Abstract
Gene expression analysis performed through comparative abundance of transcripts is facing a new challenge with the increasing need to compare samples of known cell number, such as early embryos or laser microbiopsies, where the RNA contents of identical cellular inputs can by nature be variable. When working with scarce tissues, the success of microarray profiling largely depends on the efficiency of the amplification step as determined by its ability to preserve the relative abundance of transcripts in the resulting amplified sample. Maintaining this initial relative abundance across samples is paramount to the generation of physiologically relevant data when comparing samples of different RNA content. The T7 RNA polymerase (T7-IVT) amplification is widely used for microarray sample preparation. Characterization of the reaction's kinetics has clearly indicated that its true linear phase is of short duration and is followed by a nonlinear phase. This second phase leads to modifications in transcript abundance that biases comparison between samples of different types. The impact assessment performed in this study has shown that the standard amplification protocol significantly lowers the quality of microarray data, rendering more than half of differentially expressed candidates undetected and distorting the true proportional differences of all candidates analyzed.
INTRODUCTION
In the last decade, the emergence of high-throughput technologies has increased the rate of discoveries and expanded our knowledge on molecular mechanisms and biological processes. In the field of transcriptomics, microarrays are widely utilized for comparative RNA abundance studies and have been shown to be a powerful tool for cellular functions, metabolic pathways and pathologies studies.
One of the main limitations of microarrays is the requirement for substantial RNA input to generate hybridization samples. As a consequence, many research applications with limited access to biological samples, such as laser microdissection, early mammalian development or single cell gene expression profiling, must rely on an amplification step to generate sufficient input material. For this purpose, several methods have been proposed, e.g. RiboSPIA (1), SMART PCR (2) or T7-IVT (3). The T7-IVT approach is currently the preferred method due to the linearity of its reaction kinetics. Reliability of this method has been assessed to ensure the generation of physiologically relevant microarray data. To date, the validation of this amplification method has been achieved by evaluating its ability to maintain the relative abundance between transcripts within a particular sample. By correlating the relative proportions of candidates between the unamplified and amplified aliquots of a sample (4), it was determined that this method introduces a slight bias in overall transcript representation. Indeed, higher GC contents and the presence of secondary structures found on some mRNAs are assumed to lead to lower amplification rates which results in a bias in the relative proportions of these gene transcripts relatively to the bulk of the messengers that are amplified with more ease (5–7). Nonetheless, the T7-IVT was deemed an appropriate global amplification method and has become well accepted, considering that the potential for discovery exceeds the noise contribution from these biases. Consequently, various technological platforms, including Affymetrix, have implemented the systematic use of this amplification step in their standard procedures (8,9).
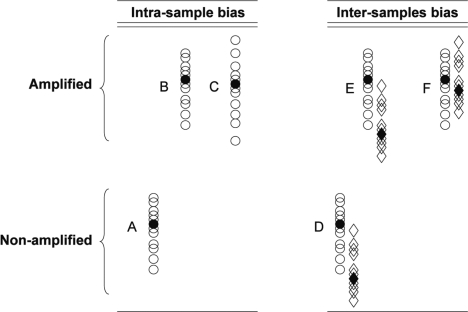
Traditionally, when working with abundant samples, fixed amounts of RNA are compared and cell numbers are accounted for through standardization using so-called housekeeping candidates, which has since been proven to be problematic as stable candidates across all treatments or cellular states, let alone all tissues, have been very difficult to find (10–14). As an alternative to housekeeping genes, normalization to spiked controls is often performed. This technique efficiently accounts for technical imprecision but does not take into consideration the parameters of the initial samples, including cell counts. In other words, in a situation of fixed RNA input, differences observed in transcript abundance become physiologically relevant only with the assumption that the cells from these samples share a similar level of overall gene expression, hence of RNA content. However, many cellular states are known for their atypical transcriptional activity. For instance, senescent cells, such as germ cells or stem cells, are known to be more quiescent, whereas actively dividing cancer cells are metabolically hyperactivated (15) and thus exhibit high transcription levels. These differences in transcription activity between samples of different cell type and/or status carry highly valuable information that could affect data analysis. The ability to preserve these differences is invaluable for comparative microarray strategies. Samples of fixed cell number offer a novel perspective by enabling the comparison of transcript abundance across samples of different type. Consequently, following the development of techniques for the isolation of individual cells from complex tissues, comparative studies with samples of fixed cell number have become increasingly common. The comparison of samples of known cell number dictates that in addition to the proportional representation of mRNA between amplified and unamplified aliquots of the same sample, an additional proportional representation must be preserved throughout the microarray sample preparation procedure. As such, the amplification step is required to preserve two types of proportions: (i) the aforementioned relative abundance between transcripts within samples, and (ii) the proportional differences in total RNA contents naturally occurring between samples of different type. Figure 1 illustrates the two types of proportional distortions where the sequence related bias is illustrated by lanes A–C, whereas the proportional representation between samples is found in lanes D–F. To date, amplification methods have not been tested for their ability to maintain the latter type of proportion.
Figure 1.
Schematic representation of the two potential biases that need to be considered when using an amplification step to produce relevant microarray data. Open circles represent individual expressed sequence tags (ESTs) that are distributed according to their relative proportions (higher position meaning more abundant). Columns A–C represent the intrasample bias where A: the original unamplified distribution; B: the amplified output with original proportions kept; and C: is the amplified output that harbors skews in the relative representations between ESTs. Columns D–F represent the intersample bias where D: relative distributions of the two original samples; E: amplified outputs with kept proportions between samples; and F: amplified outputs with skewed relative proportions between the samples but intact relative proportions within samples.
