Abstract
Massively parallel DNA sequencing is revolutionizing genomics research throughout the life sciences. However, the reagent costs and labor requirements in current sequencing protocols are still substantial, although improvements are continuously being made. Here, we demonstrate an effective alternative to existing sample titration protocols for the Roche/454 system using Fluorescence Activated Cell Sorting (FACS) technology to determine the optimal DNA-to-bead ratio prior to large-scale sequencing. Our method, which eliminates the need for the costly pilot sequencing of samples during titration is capable of rapidly providing accurate DNA-to-bead ratios that are not biased by the quantification and sedimentation steps included in current protocols. Moreover, we demonstrate that FACS sorting can be readily used to highly enrich fractions of beads carrying template DNA, with near total elimination of empty beads and no downstream sacrifice of DNA sequencing quality. Automated enrichment by FACS is a simple approach to obtain pure samples for bead-based sequencing systems, and offers an efficient, low-cost alternative to current enrichment protocols.
INTRODUCTION
DNA sequencing has revolutionized our understanding of many aspects of biology, and recent developments in high-throughput sequencing have greatly accelerated the rate at which genetic information can be obtained. For instance, one of the current ‘next generation’ DNA sequencing platforms, the Roche/454 Genome Sequencer FLX, is capable of sequencing 100–500 megabases of DNA, by massively parallel pyrosequencing in a 7-h run (1–3). However, obtaining DNA sequences from biological samples is costly and labor-intensive. For Roche/454-system based analyses this is partly due to an expensive, laborious ‘titration’ step, carried out prior to the main sequencing run.
In Roche/454 sequencing, single library fragments are annealed to DNA Capture Beads and amplified in individual micelles by emulsion PCR (emPCR), which attaches large numbers of amplified DNA molecules to the surface of sepharose beads that are deposited in the lanes of picotiter plates for sequencing. Selecting an appropriate DNA-to-bead ratio is essential in order to generate sufficient beads with amplified templates, while avoiding the production of too many beads carrying multiple amplified templates. This is usually done by sequencing-based titration, in which pilot runs are used to evaluate trials for the selection of an optimal DNA-to-bead ratio for a planned full-scale run. The standard procedure involves amplifying the samples in four parallel emulsion PCR mixtures, each with a different DNA-to-bead ratio, then sequencing the products in a picotiter plate using traditional pyrosequencing chemistry (1). Determining the optimal DNA-to-bead ratio may seem trivial but it is not straightforward, especially for sequencing facilities that analyze samples from diverse origins. For instance, variations in GC contents between species and variations due to differences in sample preparation protocols applied in different laboratories give rise to variations in fragment size distributions between libraries. In addition, many samples are not sufficiently pure to determine optimal DNA-to-bead ratios by theoretical calculations. Thus, the optimal DNA-to-bead ratio should ideally be determined for each library to be analyzed in a separate titration experiment.
Here, we describe a highly accurate flow cytometry-based method that has the potential to replace sequencing-based Roche/454 titration protocols. Flow cytometry and Fluorescence Activated Cell Sorting (FACS) have previously been used for sorting and cloning cells with particular features (4), detecting and quantifying genetic variations using beads as a solid support (5) and as a sophisticated system for in vitro expression of bead-anchored DNA for protein display (6).
The flow cytometric-based titration method presented here, using fluorescently labeled emulsion PCR products, provides a quick, cost-effective means for studying the composition of emulsion PCR products that is capable of reliably generating titration curves identical to those obtained by sequencing-based Roche/454 titration. In addition, the results demonstrate that FACS sorting can be used to enrich fractions of DNA-covered beads prior to sequencing. Collecting DNA-covered beads by FACS sorting is a straightforward approach for obtaining samples in which virtually 100% of the retained beads carry amplified template DNA. Furthermore, we show that the sequence quality is not affected by FACS sorting, and that automated FACS enrichment is capable of efficiently generating high amounts of pure DNA-carrying beads. We believe that these methods will significantly improve massively parallel sequencing efforts by reducing the costs, reagent usage and workload while increasing the sequence output and reproducibility.
