aptamer selection for a target protein in cell lysate considerably more challenging than selection of aptamers for cell-surface proteins on intact cells. Here, we demonstrate that aptamers can be selected for a protein in the cell lysate if the above-listed problems are solved. We also propose two ways of obtaining target-specific aptamers for a protein in cell lysate. In the first way, target-specific aptamers are obtained in consecutive rounds of positive selection; this is possible if the target protein is the predominant one in the cell lysate. In the second way, target-specific aptamers are obtained by a combination of regular (positive) selection with negative selection against a target-free cell lysate. The latter removes non-target-specific aptamers. For a level of 5% MutS, we conducted a complete AptaPIC process. After five consecutive rounds of positive aptamer selection, we obtained an aptamer-enriched DNA pool with high affinity to MutS and low affinity to the cell lysate. Finally, we demonstrated the affinity purification of MutS from the cell lysate using the developed aptamer pool as an affinity probe.
MATERIALS AND METHODS
Chemicals and materials
Thermostable DNA mismatch binding protein (MutS) from Thermus aquaticus was kindly donated by InterSciences (Markham, ON, Canada). Fused-silica capillaries were purchased from Polymicro (Phoenix, AZ). All solutions were prepared using the Milli-Q-quality deionized water and filtered through a 0.22 µm filter (Millipore, Nepean, ON, Canada). Recombinant Taq DNA polymerase, buffer components and all other chemicals were from Sigma-Aldrich (Oakville, ON, Canada) unless otherwise stated.
DNA library, primers, scrambled DNA and masking DNA
The naive DNA library contained a central randomized sequence of 40 nucleotides (nt) flanked by 20-nt primer hybridization sites (5′-/FAM/-CTC CTC TGA CTG TAA CCA CG-(N)40-GCA TAG GTA GTC CAG AAG CC-3′). A 6-carboxyfluorescein-labeled 5′-primer (5′-/FAM/-CTC CTC TGA CTG TAA CCA CG-3′) and a biotinylated 3′-primer (5′-/Bio/-GGC TTC TGG ACT ACC TAT GC-3′) were used in PCR reactions for the synthesis of double-labeled double-stranded DNA molecules. For the purpose of biotinylating aptamers in the pool, biotin-labeled 5′-primer (5′-/Bio/-CTC CTC TGA CTG TAA CCA CG-3′) and non-labeled 3′-primer (5′-GGC TTC TGG ACT ACC TAT GC-3′) were used in the PCR reaction. The DNA library, PCR primers, scrambled DNA (5′-/FAM/-CTT CTG CCC GCC TCC TTC CTG GTA AAG TCA TTA ATA GGT GTG GGG TGC CGG GCA TTT CGG AGA CGA GAT AGG CGG ACA CT-3′), single stranded masking DNA (5′-CAA AAA ATG AGT CAT CCG GA-3′) and double stranded masking DNA (5′-ATT ATT ATT ATT ATT ATT ATT ATT ATT ATT ATT ATT ATT ATT ATT ATT ATT ATT AT-3′ and the complementary strand) were custom-synthesized by IDT (Coraville, IA).
Cell lysate preparation
E. coli BL21(DE3) were grown at 37°C to a density corresponding to an absorbance of 1.5–1.6 at 600 nm measured in a cuvette with an optical path-length of 1 cm. Cells were harvested by centrifugation at 5000 × g for 10 min at 4°C. Pelleted cells were resuspended in the sonication buffer: 50 mM Tris–HCl, 2.5 mM MgCl2, 5 mM KCl at pH 8.3 containing the protease inhibitors (PI) cocktail as instructed by the manufacturer (Sigma Aldrich, Oakville, ON, Canada). Bacterial lysates were prepared by sonication on ice with 5 s ‘on’/15-s ‘off’ intervals for a total of 15 min. Cell debris was pelleted by centrifugation at 15 000 × g for 20 min at 4°C and removed. Cell lysates were aliquoted and stored at –20°C. The concentration of ‘total’ protein in the cell lysate was measured by Bradford assay, using bovine serum albumin as a standard.
Inhibition of proteolysis
To monitor the proteolysis of MutS in the cell lysate, the lysate was prepared as described above, without addition of the protease (PI) cocktail into the sonication buffer. Sample containing 54 µg/ml MutS, cell lysate (containing 972 µg/ml of the total protein) and 12 µg/ml trypsin was incubated 3 h at 45°C with or without the addition of the PI cocktail as instructed by the manufacturer. The samples were analyzed on 15% SDS–PAGE with Coomassie Blue staining.
Inhibition of DNA degradation, gel-capillary electrophoresis (Gel-CE)
Samples containing 30 nM fluorescently labeled scrambled DNA and cell lysate (containing 365 µg/ml of the total protein) were incubated for 15 min at 25°C with or without addition of EGTA and 300 nM masking DNA. The samples were analyzed by gel capillary electrophoresis (eCAP ssDNA 100-R Kit, Beckman Coulter, Mississauga, ON, Canada) using a P/ACE MDQ apparatus (Beckman Coulter, Mississauga, ON, Canada) equipped with a fluorescence detector; a 488 nm line of an Ar-ion laser was utilized to excite fluorescence.
