Abstract
In vitro antibody-display technologies are powerful approaches for isolating monoclonal antibodies from recombinant antibody libraries. However, these display techniques require several rounds of affinity selection which is time-consuming. Here, we combined mRNA display with a microfluidic system for in vitro selection and evolution of antibodies and achieved ultrahigh enrichment efficiency of 106- to 108-fold per round. After only one or two rounds of selection, antibodies with high affinity and specificity were obtained from naïve and randomized single-chain Fv libraries of ∼1012 molecules. Furthermore, we confirmed that not only protein–protein (antigen–antibody) interactions, but also protein–DNA and protein–drug interactions were selected with ultrahigh efficiencies. This method will facilitate high-throughput preparation of antibodies and identification of protein interactions in proteomic and therapeutic fields.
INTRODUCTION
Rapid preparation of monoclonal antibodies with high affinity and specificity is required in diverse fields from fundamental molecular and cellular biology to drug discovery and diagnosis (1). In addition to classical hybridoma technology, in vitro antibody-display technologies (2–7) are powerful approaches for isolating single-chain Fv (scFv) antibodies from recombinant antibody libraries. However, these display techniques require several rounds of affinity selection (typically, library size is 107–1012, while enrichment efficiency is 10- to 103-fold per round). Recently, microfluidic systems have been developed for high-throughput protein analysis (8), because they offer the advantages of very low sample volumes, rapid analysis and automated recovery of captured analytes for further characterization. However, there has been little attempt to combine microfluidic systems with in vitro antibody-display technologies so far. Previously we have developed an mRNA display system named in vitro virus (IVV) (9), in which an in vitro-translated full-length protein (phenotype) is covalently attached to its encoding mRNA (genotype) through puromycin (9–11). Here, we show that a microfluidic system can be applied to the mRNA display selection of naïve and randomized scFv libraries, unexpectedly resulting in ultrahigh efficiency of scFv selection.
MATERIALS AND METHODS
Preparation of antigen proteins
Recombinant p53 protein was purchased from Active Motif, and was diluted with a provided buffer (20 mM Tris–HCl, pH 8.0, 20% glycerol, 100 mM KCl, 0.2 mM EDTA and 1 mM DTT).
All primer sequences used for PCR are listed in Table 1. Full-length ORFs corresponding to p53 and MDM2 were amplified by PCR from cDNAs with KOD-plus DNA polymerase (Toyobo) using primers p53-NT1 and p53-R-NT02 for p53 or primers MDM2-F and MDM2-R for MDM2 (24 cycles of 30 s at 94°C, 30 s at 58°C and 2 min at 68°C). The PCR product was separated on 1% low melting temperature agarose gel (Sigma), and gel-purified using a Wizard PCR preps DNA purification kit (Promega). To add a His-tag sequence, the purified MDM2 DNA was reamplified by PCR with KOD-plus DNA polymerase using primers CACC-MDM2-F and MDM2-His10R. The PCR product was gel-purified as described above.
Table 1.
Oligonucleotide primers used for PCR
| Primer name | Sequences |
|---|---|
| p53-NT1 | ATGGAGGAGCCGCAGTCAG |
| p53-R-NT02 | GTCTGAGTCAGGCCCTTCTGTCTTG |
| MDM2-F | ATGTGCAATACCAACATGTCTGTGTCTACCGATGGTGCT |
| MDM2-R | GGGGAAATAAGTTAGCACAATCATTTGAATTGGTTGTCT ACATACTGG |
| CACC -MDM2-F | CACCATGTGCAATACCAACATGTCTGTGTCTACCG |
| MDM2- His10R | CTATCAATGGTGGTGATGATGGTGATGGTGATGATGGG GGAAATAAGTTAGCACAATCATTTGAATTGG |
| F-Bio | GGCGCCGGCACCCCGGTGAC |
| R-Bio | CGCCAGGGTCATCAGGGTGT |
| CACC- p53-NT01 | CACCATGGAGGAGCCGCAGTCAG |
| p53-Bio-link | CAAGACAGAAGGGCCTGACTCAGACGGCGCCGGCACCC CGGTGAC |
| MDM2- Bio-link | GACAACCAATTCAAATGATTGTGCTAACTTATTTCCCC GGCGCCGGCACCCCGGTGAC |
| MulgG1/2 forward | CTGGACAGGGATCCAGAGTTCCA |
| MulgG3 forward | CTGGACAGGGCTCCATAGTTCCA |
| MuCK forward | CTCATTCCTGTTGAAGCTCTTGAC |
| HB Primers | |
| HB1 | GACTGGTGGACAGCAAATGGGTGAKGTRMAGCTTCAGGAGTC |
| HB2 | GACTGGTGGACAGCAAATGGGTGAGGTBCAGCTBCAGCAGTC |
| HB3 | GACTGGTGGACAGCAAATGGGTCAGGTGCAGCTGAAGSASTC |