MATERIALS AND METHODS
Library preparation and emulsion PCR
An amplicon library generated by amplifying human genomic p53 exon 2 (target size 193 bp) with fusion primers consisting of a target-specific sequence with a 19-mer A/B handle sequence at the 5′-end (upstream primer with A sequence, 5′-GCCTCCCTCGCGCCATCAG-3′; downstream primer with B sequence, 5′-GCCTTGCCAGCCCGCTCAG-3′) and a shotgun whole-genome bacterial library (genome size 2 Mb) with universal shotgun 44-mer A/B Roche/454 adaptors were prepared and amplified by emulsion PCR according to the Roche/454 manufacturer's instructions (Roche, Basel, Switzerland). This procedure yields dsDNA sequences coupled to DNA Capture Beads through the immobilized shotgun or amplicon B probes with a biotin moiety at the 5′-end of the protruding strand.
For the analysis of the p53 amplicon library, emPCR reactions were performed with a DNA-to-bead ratio of two copies per bead (cpb), which had been found to be optimal in preliminary sequencing titrations. The emulsions were broken and the beads were recovered by washing them in isopropanol and ethanol, then second strand removal was carried out by washing the beads twice in 0.1 M NaOH and then twice in 1× Annealing Buffer (1×AB). For the shotgun bacterial library emPCR samples for each of the DNA-to-bead ratios 0.5, 2, 4 and 16 were prepared. In addition, for sequence quality evaluation and to investigate the possible impact of bead density per sequencing lane additional samples with four cpb were prepared. In all shotgun samples, the second strands were retained. The beads and AB were supplied by Roche, and all of these procedures were performed in accordance with Roche's recommendations.
Labeling, FACS analysis and sorting
Following strand separation, the single-stranded DNA attached to the amplicon library beads was hybridized with a probe (5′-GCCTCCCTCGCGCCATCAG-3′) supplied by MWG (Edersberg, Germany) complementary to the projecting A′ sequence. Parallel samples were labeled with probes carrying Alexa647 fluorescent dye labels or with unlabeled probes in combination with SYBR Green I Nucleic Acid Gel Stain (Molecular Probes, Inc., Eugene, OR, USA) for FACS analysis of beads carrying amplified DNA template. For the SYBR Green labeling, 20 pmol of the unlabeled amplicon A probe supplied by MWG was added to approximately 300 000 beads from the amplicon library and 1 μl of 50× SYBR Green in a solution containing 1×AB in a total volume of 28 μl. The samples were heated to 92°C for 20 s, and then incubated for 20 min at 40°C in the dark, with shaking. As a negative control 15 000 nonreacted DNA Capture Beads (Roche, Basel, Switzerland) were labeled in the same manner. For the Alexa647 labeling, 20 pmol of Alexa647 fluorescent dye-conjugated A-probe was added to approximately 300 000 beads in a solution containing 1×AB, in a total volume of 27 μl. The resulting mixture was heated to 92°C for 20 s, then incubated for 20 min at 40°C in the dark, with shaking. As a negative control a duplicate sample was labeled using the same concentration of a noncomplementary Alexa647-conjugated probe (5′-GCCTTGCCAGCCCGCTCAG-3′), also supplied by MWG. The labeled samples were separately transferred to 5 ml Polystyrene Round-Bottom FACS Tubes (BD Biosciences, Franklin Lakes, NJ, USA) and diluted by adding 500 μl phosphate-buffered saline (PBS) containing 0.1% Pluronic F108 NF surfactant (PBSP) (BASF Corporation, Mount Olive, NJ, USA).
The recovered shotgun library samples with biotinylated dsDNA immobilized on the bead surfaces, obtained from emPCR reactions for each titrated DNA-to-bead ratio, were directly labeled by adding 8 μl streptavidin-conjugated Alexa647 dye (0.2 mg/ml, Invitrogen, Carlsbad, CA, USA) and 1×AB to a total volume of 100 μl. A negative control was also prepared, consisting of 15 000 nonreacted DNA Capture Beads labeled with 2 μl streptavidin-conjugated Alexa647 dye (0.2 mg/ml) in 1×AB, in a total volume of 27 μl. Samples were incubated at room temperature for 10 min in the dark, then transferred to FACS tubes and diluted by adding 500 μl PBSP for subsequent flow cytometric analysis. To evaluate the washing efficiency, following FACS analysis fractions of the samples were transferred to Ultrafree®-MC Centrifugal Filter Units (Millipore, Billerica, MA, 0.65 μm pore size) and the labeling and second strands were removed by washing twice in 150 μl 0.1 M NaOH for 5 min, and then three times in 150 μl 1×AB. Thereafter, a second round of labeling with streptavidin-conjugated Alexa647 and FACS analysis was performed as described above.