Non-equilibrium capillary electrophoresis of equilibrium mixtures (NECEEM)
All NECEEM procedures were performed using the following instrumental setup. CE was carried out with a P/ACE MDQ apparatus (Beckman Coulter, Mississauga, ON, Canada) equipped with a fluorescence detector; a 488 nm line of an Ar-ion laser was utilized to excite fluorescence. Uncoated fused silica capillaries with an inner diameter of 75 µm and outer diameter of 360 µm were used. For the aptamer selection, an 80 cm-long (70 cm to the detection window) capillary was utilized. NECEEM-facilitated analysis of enriched DNA libraries was carried in a 50 cm-long (40 cm to the detection window) capillary.
Both the inlet and the outlet reservoirs contained the electrophoresis run buffer (50 mM Tris–acetate at pH 8.3). The samples were injected into the capillary, pre-filled with the run buffer, by a pressure pulse of 16 s × 1 psi (13.4 kPa) and 5 s × 0.5 psi (3.4 kPa) for aptamer selection and for NECEEM-facilitated affinity analysis, respectively. The length and the volume of the corresponding sample plugs were 23 mm and 101 nl for aptamer selections and 6 mm and 26 nl for NECEEM-facilitated affinity analysis, respectively. Electrophoresis was carried out with a positive electrode at the injection end of the capillary; the direction of the electroosmotic flow was from the inlet to the outlet reservoir. Separation for aptamer selection was carried out by an electric field of 250 V/cm; NECEEM affinity analysis was performed at 400 V/cm. The temperature of the capillary during the separation was maintained at 20°C. When needed, fractions were collected in an automated mode by replacing the regular outlet reservoir with a fraction collection vial containing 5 µL of deionized water.
The capillary was rinsed with the run buffer solution for 2 min prior to each run. At the end of each run, the capillary was rinsed with 100 mM HCl for 2 min and 100 mM NaOH for 2 min, followed by a rinse with deionized water for 2 min. After the fraction collection step, the capillary was rinsed with RNase-away solution for 1 min to remove any remnants of DNA from the capillary.
Selection of aptamers for protein target in cell lysate by consecutive rounds of positive selection
The equilibrium mixture for NECEEM-based selection of aptamers was prepared in the incubation buffer (50 mM Tris–HCl, 2.5 mM MgCl2, 5 mM KCl and 5 mM EGTA at pH 8.3) using the following two-step procedure. First, a DNA library (an 88 µM solution in the selection buffer) was denatured by heating at 99°C for 5 min and immediately transferred on ice. Second, 1 µl of 88 µM naive DNA library was mixed with 40 µM single stranded masking DNA, 50 nM fluorescein, cell lysate (containing 178 µg/ml of the total protein) and a known concentration of MutS (see section ‘Selection of Aptamers for MutS Present in Cell Lysate at Different Levels’). Finally the mixture was incubated at 25°C for 15 min to obtain the equilibrium mixture.
A 23 mm-long (101 nl) plug of the equilibrium mixture was injected into the capillary pre-filled with the run buffer; the plug contained approximately 5 × 1012 molecules of DNA. The injected equilibrium mixture was subjected to NECEEM. In the beginning of NECEEM, the electrophoresis was carried out with the outlet reservoir containing 5 µl of deionized water to collect bound fraction of DNA. Fluorescein was used as a marker of the right-hand side boundary of the fraction collection window; the outlet reservoir was changed back to the run buffer as soon as fluorescein eluted from the capillary. A fraction of DNA of approximately 4 µl was collected and amplified by PCR. In addition to the collected DNA template, the PCR mixtures contained 50 mM KCl, 10 mM Tris–HCl (at pH 8.6), 2.5 mM MgCl2, all four dNTPs (200 µM each), primers (300 nM each), and 0.05 unit/µl Taq DNA polymerase. The total volume of the PCR reaction mixture was 50 µl. The optimum number of cycles for amplification was determined by real-time PCR as described elsewhere (22). The optimum number of PCR cycles (14 cycles for the first round of aptamer selection) was conducted, with every cycle consisting of melting at 94°C for 30 s, annealing at 56°C for 15 s, and extension at 72°C for 15 s. The product of symmetrical PCR was used as a template in asymmetrical PCR to produce single stranded DNA. The asymmetrical PCR mixture contained 50 mM KCl, 10 mM Tris–HCl (at pH 8.6), 2.5 mM MgCl2, all four dNTPs (200 µM each), 50 nM biotinylated reverse primer, 1 µM fluorescently labeled forward primer, 0.05 unit/µl Taq DNA polymerase, and 3 µl of the symmetrical PCR product. The total volume of the PCR reaction mixture was 50 µl. Fifteen cycles of PCR were carried out following the same temperature protocol as for symmetrical PCR. Double-stranded DNA produced in asymmetric PCR was removed using streptavidin-coated magnetic beads (Pierce, Rockford, IL). After pulling down the magnetic beads, the supernatant was transferred onto a 30 kDa molecular cut-off filter (Millipore, Nepean, ON, Canada) to separate the excess of primers from the PCR product. The obtained enriched DNA libraries were subjected to NECEEM-facilitated evaluation of their affinity towards MutS-free cell lysate and MutS.
The subsequent rounds of aptamer selection were carried out following the same procedure with 1 µl of enriched DNA library (concentration ∼200 nM) used instead of the naive DNA library and concentration of masking DNA lowered to 300 nM (instead of 40 µM) in the equilibrium mixture.