| HB4 | GACTGGTGGACAGCAAATGGGTGAGGTCCARCTGCAACARTC |
| HB5 | GACTGGTGGACAGCAAATGGGTCAGGTYCAGCTBCAGCARTC |
| HB6 | GACTGGTGGACAGCAAATGGGTCAGGTYCARCTGCAGCAGTC |
| HB7 | GACTGGTGGACAGCAAATGGGTCAGGTCCACGTGAAGCAGTC |
| HB8 | GACTGGTGGACAGCAAATGGGTGAGGTGAASSTGGTGGAATC |
| HB9 | GACTGGTGGACAGCAAATGGGTGAVGTGAWGYTGGTGGAGTC |
| HB10 | GACTGGTGGACAGCAAATGGGTGAGGTGCAGSKGGTGGAGTC |
| HB11 | GACTGGTGGACAGCAAATGGGTGAKGTGCAMCTGGTGGAGTC |
| HB12 | GACTGGTGGACAGCAAATGGGTGAGGTGAAGCTGATGGARTC |
| HB13 | GACTGGTGGACAGCAAATGGGTGAGGTGCARCTTGTTGAGTC |
| HB14 | GACTGGTGGACAGCAAATGGGTGARGTRAAGCTTCTCGAGTC |
| HB15 | GACTGGTGGACAGCAAATGGGTGAAGTGAARSTTGAGGAGTC |
| HB16 | GACTGGTGGACAGCAAATGGGTCAGGTTACTCTRAAAGWGTSTG |
| HB17 | GACTGGTGGACAGCAAATGGGTCAGGTCCAACTVCAGCARCC |
| HB18 | GACTGGTGGACAGCAAATGGGTGATGTGAACTTGGAAGTGTC |
| HB19 | GACTGGTGGACAGCAAATGGGTGAGGTGAAGGTCATCGAGTC |
| LB Primers | |
| LB1 | GTGGCAGCATTGAGGGTCGCGAYATCCAGCTGACTCAGCC |
| LB2 | GTGGCAGCATTGAGGGTCGCGAYATTGTTCTCWCCCAGTC |
| LB3 | GTGGCAGCATTGAGGGTCGCGAYATTGTGMTMACTCAGTC |
| LB4 | GTGGCAGCATTGAGGGTCGCGAYATTGTGYTRACACAGTC |
| LB5 | GTGGCAGCATTGAGGGTCGCGAYATTGTRATGACMCAGTC |
| LB6 | GTGGCAGCATTGAGGGTCGCGAYATTMAGATRAMCCAGTC |
| LB7 | GTGGCAGCATTGAGGGTCGCGAYATTCAGATGAYDCAGTC |
| LB8 | GTGGCAGCATTGAGGGTCGCGAYATYCAGATGACACAGAC |
| LB9 | GTGGCAGCATTGAGGGTCGCGAYATTGTTCTCAWCCAGTC |
| LB10 | GTGGCAGCATTGAGGGTCGCGAYATTGWGCTSACCCAATC |
| LB11 | GTGGCAGCATTGAGGGTCGCGAYATTSTRATGACCCARTC |
| LB12 | GTGGCAGCATTGAGGGTCGCGAYRTTKTGATGACCCARAC |
| LB13 | GTGGCAGCATTGAGGGTCGCGAYATTGTGATGACBCAGKC |
| LB14 | GTGGCAGCATTGAGGGTCGCGAYATTGTGATAACYCAGGA |
| LB15 | GTGGCAGCATTGAGGGTCGCGAYATTGTGATGACCCAGWT |
| LB16 | GTGGCAGCATTGAGGGTCGCGAYATTGTGATGACACAACC |
| LB17 | GTGGCAGCATTGAGGGTCGCGAYATTTTGCTGACTCAGTC |
| LBλ | GTGGCAGCATTGAGGGTCGCGATGCTGTTGTGACTCAGCAATC |
| HF Primers | |
| HF1 | GATGGCCGAGACGTTTTGGCCGAGGAAACGGTGACCGTGGT |
| HF2 | GATGGCCGAGACGTTTTGGCCGAGGAGACTGTGAGAGTGGT |
| HF3 | GATGGCCGAGACGTTTTGGCCGCAGAGACAGTGACCAGAGT |
| HF4 | GATGGCCGAGACGTTTTGGCCGAGGAGACGGTGACTGAGGT |
| LF Primers | |
| LF1 | CTTGTCGTCGTCGTCCTTGTAGTCACGTTTGATTTCCAGCTTGG |
| LF2 | CTTGTCGTCGTCGTCCTTGTAGTCACGTTTTATTTCCAGCTTGG |
| LF3 | CTTGTCGTCGTCGTCCTTGTAGTCACGTTTTATTTCCAACTTTG |
| LF4 | CTTGTCGTCGTCGTCCTTGTAGTCACGTTTCAGCTCCAGCTTGG |
| LFλ | CTTGTCGTCGTCGTCCTTGTAGTCACCTAGGACAGTCAGTTTGG |
| VH-back | GACTGGTGGACAGCAAATGGGT |
| VH-forward | GATGGCCGAGACGTTTTGGC |
| VL-back | GTGGCAGCATTGAGGGTCGC |
| FLAG-R | CTTGTCGTCGTCGTCCTTGTAGTC |
| SP6-omegaF | ATTTAGGTGACACTATAGAACAACAACAACAACAAACAACAA CAAAATGGCTAGCATGACTGGTGGACAGCAAATGGGT |
| McD-Linker + | GCCAAAACGTCTCGGCCATCTGGTGGAGGCGGTTCAGGCGGA GGTGGCTCTGGCGGTGGCGGATCCGGAGGTGGTGGCAGCAT |
| TGAGGGTCGC | |
| McD-3′- UTR (HisTag) | TTTTTTTTATGGTGATGGTGGTGATGGTGCAACCTGACTCG ACTCGTACGCTTGTCGTCGTCGTCCTTGTAGTC |
| SP6-F | ATTTAGGTGACACTATAGAACAACAACAACAACAAACAAC |
| McD-R (HisTag) | TTTTTTTTATGGTGATGGTGGTGATGGTGCAACCTGACTCG ACTCGTACG |
| McD-R-His | ATGGTGATGGTGGTGATGGTGCAACCTGACTCGACTCGTACG |
| T7-long-F | ATGGCTAGCATGACTGGTGGACAGCAAATGGGT |
| McD- R(HisTag)- stop | TCAATGGTGATGGTGGTGATGGTGCAACCTGACTCGACTCG TACG |
R = G or A; Y = T or C; M = A or C; K = G or T; S = G or C; W = A or T; H = A or C or T; B = G or T or C; V = G or C or A and D = G or A or T.
In order to attach the antigens on a streptavidin-conjugated solid resin, an ELISA plate or a sensor chip, a Bio-tag sequence capable of attaching biotin in Escherichia coli was introduced at the C-terminus of p53 and MDM2. The Bio-tag sequence encoding a 72 amino acid peptide from oxaloacetate decarboxylase of Klebsiella pneumoniae was amplified by PCR from the BioEase™ plasmid (Invitrogen) using primers F-Bio and R-Bio. The PCR product was agarose gel-purified. To add the Bio-tag, the purified p53 or MDM2 gene fragment described above was mixed with the Bio-tag fragment, and was reamplified by PCR (100 µl) with 5 U of KOD-plus DNA polymerase using CACC-p53-NT01 primer (3 pmol), R-Bio primer (3 pmol) and p53-Bio-link oligonucleotide (0.1 pmol) for p53, or CACC-MDM2-F primer (3 pmol), R-Bio primer (3 pmol) and MDM2-Bio-link oligonucleotide (0.1 pmol) for MDM2 (8–12 cycles of 30 s at 94°C, 30 s at 58°C and 2 min at 68°C).