Fluorescence signals from the SYBR Green and Alexa647 dye labeled beads were detected at 530/30 nm and 660/20 nm, respectively, using a FACSVantage SE flow cytometer (BD Biosciences) with 488 nm and 633 nm excitation lasers, fitted with a 100 μm nozzle. FACS enrichment of DNA-carrying beads was carried out by collecting beads from the fluorescent population into a microcentrifuge tube containing 150 μl 1×TE (10 mM Tris–HCl, pH 7.5, 1 mM EDTA). To avoid beads sticking to and drying on the walls of the tubes, the tubes were regularly flushed using 50 μl 1×TE buffer. The sorted beads were washed in NaOH as previously described and beads were resuspended in 250 μl 1×AB. The flow-cytometry data were analyzed using CELLQUEST software (BD Biosciences).
Sequencing using the Roche/454 system
All of the titration and sequencing by the Genome Sequencer GS FLX (454 Life Sciences, Branford, CT, USA) steps were performed according to the manufacturer's instructions. Briefly, the shotgun library was sequencing-titrated by loading 24 000 beads from samples with 0.5, 2, 4 and 16 cpb into separate lanes of a picotiter plate (PTP) fitted with a 16-lane gasket and sequenced using an LR70 kit according to the manufacturer's instructions. For sequence quality assessment 5650 FACS-enriched beads from the 4 cpb emPCR products of the shotgun library were loaded and sequenced using an LR70 kit in one lane of a 16-lane PTP. In addition, to investigate bead density impact on sequence quality 15 000, 30 000, 40 000 and 50 000 FACS-enriched beads from the same library were loaded and sequenced in separate lanes of a 16-lane PTP as described above.
RESULTS
Overview
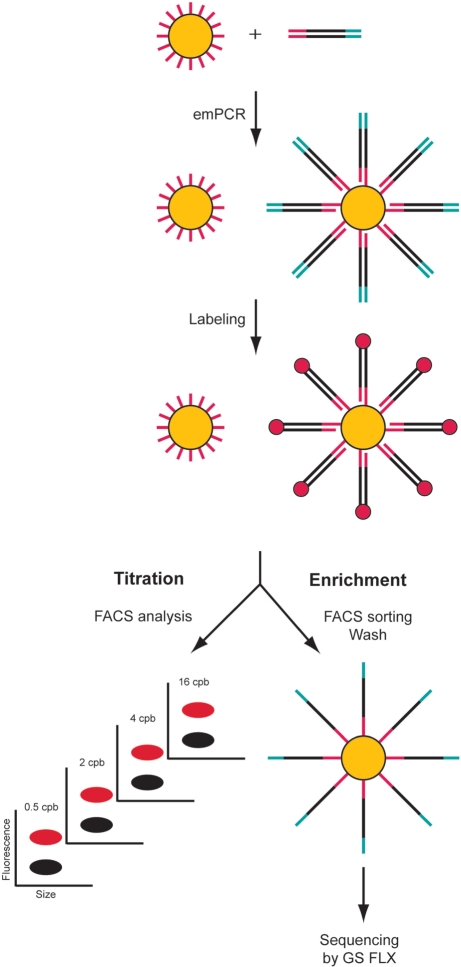
The aim of this study was to demonstrate the feasibility of using a rapid, highly efficient flow cytometric approach for preparing DNA-carrying beads to be used in massively parallel DNA sequencing. The results show that this strategy for identifying beads carrying amplified DNA provides a means for efficient enrichment of DNA-carrying beads for sequencing using the Roche/454 system, as well as a highly accurate and convenient alternative to titration by sequencing. FACS titration and enrichment is straightforward, as depicted in Figure 1. Emulsion PCR reactions are carried out with target sequences flanked by Roche/454 A and B primers, introduced either directly by PCR or by linker ligation. The sequences are captured and amplified on the surface of DNA Capture Beads via the immobilized B probe and an A primer in solution carrying a biotin molecule at its 5′-end. DNA-carrying and empty beads are distinguished by fluorescent labeling of the DNA and fluorescent beads are quantified (titration) and/or sorted (enrichment) using FACS. Labeling is removed prior to loading onto the PTP for sequencing.