NECEEM-facilitated analysis of obtained enriched DNA libraries
Progression of aptamer selection was monitored by NECEEM-facilitated analysis of affinities of the enriched DNA libraries to MutS or MutS-free cell lysate. One microliter of the obtained enriched DNA library (at a concentration of approximately 200 nM) was mixed with 300 nM single-stranded masking DNA, the indicated concentration of MutS or cell lysate (containing 391 µg/ml of the total protein) in a total volume of 5 µl. The equilibrium mixture was incubated for 15 min at 25°C. A 5 mm-long (26 nl) plug of the equilibrium mixture was injected by pressure into the capillary pre-filled with the run buffer and was subject to electrophoresis at 400 V/cm. Affinities of the obtained enriched DNA libraries for MutS were calculated as described elsewhere (23) (see Supplementary Data).
Selection of aptamers for MutS present in cell lysate at different levels
In order to understand what was the lowest ratio of target protein to total protein in the sample that could still allow selection of aptamers without the need of negative selection, a series of aptamer selections were performed for samples containing a fixed amount of the cell lysate and different amounts of MutS. The equilibrium mixtures were prepared as described above with addition of 100, 60, 40, 20, 16, 10, 1 or 0.1 nM MutS to result in 5%, 3%, 2%, 1%, 0.8%, 0.5%, 0.05% or 0.005% of the target protein from total sample protein, respectively. The aptamer selections and analysis of the obtained enriched DNA libraries were carried out essentially as described in the section ‘Selection of Aptamers to Protein Target in Cell Lysate by Consecutive Rounds of Positive Selection’.
Protein isolation from cell lysate
The aptamer pool obtained from the fifth round of aptamer selection was used to purify MutS from the cell lysate. The aptamer pool was biotinylated in asymmetric PCR amplification in a mixture similar to that indicated above but containing 1 µM biotinylated forward primers and 50 nM non-labeled reverse primers. To each 150 µl of PCR mixture, 9 µl of the aptamer pool was added; 15 cycles of amplification were carried out as described above. For the control experiment, biotinylated naive DNA library was prepared by amplification of 9 µl of 50 nM initial naive library in the same procedure. Products of asymmetrical PCR were purified from primers on 30 kDa molecular weight cut-off filters. Because the double stranded product of PCR was also biotinylated, we could not purify the biotinylated ssDNA product from dsDNA.
The entire product of PCR was mixed with 40 µM single-stranded masking DNA, 5 µM double-stranded masking DNA, cell lysate (containing 91.25 µg/ml of the total protein), and 200 nM MutS in the incubation buffer. The mixture was incubated for 15 min at 25°C. Thirty microliters of 5 mg/ml streptavidin-coated magnetic beads were added to each of the samples and the mixture was incubated for another 10 min at 25°C. The magnetic beads with the attached aptamers and proteins were pulled down using a magnet and washed 3 times with the incubation buffer. The unbound fraction was combined with all three fractions collected after washings, concentrated by evaporation of water in a vacufuge (Vacufuge Concentrator 5301, Eppendorf, Germany) and analyzed on SDS–PAGE with Coomassie Blue staining. To elute the bound protein from aptamers, the bound fraction was re-suspended in the incubation buffer containing 6 M urea and incubated 15 min at 25°C. After the incubation, magnetic beads were pulled down by a magnet and supernatants of bound fractions were purified from urea by centrifugation on a 5 kDa molecular cut-off filter at 7000 r.p.m. for 15 min at 4°C and analyzed on 4–20% SDS–PAGE.
RESULTS AND DISCUSSION
Degradation of target protein and DNA in cell lysate
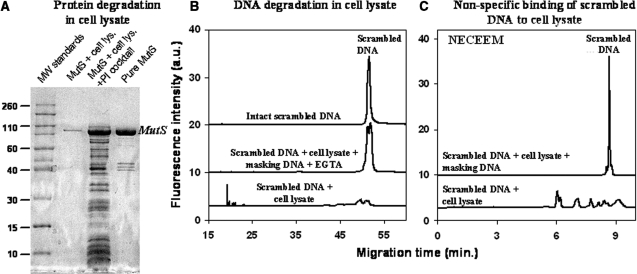
We first studied protein and aptamer degradation in the cell lysate. To monitor the integrity of MutS, we performed SDS–PAGE of MutS after its incubation with a cell lysate containing active and inhibited proteases. To monitor degradation of DNA by nucleases we used a fluorescently labeled synthetic 80-base-long oligonucleotide (the same length as aptamers) with a scrambled sequence. The integrity of the DNA was monitored by Gel-CE, which separates the original DNA molecules from their degradation products. We also artificially enhanced the proteolytic activity in the cell lysate by externally adding trypsin to the analyzed mixture. This was done to mimic a scenario more challenging than proteolytic degradation of MutS in the cell lysate. We showed that the addition of a commercially available PI cocktail (Sigma-Aldrich) to the cell lysate preserved MutS from degradation for 3 h, even in the presence of the external protease (Figure 2A). This time was enough to complete a round of aptamer selection; thus, the problem of protein degradation was resolved.
Figure 2.