The resulting PCR products (MDM2-His-tag, p53-Bio-tag and MDM2-Bio-tag) were gel-purified and then cloned into the vector pET101/D-TOPO (Invitrogen) using One Shot TOP 10 chemically competent E. coli cells (Invitrogen). The orientation and sequence of the cloned genes were verified by sequencing the isolated plasmids. The plasmids were then used to transform E. coli BL21Star (DE3) One Shot cells (Invitrogen). The transformed cells were cultured at 37°C in 400 ml of TB medium containing 100 µg/ml carbenicillin (Sigma) until the OD660 reached 0.5–0.6, then isopropylthio-ß-d-galactoside (Nacalai tesque) was added to a final concentration of 1 mM, and the cells were harvested 4–6 h later.
For purification of proteins, the cells were collected by centrifugation, and resuspended in 20 ml of TBS (20 mM Tris–HCl buffer, pH 7.5, 138 mM NaCl) containing 8 U of DNase I (Invitrogen), 40 µl of EDTA-free protease inhibitor cocktail (Nacalai tesque) and 1 mM 2-mercaptoethanol (Nacalai tesque). The cells were lyzed by sonication using a Bioruptor UCW-201 (Cosmo Bio) twice for 15 min at 30-s intervals. The crude extracts were centrifuged for 20 min at 8500 r.p.m. The precipitates were suspended in 20 ml of TBS containing 8 M urea and 8 U of DNase I, 40 µl of EDTA-free protease inhibitor cocktail and 1 mM 2-mercaptoethanol, and then recentrifuged for 20 min at 8500 r.p.m. The supernatants containing the histidine-tagged proteins in denatured form were immobilized on the TALON superflow metal affinity resin (Clontech), and the columns were washed with 10 volumes of TBS containing 10 mM imidazole and 6 M urea, 10 volumes of TBS containing 1 M NaCl and 10 volumes of TBS, to allow refolding of the bound proteins on the columns. The refolded proteins were then eluted in three fractions of 2 ml TBS containing 250 mM imidazole. Subsequently, the proteins were separated by size exclusion chromatography using Sephadex G-75 10/300 GL (Amersham Biosciences) on an AKTA explorer 10S (Amersham Biosciences) equilibrated with HBS-EP buffer (10 mM HEPES–NaOH, pH 7.4, 150 mM NaCl, 3 mM EDTA and 0.005% Tween-20) at a flow rate of 0.5 ml/min. The purified proteins were analyzed by SDS–PAGE followed by staining with SimplyBlue™ (Invitrogen).
Construction of scFv DNA libraries
The mouse scFv DNA library was constructed as previously described by Marks et al. (2) with the following modifications. First-strand cDNA was synthesized from 0.55 µg of mouse spleen poly(A)+ RNA (Clontech) using immunoglobulin-specific primers MulgG1/2 forward, MulgG3 forward or MuCK forward (Table 1) with ReverTraAce (Toyobo) according to the manufacturer's protocol. The products were amplified by PCR with 0.625 U of KOD-dash DNA polymerase (Toyobo) using 2.5 pmol each of 19 different HB primers and 1.25 pmol of reverse primers MulgG1/2 forward and MulgG3 forward for VH (heavy-chain) or 18 different LB primers and 2.5 pmol MuCK forward for VL (light-chain) [25 cycles of 30 s at 96°C, 30 s at 50°C (for VH) or 48°C (for VL) and 1 min at 72°C]. Each VH and VL fragment was generated by nested PCR as described above, except for the use of an equimolar mixture of HF primers (for VH) or an equimolar mixture of LF primers (for VL). The resulting VH and VL genes were agarose gel-purified.
Random point mutations were further introduced into the VH and VL fragments using a GeneMorph II Random Mutagenesis kit (Stratagene) according to the manufacturer's protocol. Briefly, PCRs were performed in 100 µl volumes containing 0.04 fmol of the VH or VL fragment as a template, 2 pmol each of forward and reverse primers (VH-back and VH-forward primers for VH; VL-back and FLAG-R primers for VL), 0.2 mM dNTPs and 2 U Mutazyme II DNA polymerases (30 cycles of 30 s at 95°C, 30 s at 58°C and 1 min at 72°C). The resulting VH and VL mutated genes were gel-purified.
Finally, a mouse scFv DNA library was constructed by assembly of DNA fragments containing an SP6 promoter, the translational enhancer from tobacco mosaic virus (12,13), a T7·tag, the mutated VH and VL gene fragments, a (Gly4Ser)4 linker, a FLAG-tag and poly(A) sequence by an overlap extension PCR: 915 µl volumes containing 2 pmol each of VH and VL fragments, 2 pmol each of primers, SP6-omegaF, McD-linker + and McD-3′-UTR(HisTag), 0.5 mM dNTPs and 5 U KOD-dash DNA polymerase (10 cycles of 30 s at 96°C, 30 s at 58°C and 1 min at 72°C). The products were reamplified by PCR using 40 pmol each of primers SP6-F and McD-R(HisTag) that were added to a total volume of 1000 µl (10 cycles of 30 s at 96°C, 30 s at 58°C and 1 min at 72°C). The resulting scFv DNA library was finally gel-purified.
mRNA display selection on a microfluidic chip
The biotinylated antigen proteins were immobilized on SA sensor chips (Biacore) by passing HBS-EP buffer at a flow rate of 10 µl/min on a Biacore 3000 instrument (Biacore). Amounts of p53 proteins immobilized on the chips in flow cells Fc-1, Fc-2, Fc-3 and Fc-4 were 1290, 999, 1424 and 1521 RU, respectively, and those of MDM2 proteins were 677, 537, 632 and 835 RU, respectively.