Figure 1.
Schematic picture of the FACS titration and FACS enrichment work process. Following emulsion PCR, beads carrying amplified target DNA are fluorescently labeled to distinguish them from the nonreacted beads. The proportions of DNA-covered and nonreacted beads in titration samples are then determined by flow cytometric analysis, and used to create a titration curve. In addition, fluorescent DNA-carrying beads for subsequent sequencing can be enriched by FACS sorting. The fluorescent labels are removed by washing in NaOH and massive sequencing is carried out using the Roche/454 system.
Labeling
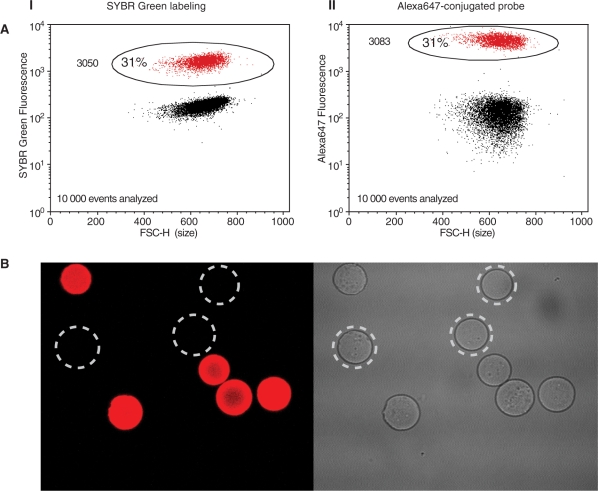
Two strategies (nonspecific labeling using SYBR Green or hybridization-specific labeling using an Alexa647 dye conjugated probe; Figure 2, left and middle panels, respectively) were used to label a model system consisting of a p53 exon 2 amplicon library conjugated to sepharose beads, which were then analyzed by flow cytometry. Figure 3A shows that both labeling strategies effectively label DNA-carrying beads, and both strategies yielded estimates of the fraction of beads carrying DNA template of 31%. Both SYBR Green and the Alexa647 fluorescent dye conjugated probe adhere to some degree to the empty DNA Capture Beads, causing a shift in the fluorescence of the beads compared to unlabeled beads. However, the fluorescence intensity of the empty beads is still weak in comparison to that of beads carrying amplified DNA, hence the two populations can be readily distinguished. The microscopic images in Figure 3B show that labeled beads retain their size and fluorescence properties after flow cytometric sorting, further demonstrating that FACS sorting does not affect the physical properties of the beads, even at high sort speeds.
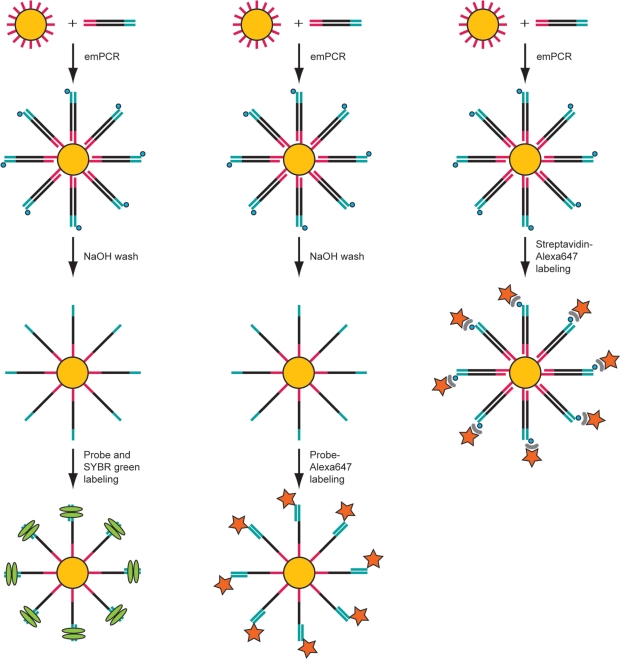
Figure 2.