Three processes that interfere with aptamer development for a target protein in cell lysate: protein degradation (A), DNA degradation (B) and binding of DNA by non-target components of cell lysate (C). (A) MutS degradation was analyzed by SDS–PAGE with Coomassie staining. The four lanes (from left to right) in the gel image correspond to: (i) molecular weight standards (×1000), (ii) MutS degraded by proteases in the cell lysate, (iii) MutS rescued from proteolysis in the cell lysate by a protease inhibitor (PI) cocktail, and (iv) pure MutS. The samples analyzed contained 972 µg/ml of cell lysate protein and 54 µg/ml of MutS. (B) DNA degradation was analyzed by gel capillary electrophoresis (Gel CE) using a scrambled 80-nt DNA as an experimental model. The three traces (from bottom to top) correspond to: (i) scrambled DNA degraded in cell lysate, (ii) scrambled DNA rescued from the degradation in the cell lysate by masking DNA and EGTA and (iii) scrambled DNA as a control. (C) Binding of DNA to non-target components of the cell lysate was studied by NECEEM using the scrambled DNA. The two traces (from bottom to top) in (C) correspond to: (i) scrambled DNA bound to non-target components of the cell lysate and/or partially degraded DNA and (ii) scrambled DNA rescued from binding and degradation by masking DNA. NECEEM was carried out in a 50-cm-long capillary at an electric field of 400 V/cm with 50 mM Tris–acetate at pH 8.3 as a run buffer.
Nuclease activity of the lysate, on the other hand, cannot be inhibited by standard commercial nuclease inhibitors. Standard nuclease inhibitors are not suitable, because they contain proteases or protein modifying chemicals that can degrade the target protein (in our aptamer selection, we need to protect from degradation both the target protein and DNA). An alternative way to inhibit nucleases is the addition of a chelating agent such as EGTA, and an excess of a masking DNA. EGTA suppresses nuclease activity by chelating Ca2+, which some nucleases require for their activity (24). The masking DNA is DNA without a fluorescent label and PCR primer regions that competitively protects aptamers from degradation by nucleases (17,25). When used together, EGTA and masking DNA kept about 90% of the aptamer intact for 3 h of incubation with cell lysate (Figure 2B). This 90% included DNA molecules that were missing only 1–2 nt (the split peak in Figure 2B, second trace from the top). The missing 1–2 nt do not prevent the annealing of the primers to the partially degraded DNA template; thus, this partially degraded DNA is amplifiable by PCR and can be used in aptamer selection. The nuclease activity can also be inhibited by solely using masking DNA if the addition of a chelating agent such as EGTA is undesirable; however, masking DNA should be taken in a greater excess to the library than demonstrated here.
Non-specific DNA binding in cell lysate
We studied binding of the scrambled DNA by non-target components of the cell lysate such as DNA-binding proteins. This binding could interfere with selection of target-specific aptamers as DNA molecules bound to non-target components of the cell lysate would be collected as ‘binders’ and amplified along with aptamers. The scavenging of aptamer by DNA-binding proteins can be suppressed by using of an excess of the masking DNA (17,25). The purpose of having an excess of the masking DNA is to ‘pre-occupy’ DNA-binding proteins and, thus, competitively suppress this non-specific binding. To monitor the DNA binding we analyzed a mixture of the scrambled DNA and MutS-free cell lysate by NECEEM (26) (Figure 2C). NECEEM separates molecular species based on their charge to size ratios. While being a powerful method for studying protein–DNA binding, NECEEM does not distinguish between products of DNA binding and products of DNA degradation as they may have charge to size ratios different from that of free/intact DNA. Accordingly, the multiple peaks observed in Figure 2C to the left of the free/intact DNA peak cannot be unambiguously assigned to complexes of DNA with non-target proteins from the cell lysate. They can also be products of DNA degradation. Nevertheless, the addition of a 10-time excess of masking DNA over the scrambled DNA suppressed DNA binding/degradation to the undetectable level.
Potential aptamer selection for other proteins in cell lysate
The last problem that can arise when selecting aptamers to a target protein in cell lysate is the possibility of aptamer selection for non-target molecules and obtaining an aptamer pool with poor affinity and specificity for the target protein. This problem is usually solved by alternating regular selection steps for a complex mixture containing the target protein with negative selection (counter-selection) steps for the sample containing all other components without the target protein (17). Essentially, negative selection eliminates aptamers for non-target molecules leaving only aptamers for the target molecule. Aptamers are known to be more readily developed for the most abundant proteins in the sample (27). Thus, we hypothesized that the enrichment of a DNA library will take place primarily with respect to the over-expressed protein and negative selection may not be required to achieve a desired specificity of aptamer pool to the target molecule. To test this hypothesis, we conducted five rounds of aptamer selection for the cell lysate containing 5% of MutS over the total protein.