The scFv DNA library was transcribed using RiboMAX large-scale RNA production systems-SP6 (Promega) in a total volume of 20 µl containing 80 mM HEPES-KOH, pH 7.5, 2 mM spermidine, 40 mM DTT, 32 mM MgCl2, 5 mM each of ATP, CTP and UTP, 1 mM GTP, 5 mM m7G(5′)ppp(5′)G RNA capping analog (Invitrogen), 1 pmol of scFv DNA library and 2 µl of SP6 RNA polymerase (Promega). After transcription reaction at 37°C for 3 h, the DNA template was digested with DNase I (Promega) at 37°C for 1 h. The resulting RNA was purified with an RNeasy mini kit (Qiagen) and was ligated with a PEG-puromycin spacer [p(dCp)2-T(fluorescein)p-PEGp-(dCp)2-puromycin] (11,13) using T4 RNA ligase at 15°C for 15–40 h in a total volume of 50 µl containing 50 mM Tris–HCl, pH 7.5, 10 mM MgCl2, 12 mM DTT, 1.4 mM ATP, 5% DMSO, 0.002% BSA, 40 U of RNase inhibitor (Toyobo), 0.2 mM PEG-puromycin spacer, 0.6 mM PEG2000 (Nacalai tesque), 50 pmol of RNA and 200 U of T4 RNA ligase (Takara). The resulting RNA-PEG-puromycin library was purified with the RNeasy mini kit, and could be stored at −80°C for more than 1 year without any degradation.
In vitro translation was performed in a total volume of 100 µl containing 10 pmol of the RNA-PEG-puromycin library, 20 µl of wheat germ cell-free extract (Zoegene), 40 µg of creatine kinase and 20 µl of a translation buffer (Zoegene) at 26°C for 5 min. The reaction mixture was subjected to gel filtration on Sephadex G200 (Amersham Biosciences) using a 0.8 × 4 cm column (Bio-Rad) with TBS containing 10 mM EDTA, and 2-drop fractions were collected. The fluorescence of mRNA-displayed proteins in eluate fractions was monitored with a Multi-detection microplate reader Powerscan HT (Dainippon Pharmaceutical). The 4th to 7th fractions containing mRNA-displayed proteins were diluted with HBS-EP buffer to 300 µl and injected onto the antigen-immobilized sensor chip. The selection experiments were performed at 25°C with the Biacore 3000 using HBS-EP buffer at a flow rate of 20 µl/min. After association for 250 s and dissociation for 3000 s, the sensor surfaces were washed 10 times with HBS-EP buffer for 1200 s. The bound molecules were eluted competitively from the sensor surface with 7 µl of 0.1 nM free antigen proteins for 1200 s. Then 7 µl of recovered solution was subjected to RT-PCR in a total volume of 100 µl containing 1 mM dNTPs, 50 mM Tris–HCl, pH 8.0, 75 mM KCl, 3 mM MgCl2, 10 mM dithiothreitol, 200 U of RNase inhibitor (Toyobo), 500 U of ReverTraAce (Toyobo) and 50 pmol McD-R-His primer (Table 1), at 50°C for 30 min and heated at 99°C for 5 min. The RT product was amplified by PCR with KOD-plus DNA polymerase using primers T7-long-F and McD-R-His (24–36 cycles of 30 s at 94°C, 30 s at 58°C and 2 min at 68°C). DNA used for next round was reconstructed by overlap extension PCR with KOD-plus DNA polymerase using primers SP6-omegaF and McD-R-His (8–12 cycles of 30 s at 94°C, 30 s at 58°C and 2 min at 68°C). The resulting DNA was purified using the Wizard PCR preps DNA purification kit.
Finally, selected DNAs were cloned using a TOPO TA cloning kit (Invitrogen) and sequenced with a CEQ 2000 DNA analysis system (Beckman Coulter). Genetyx-mac 13.0.10 sequence analysis software and ClustalX 1.83 were used for alignment and sequence manipulations.
In vitro evolution of a selected scFv
Random point mutations were introduced into M1-19 scFv using a GeneMorph II Random Mutagenesis kit (Stratagene) according to the manufacturer's protocol. Briefly, PCRs were performed in 50 µl volumes containing 0.0003 fmol of M1-19 scFv as a template, 1 pmol each of forward and reverse primers (T7-long-F and McD-R-His), 0.2 mM dNTPs and 1 U Mutazyme II DNA polymerases (34 cycles of 30 s at 95°C, 30 s at 58°C and 1 min at 72°C). The resulting mutated DNA was reconstructed by overlap extension PCR with KOD-plus DNA polymerase using primers SP6-omegaF and McD-R(HisTag) (8 cycles of 30 s at 94°C, 30 s at 58°C and 2 min at 68°C]. The resulting mutated M1-19 scFvs were gel-purified.
In vitro transcription, translation and microfluidic selection were performed as described in the previous section, with the exception that the flow rate was increased to 100 µl/min for on-rate selection and the washing time was increased 20-fold for off-rate selection.
Synthesis and purification of selected scFvs
The scFv DNA template having a stop codon was amplified by PCR from each cloned plasmid with KOD-plus DNA polymerase using primers SP6-omegaF and McD-R(HisTag)-stop (20–30 cycles of 30 s at 94°C, 30 s at 58°C and 2 min at 68°C). The DNA was in vitro-transcribed into mRNA using RiboMAX large-scale RNA production systems-SP6 at 37°C for 3 h, DNase I was added, and then incubation was continued for a further 1 h at 37°C. The resulting mRNA was purified using the RNeasy mini kit, and was in vitro-translated in the wheat germ cell-free translation system using a dialysis cup (molecular size cutoff of 12 000 Da; Daiichi Pure Chemicals) at 26°C for 2–4 h.