Schematic picture of the different labeling strategies. Samples from which the second strand has been removed by NaOH washing can be indirectly labeled using a nonlabeled probe in combination with SYBR Green (green ellipse) (left panel) or directly by hybridization of an Alexa647-coupled probe (middle panel). Direct sample labeling of beads with the second strand retained can be carried out by binding streptavidin-conjugated Alexa647 dye (star) to the biotin moiety (blue circle) on the protruding DNA strand (right panel).
Figure 3.
Flow cytometric analysis providing distinct separation of the two bead populations, and confocal micrograph of labeled emPCR samples. (A) Dotplots from FACS analysis of amplicon library emulsion PCR products labeled with (I) SYBR Green, and (II) Alexa647-conjugated probe, with fluorescence on the y-axis and FSC-H (Forward Scatter Height, size) on the x-axis, providing clear discrimination between the populations of highly fluorescent DNA-carrying (circled) and weakly fluorescent empty DNA Capture Beads in each sample. The numbers of labeled beads are listed to the left in each dotplot. (B) A confocal microcrograph taken using a LSM 5 Pascal instrument (Carl Zeiss, Germany) with a 40× objective of an emPCR sample, in which the DNA-covered beads are labeled with streptavidin-conjugated Alexa647 dye.
To assess the utility of this protocol for enrichment, we also investigated the possibility of removing the labeling from the DNA capture beads using NaOH to eliminate both probes and dyes. The signal was effectively eliminated from beads labeled with the Alexa647 dye-conjugated probe but not from SYBR Green-labeled beads, therefore we used Alexa647-labeling in further analyses (data not shown). For practical reasons, we also switched to a streptavidin-conjugated Alexa647 dye, which couples to the biotin molecule on the amplified dsDNA extending from the surface of the beads without the need for NaOH-mediated strand separation (Figure 2, right panel).
Titration
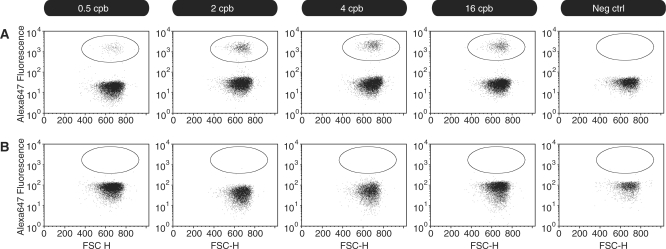
The initial results prompted us to analyze a more complex sample, a bacterial shotgun library, which was initially analyzed by Roche/454 sequencing-titration. The library was emPCR-amplified using DNA copy-to-bead ratios of 0.5, 2, 4 and 16, the products were labeled with streptavidin-conjugated Alexa647, and the labeled samples were analyzed immediately by flow cytometry. Figure 4A exemplifies that the fractions of DNA-carrying beads and empty beads can be clearly distinguished, confirming previous observations from the p53 amplicon data.
Figure 4.
FACS-titration analysis of samples from a shotgun library. (A) emPCR-amplified samples with four different DNA-to-bead ratios were labeled with streptavidin-conjugated Alexa647 dye and analyzed by flow cytometry. The fluorescence is shown on the y-axis and FSC-H (Forward Scatter Height), reflecting objects' sizes, on the x-axis. (B) Only the nonfluorescent population remained after removing second strands by washing in NaOH and relabeling with streptavidin-conjugated Alexa647 dye.