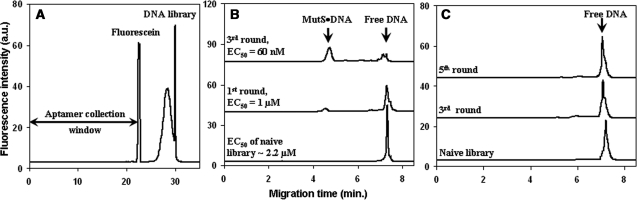
Aptamer selection starts with a naive DNA library. In our case, the fluorescently labeled 80-nt DNA library consisted of 40-nt random region flanked by 20-nt primer hybridization regions. The naive library was mixed with the cell lysate containing 5% MutS and incubated in the presence of the excess of masking DNA. The equilibrium mixture was subjected to NECEEM separation. We used NECEEM for the partitioning of bound DNA from unbound DNA. The separation mode of NECEEM used here was free zone capillary electrophoresis in an uncoated capillary in which both DNA and protein are moved towards the negative electrode by the electroosmotic flow (EOF). In addition to the EOF, charged molecules will have electrophoretic mobilities. The direction of the electrophoretic mobility of positively charged molecules is co-directed with that of the EOF. The direction of the electrophoretic mobility of negatively charged molecules is opposite to that of the EOF. DNA is heavily negatively charged due to its sugar-phosphate backbone and as a result, in free zone electrophoresis with positive electrode at the inlet, DNA migrates slower than the majority of proteins. Moreover, protein–DNA complexes also migrate between the free protein and unbound DNA. We used fluorescein as a marker of the right-hand boundary of the fraction collection window. Fluorescein is an ideal marker for this purpose because the unbound DNA library migrates slightly slower than fluorescein. Collecting a fraction up to fluorescein allows as wide a fraction collection window as possible ensuring, however, that little unbound DNA is collected. The entire fraction migrating faster than fluorescein was collected to obtain the bound DNA (Figure 3A). The collected DNA fraction was amplified. For this purpose, we used tandem PCR. The first PCR was symmetrical PCR with equal amounts of primers, the second—asymmetrical PCR where a forward primer was in 20-time excess over a reverse primer. After that, the PCR product was purified from double stranded DNA, primers, polymerase and dNTPs. This was the end of the first round of aptamer selection. In the second round, the enriched DNA library obtained in the first round was incubated with the MutS-containing cell lysate and the procedure described above was repeated. Five rounds of library enrichments were carried out. The enriched libraries obtained in every round were separately evaluated for their binding to pure MutS and to the components of MutS-free cell lysate using NECEEM. To characterize the affinity of the enriched DNA libraries to MutS, we used EC50 (an effective concentration of DNA at which a half of it is bound to the target protein) (17) instead of a conventional equilibrium dissociation constant, Kd. The latter can only be used for a single interacting pair. As EC50 is not a constant, it should always be accompanied by the concentration of a target protein at which it was measured.
Figure 3.
Selection of aptamers for MutS in the E. coli cell lysate by consecutive rounds of positive selection. (A) Aptamer collection window for NECEEM-based selection of aptamers. The equilibrium mixture was injected into the capillary and electrophoresis was carried out in an 80-cm-long capillary at a 250 V/cm electric field with 50 mM Tris–acetate at pH 8.3 as a run buffer. The entire fraction migrating before fluorescein was collected for PCR amplification of DNA contained in it. (B) Progression of selection with regards to MutS. A naive DNA library and enriched aptamer pools were incubated with pure MutS and NECEEM was used to estimate EC50 values for 220 nM MutS. The three traces (from bottom to top) correspond to the mixture of MutS with (i) naive library, (ii) enriched DNA library after one round of aptamer selection and (iii) enriched DNA library after three rounds of aptamer selection. (C) Progression of selection with regards to the MutS-free cell lysate. The naive DNA library and enriched aptamer pools were incubated with MutS-free cell lysate and analyzed by NECEEM. The three traces (from bottom to top) correspond to mixtures of MutS-free cell lysate with: (i) naive library, (ii) enriched DNA library after three rounds of aptamer selection and (iii) enriched DNA library after five rounds of aptamer selection. In (B) and (C), NECEEM was carried out in a 50-cm-long capillary at an electric field of 400 V/cm with 50 mM Tris–acetate at pH 8.3 as a run buffer.
In only three rounds of aptamer selection, the affinity of the enriched DNA library to MutS improved 40 times compared to that of the naive library (Figure 3B). Two additional rounds of enrichment did not further improve the affinity of the enriched libraries to MutS (results not shown). Interestingly, the affinity of the enriched library to the non-target components of the cell lysate remained unchanged even after five consecutive rounds of positive selection (Figure 3C). These results suggest that while we were able to develop aptamers for the target protein MutS, there was no accumulation of aptamers for non-target components of the cell lysate. The demonstrated experimental results indicate that when the target protein is present at an over-expressed level, counter-selection against the target-free cell lysate may be omitted because no enrichment of the library takes place with regards to the cell lysate components. One possible explanation for this observation is that the concentration of MutS is at least 10 times higher than that of the most abundant protein in the cell lysate (according to SDS–PAGE). Thus, MutS becomes a ‘leading’ target in the aptamer selection process while the contribution of non-target molecules to the obtained aptamer-enriched library becomes negligible.
The second way of obtaining target-specific aptamers is the introduction of a step of negative selection against a target-free cell lysate (17). Although for the specific case presented in this work, the step of negative selection was not required, as no enrichment of the library towards components of the cell lysate took place, this could be not true for different targets at different levels of expression. For cases when the amount of the target protein is not sufficiently high, it may be required to introduce the negative selection step in order to obtain target-specific aptamers (Supplementary Data).
Protein isolation from cell lysate
As the affinity of the enriched library did not significantly improve in the last two rounds of aptamer selection, the aptamer pool obtained after the fifth round of the selection was used for aptamer-mediated protein purification. Conventionally, after the aptamer pool is obtained, individual aptamer clones are isolated, analyzed for their affinity to the target molecule, and a clone with the best affinity is chosen for further applications. The cloning and screening steps are time-consuming (1–2 weeks) and expensive. Screening one hundred clones could cost more than a thousand dollars. Here, we propose skipping these steps and using the entire enriched aptamer pool for affinity purification of the target protein instead of individual aptamers.