The translation products were purified on TALON superflow metal affinity resin with a running buffer (20 mM Tris–HCl buffer, pH 7.5, 138 mM NaCl and 10 mM imidazole). The columns were washed with 10 volumes of the same buffer, and then eluted with 1 volume of the buffer containing 250 mM imidazole. Subsequently, the monomeric scFvs were separated by size exclusion chromatography using Sephadex G-75 as described above. The column was calibrated in the HBS-EP buffer with standards (Amersham Pharmacia Biotech): blue dextran 2000 (Mr 2 000 000), albumin (Mr 64 700), ovalbumin (Mr 45 800), chymotrypsinogen A (Mr 19 900) and ribonuclease A (Mr 15 400).
Pull-down assays
Streptavidin MagneSphere Paramagnetic Particles (500 µl, Promega) were incubated with 1 µM of biotinylated p53, biotinylated MDM2 or BSA-biotin (Sigma) in 500 µl of TBST-BSA (20 mM Tris–HCl buffer, pH 7.5, 138 mM NaCl, 0.1% Tween-20 and 0.1% BSA) for 2–6 h at 25°C, and washed with TBST-BSA. The antigen-immobilized beads were incubated with 5 µl of translation products using wheat germ extracts in TBST-BSA. After incubation for 1 h at 25°C, the beads were washed extensively with TBST-BSA and boiled in a lithium dodecyl sulfate sample buffer (Invitrogen) containing 0.1 M DTT at 70°C for 10 min. ScFv proteins were separated by SDS–PAGE on 4–12% Bis–Tris NuPAGE gels in MES running buffer (Invitrogen) and transferred to a nitrocellulose membrane using the iBlot dry blotting system (Invitrogen). The membrane was blocked with Blocking One Buffer (Nacalai tesque) in TBST (20 mM Tris–HCl buffer, pH 7.5, 138 mM NaCl and 0.1% Tween-20), and then probed with horseradish peroxidase (HRP)-conjugated mouse anti-FLAG-tag monoclonal antibody (Sigma) and an ECL western blotting kit (Amersham Biosciences).
Competitive ELISA
Streptavidin transparent C96 plates (Nunc) were incubated with 1 nM biotinylated p53 or biotinylated MDM2 in 100 µl of TBST-BSA per well for 2–6 h at 25°C and washed with TBST-BSA. ScFvs were diluted with blocking solution in the presence and absence of free p53 or MDM2 protein as a competitor (1–200 nM), and incubated for 1 h at 25°C. Then, the protein mixture solution (100 µl) was transferred into wells on a plate with immobilized antigen and the plate was incubated for 1 h at 25°C. The plate was washed five times with TBST-BSA, then further incubated for 1 h with HRP-conjugated mouse anti-T7·tag antibody (Novagen; 1:10 000 dilution in TBST-BSA; 100 µl per well). The plate was washed five times with TBST-BSA and then 100 µl of 3,3′,5,5′-tetramethylbenzidine substrate (Zymed) was added per well. The reaction was quenched with 1 N HCl (100 µl per well) and the absorbance at 450 nm was measured (reference wavelength at 630 nm). For each clone, the relative binding activity was calculated as the ratio of ELISA signal in the presence of competitor to that in the absence of competitor.
Western blot analysis
HEK-293 (human embryonic kidney cells) whole-cell lysate (non-denatured) was purchased from Abnova. Cell lysates and recombinant proteins were separated by SDS–PAGE on 4–12% Bis–Tris NuPAGE gels in MOPS running buffer (Invitrogen) and transferred to the nitrocellulose membrane using the iBlot dry blotting system. The membrane was blocked with Blocking One Buffer in TBST, and probed with scFv antibodies with the FLAG-tag in TBST containing 0.1% BSA. Proteins were detected using anti-FLAG-M2 antibody (Sigma) and goat polyclonal anti-mouse IgG conjugated with alkaline phosphatase (Bio-Rad). In a control experiment, membranes were probed with anti-MDM2 antibody 2A10 (Abcam) and goat polyclonal anti-mouse IgG conjugated with alkaline phosphatase. Both membranes were visualized using Immobilon Western AP substrate (Millipore).
Surface plasmon resonance analysis
Binding kinetics was determined by Surface plasmon resonance (SPR) with the Biacore 3000. All experiments were performed at 25°C using TBST. Biotinylated p53 (55 kDa), biotinylated MDM2 (66 kDa) or BSA-biotin was immobilized onto the SA sensor chip. The measurements were performed under conditions of 390 (p53) and 450 (MDM2) resonance units of the ligand and at a flow rate of 60 µl/min. To determine dissociation constants, three different concentrations of the purified monomeric scFvs, M1-19 (32 kDa) and P1-93 (31 kDa) were injected. The injection periods for association and dissociation were 60 and 300 s, respectively. After each measurement, the chip surface was regenerated with 10 µl of Glycine 2.0 (Biacore), and 10 µl of 50 mM NaOH, 1 M NaCl. The binding data were analyzed with the 1:1 binding with mass transfer model in the BIAevaluation software ver. 4.1 (Biacore).
Protein–DNA and protein–drug interaction analysis
Materials and methods for mRNA display selection of DNA-binding protein complexes c-fos and c-jun and an FK506-binding protein fkbp12 were described previously (14,15). Briefly, a mixture of three genes encoding c-Fos (118–212 aa), c-Jun (237–334 aa) and GST (full length; negative control), and a mixture of two genes encoding FKBP12 (full length) and p53 (15–29 aa; negative control) were used as model libraries. The mRNA display selection on the SA sensor chip was performed as described above, with the exception that biotinylated bait DNA containing six repeats of the TPA-responsive element (TGAC/GTCA) (14) or biotinylated FK506 (15) was immobilized on the sensor chip. Amounts of DNA (Mr 60,634) immobilized on the chips in flow cells Fc-1, Fc-2, Fc-3 and Fc-4 were 1314, 1433, 1920 and 1874 RU, respectively, and those of FK506 (Mr 1066) were 683, 650, 655 and 600 RU, respectively.