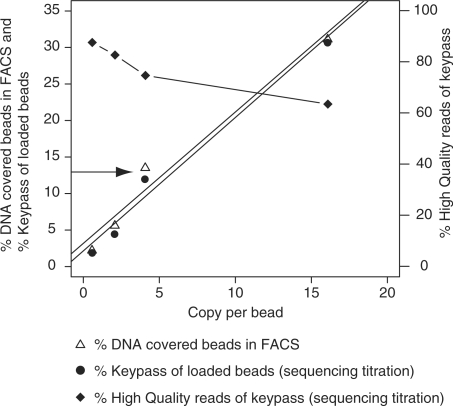
The proportion of DNA-covered beads increase as the DNA-to-bead ratio is increased and good agreement can be demonstrated between FACS analysis data and Roche/454 shotgun sequencing-titration data (Figure 5, Supplementary Material 1). For each sample, the proportion of DNA-covered beads determined by flow cytometry was nearly identical to the fraction of beads with a readable key sequence in the sequencing-titration analyses. These results corroborate the finding that titration by FACS can be used as a convenient alternative to sequencing-titration to identify the most suitable DNA-to-bead ratio.
Figure 5.
Comparison of data obtained by FACS titration and sequencing-titration of a shotgun library. Plot showing the percentage of DNA-carrying beads according to FACS analysis (triangles, left y-axis), obtained from emPCR-amplified samples from a shotgun library with four different DNA-to-bead ratios. Also depicted is the overlapping proportion of keypass reads out of loaded beads (circles, left y-axis) and the percentage of HQ reads out of loaded beads (diamonds, right y-axis) generated in Roche/454 sequencing-titration of the duplicated samples. The arrow indicates the 12.5% limit of keypass reads out of loaded beads, which is used for selecting the optimal DNA copy-to-bead ratio (4 cpb).
Towards automated enrichment
We proceeded to investigate if FACS sorting could be used to enrich the DNA-carrying beads without adversely affecting their ability to act as high-quality (HQ) sequencing templates. For this, we first verified the previous observations that NaOH treatment efficiently removes the dye and the nonimmobilized complementary strands from the bead surfaces. Here, the obtained immobilized single-stranded targets were treated with an additional round of control labeling with streptavidin-conjugated Alexa647 dye to identify any residual double-stranded DNA templates. As shown in Figure 4B, the biotinylated second strand was completely removed by this procedure, in experiments with all tested DNA concentrations, without affecting the characteristics of the bead population.
The quality of FACS-enriched beads was analyzed by sequencing 5650 FACS-sorted DNA-positive beads from the shotgun library using the Roche/454 system. The resulting data was compared to sequencing data obtained from a lane loaded with 24 000 nonenriched emPCR beads from the same original sample. The proportion of HQ reads relative to the total number of keypass reads was in both cases 75% (Table 1). In order to qualify as a HQ sequence, beads with a readable key sequence must pass through several other filters, excluding beads carrying more than one extended template (and thus yielding mixed signals) and those with either too complex or too short sequences. Such flaws can result from either use of inappropriate DNA-to-bead ratios or variations in the sequence contents of libraries (which cannot be readily avoided using current sample preparation techniques). The average read length of FACS enriched beads and nonenriched beads was 240 and 247 bp, respectively, and the average GC content in both samples was 38.8% (Table 1). For the two samples both read length and GC content data followed a similar distribution pattern to that of a full shotgun data set from the same library (data not shown). Moreover, to investigate potential bias introduced by the enrichment protocol, reads were mapped to an assembly of a shotgun dataset from the same DNA library (Supplementary Material 5). No significant difference in coverage between the FACS enriched sample and the nonenriched or the full shotgun samples could be visually observed in the graph. The results show that labeling, FACS sorting and washing did not influence the beads or the DNA in a manner that affected the sequence quality or the sample coverage. Thus, the results show that labeling, FACS sorting and washing did not influence the beads or the DNA in a manner that affected the sequence quality. Indeed, sample purities as high as 100% DNA covered beads have been found in re-analyses of FACS-enriched beads (data not shown), indicating that the number of HQ reads can be further increased, while retaining (or improving) the bead/DNA quality and increasing the number of loaded beads that carry a readable sequence as compared to enrichment by standard Roche/454 protocol.
Table 1.