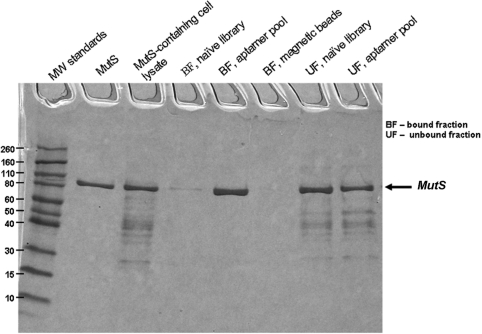
For the protein isolation, the aptamer pool was biotinylated in asymmetrical PCR amplification using a biotin-labeled forward primer. The product of PCR amplification was purified with a molecular cut-off filter. The final biotinylated pool contained ssDNA (more than 99%) and dsDNA (less than 1%). To prevent the non-specific binding of dsDNA to the components of the cell lysate in the protein purification step, we used the excess of masking dsDNA in addition to masking ssDNA. MutS-containing cell lysate was mixed with the biotinylated aptamer pool in the presence of excess masking ds/ss DNA and incubated for 15 min. Next, streptavidin-coated magnetic beads were added and the mixture was incubated for another 10 min followed by the pull-down of the beads by a magnet. The unbound fraction of the cell lysate (the waste) was removed and analyzed by SDS–PAGE. In the final step of AptaPIC, the target protein was dissociated from DNA aptamers and magnetic beads. There is no universal solution for this step. Depending on the application and the nature of the protein, the elution of the immobilized protein could be carried out in a number of ways: high temperature, treatment with high salt concentration (14), metal ion chelating agents (28), pH variation, ammonium sulfate precipitation of the protein, treatment with urea (17), nuclease degradation of aptamers, addition of an excess of a complementary strand of the aptamer and even trypsinization of the protein for its mass spectrometric identification. In our case, we chose to elute the protein using 6 M urea treatment of the aptamer-bound fraction. The bound fraction was incubated in 6 M urea for 15 min to dissociate aptamer-protein complexes. Urea was removed form the eluted fraction by size-exclusion filtration before the analysis of the fraction on SDS–PAGE (Figure 4). As a control, we showed that the biotinylated naive library did not isolate MutS from the cell lysate. The presence of the band, corresponding to MutS in the aptamer-bound fraction, demonstrated the affinity of the developed aptamers for MutS. The near absence of the bands, corresponding to all other proteins, proved that the developed aptamers were very specific towards MutS. MutS sample obtained as a result of purification with aptamers developed by AptaPIC was 99% pure; this represents 4.9-fold purification (maximum possible fold purification being 5.0). The percentage yield of MutS after one step of affinity purification using the developed aptamers was around 70%. We foresee that this percentage yield can be improved by using higher amount of aptamers in the purification step.
Figure 4.
SDS–PAGE with Coomassie staining of affinity pull-down of MutS from the E. coli cell lysate using the aptamers developed by AptaPIC. The lanes from left to right correspond to: (i) molecular weight standards (×1000); (ii) pure MutS; (iii) MutS-containing cell lysate, the sample contained 91.25 µg/ml and 18 µg/ml of MutS; (iv) MutS purification using the naive DNA library, bound fraction; (v) MutS purification using the developed aptamer pool, bound fraction; (vi) fraction bound to streptavidin-coated magnetic beads; (vii) MutS purification using the naive DNA library, unbound fraction; and (viii) MutS purification using the developed aptamer pool, unbound fraction.
While urea is a soft protein denaturant, it can denature some proteins irreversibly. To verify the activity of the purified MutS after urea treatment we tested MutS binding to its synthetic DNA aptamer that was previously obtained (29) (results not shown). Heat denatured MutS did not possess an affinity to its synthetic aptamer; MutS eluted by urea on the other hand showed the expected affinity to the synthetic aptamer, suggesting that the protein was in its native state. The aptamer-facilitated purification of MutS delivered the protein with high purity and in the native state. These results clearly demonstrate that AptaPIC can be reliably used for purification of a target protein from a cell lysate.
Selection of aptamers for protein target present at different levels in cell lysate
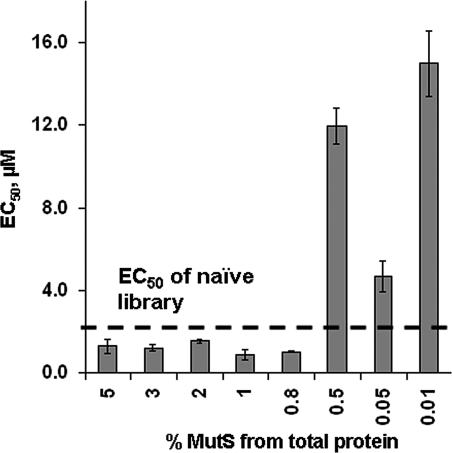
Here we demonstrated whole AptaPIC procedure on the example of 5% MutS in E. coli cell lysate without the need of introduction of the negative selection step. Omitting the step of negative selection significantly shortens the length of AptaPIC and adds some additional level of control over the procedure. However, to be able to omit the negative selection step, it is necessary that the target protein be at a high enough level of expression to become the predominant target of aptamer selection. To find out the lowest amount of MutS that allowed the development of aptamers without the need of negative selection, we performed a series of aptamer selections for cell lysate containing different amounts of MutS. Only the first round of aptamer selection (from the naive library) was performed for each sample and the affinities of the obtained enriched libraries to pure MutS were assessed by NECEEM. We considered that the development of aptamers for the target was possible if the affinity of the enriched library (obtained after one round of positive aptamer selection) to MutS was better than that of the naive library. In other words, the aptamer selection was possible if an EC50 value of the enriched library was lower than that of the naive library. The EC50 value for the naive library was approximately 2.2 µM for 220 nM MutS. The EC50 values of the enriched libraries obtained for different MutS concentrations in the cell lysate are presented in Figure 5. The lowest amount of MutS that resulted in the improvement of affinity of the enriched library to MutS compared to that of the naive library was 0.8% MutS with respect to the total protein in the cell lysate. When the ratio of MutS to total protein was further lowered, aptamer selection for MutS did not succeed. In fact, EC50 values of the enriched libraries obtained for samples with less than 0.8% MutS were higher than that of the naive library. The most likely explanation for the latter observation is that the aptamer selection took place for non-target components of the cell lysate and decreased the affinity of the obtained enriched library to the MutS protein. The results demonstrated in Figure 5 suggest that aptamers could be obtained even for a relatively poorly expressed recombinant protein without the need of negative selection given that the target protein is the most abundant in the cell lysate. It should be noted that having a low amount of the target protein in the cell lysate may result in contamination of the aptamer pool with aptamers for non-target molecules. As a result, the protein purification using such aptamer pool can be compromised.