RESULTS
In vitro selection of antibodies from a naïve scFv library
The mRNA display selection of scFv was performed on a Biacore microfluidic chip (Figure 1) as described in the Materials and methods section. Since the diversity of the mouse scFv library prepared from mouse spleen poly A+ RNAs is estimated to be 106–108, while mRNA display allows screening of ∼1012 molecules, we also introduced random point mutations into the scFv library. We chose p53 (human tumor suppressor protein) and MDM2 (human murine double minute) proteins as model antigens that were immobilized on the Biacore sensor chip. The selection experiment was performed on the microfluidic chip, and selected scFv genes were amplified by reverse transcription (RT)-PCR and identified by cloning and sequencing. Unexpectedly, the recovered anti-p53 and anti-MDM2 scFv sequences converged on a single sequence and two sequences, respectively, after only two rounds of selection. These clones revealed high affinity, but also low antigen specificity, in pull-down assays (data not shown), and so we examined the clones after a single round of selection in each case (35 clones for anti-p53 and 25 clones for anti-MDM2). When the binding activities of 29 (anti-p53) and 20 (anti-MDM2) clones with distinct sequences were examined by pull-down assays, P1-93 and M1-19 showed high specificity against the respective antigens among p53, MDM2 and BSA (Figure 2A). The amino acid sequences of P1-93 and M1-19 are shown in Figure 2B. In competitive ELISA, both clones dose-dependently inhibited the ELISA signal (Figure 2C), and Scatchard plots revealed that the KDs of P1-93 and M1-19 were 22 and 5.9 nM, respectively. The KDs of P1-93 and M1-19 were also determined by SPR as 12 and 4.3 nM, respectively (Figure 2D). The values obtained by the two different methods are similar.
Figure 1.
Schematic representation of the mRNA display selection of scFv on a microfluidic chip. Step (1): an scFv DNA library is transcribed to mRNA in vitro. Step (2): the mRNA library is ligated with a PEG-puromycin spacer. Step (3): the RNA-PEG-puromycin library is translated in a cell-free translation system. Step (4): the mRNA-displayed scFv library is injected into a sensor chip on which a target antigen is immobilized. Step (5): the bound scFv is eluted competitively with the free antigen. Step (6): the RNA is amplified by RT-PCR and used for the next round of selection or cloning and sequencing.
Figure 2.
The selected scFvs anti-p53 P1-93 and anti-MDM2 M1-19. (A) Pull-down assays of the anti-p53 scFv P1-93 (top) and anti-MDM2 scFv M1-19 (bottom) using p53-, MDM2- or BSA-immobilized beads. Recovered scFv with FLAG-tag was detected with the anti-FLAG antibody. (B) Predicted amino acid sequences of the VH (black bar) and VL (gray bar) regions of anti-p53 scFv P1-93 and anti-MDM2 scFv M1-19. Residue numbering is according to Kabat et al. (20) (C) Competitive ELISA. P1-93 or M1-19 was preincubated with a competitor (0–200 nM free antigen) and allowed to bind to antigen-immobilized plates. After washing, remaining scFvs were detected with the anti-T7·tag antibody. (D) Biacore sensorgrams of the purified P1-93 (left; 31 kDa) and M1-19 (right; 32 kDa) using a p53- (blue lines; 55 kDa) or MDM2-immobilized (red lines; 66 kDa) sensor chip. The measurements were performed under conditions of 450 RU of the ligand and at a flow rate of 60 µl/min. To determine dissociation constants, three different concentrations (35, 64 and 136 nM for P1-93 and 27, 50 and 75 nM for M1-19) of the monomeric scFvs were injected.
In vitro evolution of scFv
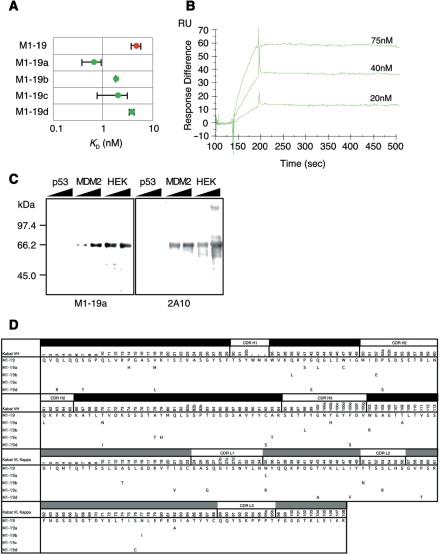
Further, we performed in vitro evolution of scFv with higher affinity against MDM2 from a randomly mutated M1-19 scFv library. We applied on-rate and off-rate selection as a selection pressure for in vitro affinity maturation with the Biacore instrument: the on-rate selection was performed by controlling flow rate, and the off-rate selection was carried out by using a prolonged washing process on the sensor chip. After one round of selection, the recovered scFv genes were cloned and sequenced, and the KDs were evaluated by competitive ELISA (Figure 3A and D). We obtained four mutants with higher affinity for MDM2 (KD = 0.7–3.8 nM) than the progenitor M1-19 from 22 distinct clones. The strongest binder M1-19a was confirmed to have a higher on-rate and lower off-rate than M1-19 by SPR (Figure 3B) and was also confirmed to recognize only antigen protein MDM2 in crude cell lysates by western blotting (Figure 3C). These results indicated that the selected scFv had high enough affinity and specificity for practical use. Although the mutations of the selected scFvs were distributed among the whole sequences and no ‘consensus’ mutations were identified, the mutation Y100bH within VH CDR3 may contribute to the improved affinity and specificity, because this region is usually important for binding with antigens.
Figure 3.
In vitro evolution of anti-MDM2 scFv M1-19. (A) Dissociation constants (KDs) of M1-19 (red circle) and the four mutant scFvs (green circles) were obtained by means of Scatchard plots of the data from competitive ELISA. (B) Biacore sensorgrams of the mutant M1-19a (amount of immobilized antigen was 450 RU). M1-19a had a higher on-rate and lower off-rate (ka = 2.5 × 105/Ms, kd = 8.6 × 105/s, KA = 3.0 × 109/M, KD = 0.34 nM) than the progenitor M1-19 (ka = 5.5 × 104/Ms, kd = 2.3 × 104/s, KA = 2.4 × 108/M, KD = 4.3 nM; Figure 2D, right, red lines). (C) Recombinant p53 and MDM2 proteins (10 and 25 ng), and HEK-293 cell lysates (1 and 2.5 µg) were analyzed by western blotting using M1-19a (left) or commercially available anti-MDM2 2A10 (right) as a control, respectively. (D) Predicted amino acid sequences of the VH (black bar) and VL (gray bar) regions of the mutant scFvs M1-19a-d. Residue numbering is according to Kabat et al. (20).