Sequence quality unaffected by FACS enrichment
| FACS enriched | Not enriched | ||
|---|---|---|---|
| Loaded beads | 5650 | 24 000 | |
| Keypass wells | 4783 | 2875 | |
| HQ wells | 3589 | 2160 | |
| HQ of key pass (%) | 75.0 | 75.0 | |
| Average Readlength (bp) | 240 | 247 | |
| GC content (%) | 38.8 | 38.8 |
Comparison of FACS-sorted beads and nonenriched beads, demonstrating that the part of HQ reads as a percentage of beads with a readable key sequence, the average read length and the GC content is equal in the two samples. Hence, the sequence quality of the beads is not affected by automated FACS enrichment.
We also performed a sequencing run with increasing numbers of FACS enriched beads up to the current standard loading density (15 000, 30 000, 40 000 and 50 000) in one region each using the 16-lane gasket on the PTP. Saturation in the frequency of HQ wells was approached as the number of loaded beads increased towards the higher end of the tested range (Supplementary Material 2). Interestingly, the fraction of short sequences increased with increasing numbers of beads contributing to a minor decrease in the percentage of HQ wells, indicating increased noise possibly due to more cross-talk between wells.
DISCUSSION
The results presented in this communication show that a straightforward FACS procedure can provide a convenient, relatively inexpensive alternative to the costly sequencing-titration in the preparation of samples for sequencing using the Roche/454 system. In conventional Roche/454 sequencing-titration, the optimal DNA concentration is determined by identifying the DNA-to-bead ratio that yields a desired number of keypass reads. The bead concentration in the sample is determined using a bead counter, then volumes containing 24 000 beads (with a range of DNA-to-bead ratios) are loaded onto a PTP and left to sediment for 10 min. After removing the supernatant the remaining beads are sequenced. The DNA concentration that gives rise to the number of keypass reads closest to 3000, or 12.5% of the number of loaded beads, is then selected for use in a full-scale sequencing run. Thus, variations in the quantities of beads loaded in the wells and/or the sedimentation in the PTP inevitably lead to inconsistencies in the titration data. In contrast, in the FACS approach there is no need to quantify the beads prior to analysis. Instead, by labeling emulsion PCR products based on an appropriate range of DNA-to-bead ratios, a titration curve can be simply generated (by scanning the beads in the samples and determining the fractions of beads carrying a fluorescent label) from which the optimal DNA-to-bead ratio for full-scale sequencing runs can be readily selected. In addition, replacing 454/Roche sequencing-titration by FACS titration would reduce the titration cost to 1/3 of the price (Supplementary Material 3).
In both the GS FLX Standard chemistry Long Paired End Sequencing protocol and the GS FLX Titanium chemistry pipeline, an alternative method for titration is applied, based on assessment of the proportion of enriched beads obtained to the total number of input beads. Briefly, in these procedures, triplicate emPCR samples with four different DNA-to-bead ratios are prepared and the samples are enriched using the standard or Titanium protocols, respectively. A target copy to bead ratio that yields ∼8–10% enriched beads is selected for the full sequencing run. This method does not require sequencing, but a total of 12 emPCR reactions for each titration must be performed for the enrichment procedure to work efficiently. Furthermore, as in the established Roche/454 sequencing-titration protocol this approach involves an indirect quantification of the DNA covered beads, which inevitably introduces bias. The active identification and quantification of DNA covered beads employed in FACS analysis also renders a sample quality assessment prior to sequencing which the other methods of titration are lacking. There are also several alternative ways to quantify libraries prior to emPCR, using either quantitative PCR (7) or digital PCR, as in the commercially available Slingshot kit from Fluidigm (San Francisco, CA, USA). Clearly, however, these indirect methods do not take into account differences in solid phase hybridization and emulsion PCR efficiency related to the characteristics of individual libraries. In addition, although such methods are well suited for determining the concentration of ancient samples, the fragment length distribution of shotgun libraries has been shown to bias the concentrations obtained by quantitative PCR (7).