Figure 5.
EC50 values for the interaction of MutS with enriched libraries obtained after one round of aptamer selection for cell lysate containing MutS at indicated percentage levels with respect to the total protein in the lysate. All EC50 values were measured for a MutS concentration of 220 nM. Errors are those of EC50 measurements and not the selection.
CONCLUSIONS
Here we introduced a method for developing aptamers for a target protein in crude cell lysate and purification of the target protein from the cell lysate using the obtained aptamers. The feasibility of the method was demonstrated on T. aquaticus MutS in E. coli cell lysate, however, we foresee that this method can be extended to application for any other protein for which the development of aptamers is possible. To conclude, we outline major features of AptaPIC. Using the AptaPIC procedure, aptamers can be developed for a protein, which is not available in a pure form. The developed aptamers can be used for purification of the protein from a complex mixture. For the successful generation of aptamers in a cell lysate, we found solutions for: (i) the protection of the target protein and potential aptamers from degradation by cell lysate proteases and nucleases and (ii) the suppression of non-specific DNA binding of lysate proteins to aptamers. As demonstrated here, if the target protein is present at the level of over-expression, aptamers can be successfully generated in consecutive rounds of positive selection without negative selections. If the amount of the target protein is not sufficiently high, the step of negative selection can be added into the procedure. The developed aptamers can then be used in the affinity purification of the target protein from the cell lysate. Considering the extreme complexity of cell lysate, AptaPIC can be extended to potentially any biological mixture such as blood, urine and others. AptaPIC facilitates the faster generation of new recombinant proteins in pure form for their structural and functional studies as well as for other biomedical applications. Moreover, due to easier and lower cost of their production, the aptamers obtained by AptaPIC can be used as antibody surrogates in applications such as western analysis, fluorescent microscopy, and flow cytometry.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Biswas I, Hsieh P. Identification and characterization of a thermostable MutS homolog from Thermus aquaticus. J. Biol. Chem. 1996;271:5040–5048. doi: 10.1074/jbc.271.9.5040. [DOI] [PubMed] [Google Scholar]
- 2.Koles K, Irvine KD, Panin VM. Functional characterization of Drosophila sialyltransferase. J. Biol. Chem. 2004;279:4346–4357. doi: 10.1074/jbc.M309912200. [DOI] [PubMed] [Google Scholar]
- 3.Arnau J, Lauritzen C, Petersen GE, Pedersen J. Current strategies for the use of affinity tags and tag removal for the purification of recombinant proteins. Protein Express. Purif. 2006;48:1–13. doi: 10.1016/j.pep.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 4.Blecher SR, Howie R, Li S, Detmar J, Blahut LM. A new approach to immunological sexing of sperm. Theriogenology. 1999;52:1309–1321. doi: 10.1016/s0093-691x(99)00219-8. [DOI] [PubMed] [Google Scholar]
- 5.Ellington AD, Szostak JW. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 6.Wochner A, Menger M, Orgel D, Cech B, Rimmele M, Erdmann VA, Glokler J. A DNA aptamer with high affinity and specificity for therapeutic anthracyclines. Anal. Biochem. 2008;373:34–42. doi: 10.1016/j.ab.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 7.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 8.Stojanovic MN, Kolpashchikov DM. Modular aptameric sensors. J. Am. Chem. Soc. 2004;126:9266–9270. doi: 10.1021/ja032013t. [DOI] [PubMed] [Google Scholar]
- 9.Huang YC, Ge B, Sen D, Yu HZ. Immobilized DNA switches as electronic sensors for picomolar detection of plasma proteins. J. Am. Chem. Soc. 2008;130:8023–8029. doi: 10.1021/ja8011066. [DOI] [PubMed] [Google Scholar]