Ultrahigh efficiencies of protein selection
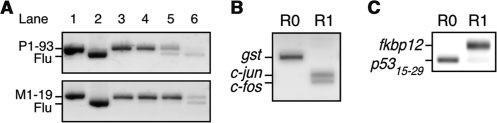
Surprisingly, our results indicated that the positive clone was efficiently enriched through only one or two rounds of selection from the large library containing ∼1012 molecules, implying ultrahigh efficiency of the method. To estimate the enrichment efficiency, we performed model experiments using a mixture of two kinds of scFv genes. The P1-93 (anti-p53) or M1-19 (anti-MDM2) gene was mixed with an anti-fluorescein scFv gene (5) (‘Flu’ as a negative control) at a ratio of 1:102, 1:104, 1:106 or 1:108, and subjected to one round of mRNA display selection on the sensor chip. The selection of the 1:106 mixture of P1-93:Flu genes and the 1:108 mixture of M1-19:Flu genes each resulted in a roughly 1:1 final gene ratio (Figure 4A), indicating enrichment efficiencies of 106- and 108-fold per round, respectively.
Figure 4.
The mRNA display selection of protein interactions on the Biacore sensor chip. (A) The P1-93 (912 bp; lane 1 top) or M1-19 (936 bp; lane 1 bottom) gene was mixed with an anti-fluorescein scFv gene (5) (Flu; 888 bp; lane 2) at a ratio of 1:102 (lane 3), 1:104 (lane 4), 1:106 (lane 5) or 1:108 (lane 6). The mixtures were subjected to mRNA display selection on the sensor chip conjugated with antigens p53 (top) or MDM2 (bottom). The RT-PCR products amplified from fractions after one round of selection were analyzed by agarose gel electrophoresis. (B) In vitro selection of protein–DNA interactions was performed using a mixture of three genes with N-terminal T7·tag and C-terminal FLAG-tag coding sequences; c-fos (349 bp), c-jun (394 bp), gst (597 bp). The template RNAs of c-fos, c-jun and gst (negative control) were mixed at a ratio of 1:1:106. The mixtures were translated and the resulting mRNA-displayed protein libraries were selected on the sensor chip conjugated with bait DNA (AP-1) (14). The RT-PCR products amplified from fractions before (R0) or after one round (R1) of selection were analyzed by agarose gel electrophoresis. (C) In vitro selection of protein–drug interactions was performed using a mixture of two genes with N-terminal T7·tag and C-terminal FLAG-tag coding sequences; fkbp12 (448 bp) and a p53 (15–29 aa) fragment (175 bp). The template RNAs of fkbp12 and the p53 fragment (negative control) were mixed at a ratio of 1:106. The mixtures were translated and the resulting mRNA-displayed protein libraries were selected on the sensor chip conjugated with bait drug (FK506) (15). The RT-PCR products amplified from fractions before (R0) or after one round (R1) of selection were analyzed by agarose gel electrophoresis.
Since we have applied mRNA display to screening of protein–protein (13), protein–DNA (14) and protein–drug (15) interactions so far, it is interesting whether the ultrahigh efficiency by the combination of mRNA display and microfluidics is also achievable for these applications. Consequently, we confirmed that not only protein–protein (antigen–antibody) interactions, but also protein–DNA and protein–drug interactions were selected by our method with high-enrichment efficiencies of >106-fold (Figure 4B and C). Since the enrichment efficiencies of these model experiments with a usual agarose resin were only 10- to 103-fold per round [Refs (14) and (15); data not shown], the enrichment efficiency was improved 103- to 105-fold over previous methods.
DISCUSSION
Although Biacore instruments have so far been utilized mainly to analyze biomolecular interactions by SPR, a few researchers have used this approach to fish for affinity targets from a randomized DNA library (16), phage-displayed protein libraries (17,18) or a ribosome-displayed antibody library (19). However, the enrichment efficiency in these applications was not high. Why, then, was ultrahigh efficiency achieved in the present protein selection by mRNA display? The mRNA-displayed protein is a relatively small object pendant from its encoding RNA moiety, which is about 10 times larger. Thus, nonspecific adsorption of RNA on solid surfaces is potentially significant. The matrix of the Biacore sensor chip consists of carboxymethylated dextran covalently attached to a gold surface, and poorly binds nucleic acid molecules, because both materials are negatively charged. In contrast, phage display and ribosome display involve large protein moieties (coat proteins or ribosome), so the use of the sensor chip may not improve the enrichment efficiency in these cases.
It should be noted that the ultrahigh enrichment efficiency made it difficult to set the number of selection rounds at a level that is appropriate to remove all nonbinders as well as to pick all binders with various affinities from a library. If the number of selection rounds is too small, many negative sequences will be cloned; on the other hand, excess rounds of selection will yield only a single sequence with the highest affinity. In this study, we obtained 20–30 different sequences, including P1-93 and M1-19, with high antigen specificity after a single round of selection, while we obtained only one or two negative sequences with high affinity but low antigen specificity from 106–108 library after two (probably excess) rounds of selection (>1012-fold).
In summary, we achieved ultrahigh efficiencies (106–108-fold per round) of protein selection by mRNA display with the microfluidic system. We obtained scFvs with high affinity and specificity from a naïve library by mRNA display selection for the first time. It took only three days to perform each selection experiment, including activity evaluation by ELISA. Although preparation of target materials of high quality is required, we anticipate this simple method to be a starting point for a versatile system to facilitate high-throughput preparation of monoclonal antibodies for analysis of proteome expression and detection of biomarkers, high-throughput analysis of protein–protein, protein–DNA and protein–drug interactions in proteomic and therapeutic fields, and rapid evolution of novel artificial proteins from large randomized libraries that often require 10 or more rounds of selection.