In this study, we show that FACS sorting offers a relatively rapid and highly convenient method for obtaining highly purified DNA-covered beads without affecting the sequence quality or bias the base composition, read length or coverage of the sample. Sorting a million DNA-carrying beads from a sample containing 92% empty beads and 8% DNA-carrying beads for loading onto one large region of a standard PTP could be done in an automated fashion in <1 h using a modest sort speed of 3000 beads/s. In this context, it is worth noting that current FACS sorters can achieve sorting speeds as high as 70 000 beads/s (MoFlo, Beckman Coulter, Fullerton, CA, USA). This may also increase interest for implementing the FACS procedure in the SOLiD (Applied Biosystems) pipeline, requiring preparation of larger amounts of DNA-carrying beads. We have successfully employed FACS labeling using a protocol similar to the one outlined in this communication on SOLiD beads (data not shown).
An interesting aspect to consider is the extent to which the number of HQ reads could be increased by loading solely DNA-positive beads onto the PTP. Currently, saturation in the number of HQ reads may occur when the number of DNA-covered beads loaded is increased beyond a certain limit, mainly because of light scattering between wells. However, with the introduction of a Titanium coated PTP in the GS FLX Titanium pipeline, it is anticipated that cross-talk between wells will be greatly reduced. If every plate is loaded at maximum density, FACS enrichment could ensure that every loaded bead carries an amplified template DNA, significantly increasing the sequencing efficiency, while improving and automating the upstream sample preparation. Moreover, as the capacity of the sequencing systems grows larger there is an increased need to multiplex different samples in the same run. FACS enrichment using color-coded probes opens up for tag-specific enrichment after emPCR, thus allowing loading and sequencing of an appropriate number of beads per individual project.
By using lower starting DNA-to-bead ratios the risk of obtaining beads carrying multiple target molecules in the emPCR could be minimized (Supplementary Material 1). Based on several titration sequencing runs (Supplementary Material 4), we estimate that the initial DNA concentration can be diluted by, on average 2-fold, given a FACS sort rate of 10 000 beads per second and a set sorting time of 1.5 h. These conditions would yield mean reductions in mixed reads of 40%. This possibility, in combination with higher sorting speeds, could yield very pure populations of single template DNA beads, thereby facilitating even greater increases in capacity with the latest Titanium chemistry.
In summary, we have demonstrated that FACS sorting has great potential to replace the titration step in massively parallel pyrosequencing, and that it provides a simple strategy for enriching DNA-carrying beads to extremely high purity, without affecting the sequence quality of the beads.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Scientific Council; the Swedish Foundation for Strategic Research; the Knut and Alice Wallenberg Foundation. Funding for open access charge: Swedish Scientific Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to thank Kristina Holmberg and Christian Natanaelsson for their help with sample preparation and sequencing, Hjalmar Brismar for microscopy assistance and Siv Andersson for library samples.
REFERENCES
- 1.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 2.Droege M, Hill B. The Genome Sequencer FLX System–longer reads, more applications, straight forward bioinformatics and more complete data sets. J. Biotechnol. 2008;136:3–10. doi: 10.1016/j.jbiotec.2008.03.021. [DOI] [PubMed] [Google Scholar]
- 3.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parks DR, Bryan VM, Oi VT, Herzenberg LA. Antigen-specific identification and cloning of hybridomas with a fluorescence-activated cell sorter. Proc. Natl Acad. Sci. USA. 1979;76:1962–1966. doi: 10.1073/pnas.76.4.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dressman D, Yan H, Traverso G, Kinzler KW, Vogelstein B. Transforming single DNA molecules into fluorescent magnetic particles for detection and enumeration of genetic variations. Proc. Natl Acad. Sci. USA. 2003;100:8817–8822. doi: 10.1073/pnas.1133470100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nord O, Uhlen M, Nygren PA. Microbead display of proteins by cell-free expression of anchored DNA. J. Biotechnol. 2003;106:1–13. doi: 10.1016/j.jbiotec.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 7.Meyer M, Briggs AW, Maricic T, Höber B, Höffner B, Krause J, Weihmann A, Pääbo S, Hofreiter M. From micrograms to picograms: quantitative PCR reduces the material demands of high-throughput sequencing. Nucleic Acids Res. 2008;36:e5. doi: 10.1093/nar/gkm1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.