- 10.Bruno JG, Carrillo MP, Crowell R. Preliminary development of DNA aptamer-Fc conjugate opsonins. J. Biomed. Mater. Res. A. 2008 doi: 10.1002/jbm.a.32182. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 11.McNamara JO, Kolonias D, Pastor F, Mittsler RS, Chen L, Giangrande PH, Sullenger B, Gilboa E. Multivalent 4–1BB binding aptamers costimulate CD8 + T cells and inhibit tumor growth in mice. J. Clin. Invest. 2008;118:376–386. doi: 10.1172/JCI33365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pan W, Craven RC, Qiu Q, Wilson CB, Wills JW, Golovine S, Wang JF. Isolation of virus-neutralizing RNAs from a large pool of random sequences. Proc. Natl Acad. Sci. USA. 1995;92:11509–11513. doi: 10.1073/pnas.92.25.11509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciesiolka J, Yarus M. Small RNA-divalent domains. RNA. 1996;2:785–793. [PMC free article] [PubMed] [Google Scholar]
- 14.Daniels DA, Chen H, Hicke BJ, Swiderek KM, Gold L. A tenascin-C aptamer identified by tumor cell SELEX: systematic evolution of ligands by exponential enrichment. Proc. Natl Acad. Sci. USA. 2003;100:15416–15421. doi: 10.1073/pnas.2136683100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Drabovich AP, Okhonin V, Berezovski M, Krylov SN. Smart aptamers facilitate multi-probe affinity analysis of proteins with ultra-wide dynamic range of measured concentrations. J. Am. Chem. Soc. 2007;129:7260–721. doi: 10.1021/ja072269p. [DOI] [PubMed] [Google Scholar]
- 16.Hicke BJ, Stephens AW, Gould T, Chang YF, Lynott CK, Heil J, Borkowski S, Hilger CS, Cook G, Warren S, Schmidt PG. Tumor targeting by an aptamer [see comment] J. Nucl. Med. 2006;47:668–678. [PubMed] [Google Scholar]
- 17.Berezovski MV, Lechmann M, Musheev MU, Mak TW, Krylov SN. Aptamer-facilitated biomarker discovery (AptaBiD) J. Am. Chem. Soc. 2008;130:9137–9143. doi: 10.1021/ja801951p. [DOI] [PubMed] [Google Scholar]
- 18.Baneyx F, Mujacic M. Recombinant protein folding and misfolding in Escherichia coli. Nat. Biotechnol. 2004;22:1399–1408. doi: 10.1038/nbt1029. [DOI] [PubMed] [Google Scholar]
- 19.Raddatz MS, Dolf A, Endl E, Knolle P, Famulok M, Mayer G. Enrichment of cell-targeting and population-specific aptamers by fluorescence-activated cell sorting. Angew. Chem. Int. Ed. 2008;47:5190–5193. doi: 10.1002/anie.200800216. [DOI] [PubMed] [Google Scholar]
- 20.Morris KN, Jensen KB, Julin CM, Weil M, Gold L. High affinity ligands from in vitro election: complex targets. Proc. Natl Acad. Sci. USA. 1998;95:2902–2907. doi: 10.1073/pnas.95.6.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang Z, Shangguan D, Wang K, Shi H, Sefah K, Mallikratchy P, Chen HW, Li Y, Tan W. Selection of aptamers for molecular recognition and characterization of cancer cells. Anal. Chem. 2007;79:4900–4907. doi: 10.1021/ac070189y. [DOI] [PubMed] [Google Scholar]
- 22.Okhonin V, Krylova SM, Krylov SN. Nonequilibrium capillary electrophoresis of equilibrium mixtures, mathematical model. Anal. Chem. 2004;76:1507–1512. doi: 10.1021/ac035259p. [DOI] [PubMed] [Google Scholar]
- 23.Berezovski M, Drabovich A, Krylova SM, Musheev M, Okhonin V, Petrov A, Krylov SN. Nonequilibrium capillary electrophoresis of equilibrium mixtures: a universal tool for development of aptamers. J. Am. Chem. Soc. 2005;127:3165–3171. doi: 10.1021/ja042394q. [DOI] [PubMed] [Google Scholar]
- 24.Reed KC, Bygrave FL. Methodology for in vitro studies of Ca-2 + transport. Anal. Biochem. 1975;67:44–54. doi: 10.1016/0003-2697(75)90270-5. [DOI] [PubMed] [Google Scholar]
- 25.Shangguan D, Cao ZC, Li Y, Tan W. Aptamers evolved from cultured cancer cells reveal molecular differences of cancer cells in patient samples. Clin. Chem. 2007;53:1153–1155. doi: 10.1373/clinchem.2006.083246. [DOI] [PubMed] [Google Scholar]
- 26.Berezovski M, Krylov SN. Nonequilibrium capillary electrophoresis of equilibrium mixtures – a single experiment reveals equilibrium and kinetic parameters of protein-DNA interactions. J. Am. Chem. Soc. 2002;124:13674–13675. doi: 10.1021/ja028212e. [DOI] [PubMed] [Google Scholar]
- 27.Shamah SM, Healy JM, Cload ST. Complex target SELEX. Acc. Chem. Res. 2008;41:130–138. doi: 10.1021/ar700142z. [DOI] [PubMed] [Google Scholar]
- 28.Romig TS, Bell C, Drolet DW. Aptamer affinity chromatography: combinatorial chemistry applied to protein purification. J. Chromatogr. B. 1999;731:275–284. [PubMed] [Google Scholar]
- 29.Drabovich AP, Krylova SM, Musheev MU, Krylov SN. Non-equilibrium capillary electrophoresis of equilibrium mixtures (NECEEM) as Swiss army knife for selection of aptamers. P/ACE Setter Newslett. Worldwide Newslett. Capillary Electrophoresis. 2006;10:1–5. [Google Scholar]