FUNDING
Grant-in-Aid for Scientific Research; Special Coordination Fund grant from the MEXT of Japan; the Industrial Technology Research Grant Program in ‘05 from the NEDO of Japan. Funding for open access charge: Keio University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank previous members of our laboratory, K. Kojoh, K. Ishihara, T. Matsunaga and M. Takeda for their initial contribution to this project. We also thank T. Shibui and S. Misawa (ZOEGENE Corporation) for gifts of wheat-germ cell-free extracts; S. Tateyama for the Fos/Jun-related DNA samples and comments; K. Horisawa, T. Tsuji and M. Onimaru for helpful comments and discussions.
Footnotes
Present address: Hideaki Takashima, Biomarker Research Group, Molecuence Corporation, Yokohama 227-8502, Japan
REFERENCES
- 1.Hoogenboom HR. Selecting and screening recombinant antibody libraries. Nat. Biotechnol. 2005;23:1105–1116. doi: 10.1038/nbt1126. [DOI] [PubMed] [Google Scholar]
- 2.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 3.Hanes J, Plückthun A. In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl Acad. Sci. USA. 1997;94:4937–4942. doi: 10.1073/pnas.94.10.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.He M, Taussig MJ. Antibody-ribosome-mRNA (ARM) complexes as efficient selection particles for in vitro display and evolution of antibody combining sites. Nucleic Acids Res. 1997;25:5132–5134. doi: 10.1093/nar/25.24.5132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fukuda I, Kojoh K, Tabata N, Doi N, Takashima H, Miyamoto-Sato E, Yanagawa H. In vitro evolution of single-chain antibodies using mRNA display. Nucleic Acids Res. 2006;34:e127. doi: 10.1093/nar/gkl618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reiersen H, Lobersli I, Loset GA, Hvattum E, Simonsen B, Stacy JE, McGregor D, Fitzgerald K, Welschof M, Brekke OH, et al. Covalent antibody display–an in vitro antibody-DNA library selection system. Nucleic Acids Res. 2005;33:e10. doi: 10.1093/nar/gni010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl Acad. Sci. USA. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lion N, Rohner TC, Dayon L, Arnaud IL, Damoc E, Youhnovski N, Wu ZY, Roussel C, Josserand J, Jensen H, et al. Microfluidic systems in proteomics. Electrophoresis. 2003;24:3533–3562. doi: 10.1002/elps.200305629. [DOI] [PubMed] [Google Scholar]
- 9.Nemoto N, Miyamoto-Sato E, Husimi Y, Yanagawa H. In vitro virus: bonding of mRNA bearing puromycin at the 3′-terminal end to the C-terminal end of its encoded protein on the ribosome in vitro. FEBS Lett. 1997;414:405–408. doi: 10.1016/s0014-5793(97)01026-0. [DOI] [PubMed] [Google Scholar]
- 10.Roberts RW, Szostak JW. RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl Acad. Sci. USA. 1997;94:12297–12302. doi: 10.1073/pnas.94.23.12297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyamoto-Sato E, Takashima H, Fuse S, Sue K, Ishizaka M, Tateyama S, Horisawa K, Sawasaki T, Endo Y, Yanagawa H. Highly stable and efficient mRNA templates for mRNA-protein fusions and C-terminally labeled proteins. Nucleic Acids Res. 2003;31:e78. doi: 10.1093/nar/gng078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miyamoto-Sato E, Nemoto N, Kobayashi K, Yanagawa H. Specific bonding of puromycin to full-length protein at the C-terminus. Nucleic Acids Res. 2000;28:1176–1182. doi: 10.1093/nar/28.5.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Miyamoto-Sato E, Ishizaka M, Horisawa K, Tateyama S, Takashima H, Fuse S, Sue K, Hirai N, Masuoka K, Yanagawa H. Cell-free cotranslation and selection using in vitro virus for high-throughput analysis of protein-protein interactions and complexes. Genome Res. 2005;15:710–717. doi: 10.1101/gr.3510505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tateyama S, Horisawa K, Takashima H, Miyamoto-Sato E, Doi N, Yanagawa H. Affinity selection of DNA-binding protein complexes using mRNA display. Nucleic Acids Res. 2006;34:e27. doi: 10.1093/nar/gnj025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doi N, Takashima H, Wada A, Oishi Y, Nagano T, Yanagawa H. Photocleavable linkage between genotype and phenotype for rapid and efficient recovery of nucleic acids encoding affinity-selected proteins. J. Biotechnol. 2007;131:231–239. doi: 10.1016/j.jbiotec.2007.07.947. [DOI] [PubMed] [Google Scholar]
- 16.Hao D, Ohme-Takagi M, Yamasaki K. A modified sensor chip for surface plasmon resonance enables a rapid determination of sequence specificity of DNA-binding proteins. FEBS Lett. 2003;536:151–156. doi: 10.1016/s0014-5793(03)00045-0. [DOI] [PubMed] [Google Scholar]
- 17.Yamamoto Y, Tsutsumi Y, Yoshioka Y, Nishibata T, Kobayashi K, Okamoto T, Mukai Y, Shimizu T, Nakagawa S, Nagata S, et al. Site-specific PEGylation of a lysine-deficient TNF-alpha with full bioactivity. Nat. Biotechnol. 2003;21:546–552. doi: 10.1038/nbt812. [DOI] [PubMed] [Google Scholar]
- 18.Malmborg AC, Dueñas M, Ohlin M, Söderlind E, Borrebaeck CA. Selection of binders from phage displayed antibody libraries using the BIAcoreTM biosensor. J. Immunol. Methods. 1996;198:51–57. doi: 10.1016/0022-1759(96)00159-7. [DOI] [PubMed] [Google Scholar]
- 19.Yuan Q, Wang Z, Nian S, Yin Y, Chen G, Xia Y. Screening of high-affinity scFvs from a ribosome displayed library using BIAcore biosensor. Appl. Biochem. Biotechnol. 2009;152:224–234. doi: 10.1007/s12010-008-8251-y. [DOI] [PubMed] [Google Scholar]
- 20.Kabat EA National Institutes of Health. Sequences of Proteins of Immunological Interest. 5th edn. Bethesda, MD: National Institutes of Health Publication No. 91-3242; 1991. [Google Scholar]