Here, we tested the ability of T7-IVT amplification to maintain this relative global abundance differences across samples of different RNA contents. The comparisons were performed using bovine oocytes and early embryos. It is known that in the cow, the first cell cycles occur in the absence of transcription (16,17). During this period of transcriptional silence, protein production is sustained through the use of the mRNAs stored during oogenesis. These maternal RNA pools are gradually depleted until embryonic genome activation (EGA), which in the cow occurs at the 8- to 16-cell stage (17). Therefore, prior to EGA, the number of nuclei is irrelevant as the RNA content across development represents the fraction of RNA remaining from the original oocyte pools. Consequently, to characterize the making of an early embryo prior to the EGA, it is the RNA content of the whole embryo that is compared and considered as the experimental unit. The comparisons performed herein accounted for the two extremes in RNA contents, i.e. the rich state that is the fully grown germinal stage oocyte to the lower-abundance state represented by the early 8-cell embryo. The linearity of the T7-IVT was characterized and, according to the kinetics observed, a procedure to follow antisense (aRNA) production in real time was developed. The global amplification method was tested to generate samples suited for microarray hybridization by comparing the RNA content of these developmental samples under the standard conditions and following a modified amplification procedure. Validation of the proportional RNA abundance differences was done using real-time PCR. The results clearly indicate that the modified procedure generates physiologically relevant RNA abundance data, whereas the recommended protocol introduces an important skew in the relative abundance values.
MATERIALS AND METHODS
All chemicals were obtained from Sigma–Aldrich (St. Louis, MO, USA) unless otherwise stated.
Oocyte recovery and selection
For germinal vesicle (GV) stage oocyte samples, bovine ovaries were collected at a commercial slaughterhouse and transported to the laboratory in a 0.9% NaCl aqueous solution containing an anti-mycotic agent. Cumulus–oocyte complexes (COCs) from 3- to 6-mm follicles were manually aspirated with an 18-gauge needle attached to a 10-ml syringe. Healthy COCs with at least five layers of cumulus were selected to proceed. The cumulus cells were removed mechanically by vortexing. The denuded oocytes were placed in PBS and washed generously, at least three times, to ensure the absence of contamination by cumulus cells. Pools of 10 GV oocytes collected on different days were then frozen in a minimal volume of PBS and stored at –80°C until RNA extraction.
In Vitro production of 8-cell embryos
For in vitro maturation, COCs bearing the characteristics described above underwent in vitro maturation after three washes in HEPES-buffered Tyrode lactate medium (TLH) supplemented with 5% bovine serum, 0.2 mM pyruvic acid and 50 µg/ml gentamicin. Groups of 10 COCs were placed in droplets of media under mineral oil. Each droplet consisted of 50 µl of maturation medium composed of modified synthetic oviductal fluid (SOF) medium (18,19) with 5% decomplemented fetal calf serum (Hyclone, Ottawa, ON, Canada), 1X modified Eagle's medium (MEM) nonessential amino acids (Gibco BRL, Burlington, ON, Canada), 1X MEM essential amino acids (Gibco BRL), as well as 1 mM glutamine supplemented with 0.1 µg/ml recombinant FSH and 1 µg/ml 17β-estradiol. The droplets containing COCs were incubated in a humidified atmosphere for 24 h at 38.5°C with 5% CO2.
In vitro fertilization of matured COCs were added to 48-µl droplets under mineral oil. The droplets were composed of modified TLH supplemented with 0.6% BSA fatty-acid free (Lifeblood Medical, Adelphia, NJ, USA), 0.2 mM pyruvic acid, 10 µg/ml heparin and 50 µg/ml gentamicin. Prior to transfer, the COCs were washed twice in TLH medium. Once transferred, 2 µl of PHE (1 mM hypotaurine, 2 mM penicillamine, 250 mM epinephrine) were added to each droplet ∼10 min before semen was added. The semen used consisted of a cryopreserved mixture of ejaculates from five bulls (Centre d’Insémination Artificielle du Québec, St.-Hyacinthe, QC, Canada). The semen was thawed in 37°C water for 1 min, put on a discontinuous Percoll gradient (2 ml of 45% Percoll over 2 ml of 90% Percoll) and centrifuged at 700 g for 30 min at 26°C. The pellet was resuspended in 1 ml of modified Tyrode medium and centrifuged at 250 g for 5 min at 26°C. The supernatant was discarded, an aliquot was used to determine the spermatozoa content using a hemocytometer, and the remaining fraction was resuspended in IVF medium to obtain a final concentration of 25× 106 cells/ml. Finally, 2 µl of the sperm suspension were added to each droplet and incubated in a humidified atmosphere at 38.5°C in 5% CO2 for 15–18 h.
Following fertilization, putative zygotes were mechanically denuded by repeated pipetting, washed three times in SOF1 with 5% decomplemented fetal calf serum for complete removal of cumulus cells from solution and transferred to culture droplets (50 µl) in groups of 20–30 embryos. Embryo culture was performed in SOF1 under mineral oil at 38.5°C in 5% CO2 in a reduced oxygen atmosphere (7%) with high humidity. SOF1 medium was replaced after 72 h by SOF2 to prevent toxicity due to ammonium accumulation resulting from amino acid degradation. The SOF1 medium contained 5% decomplemented fetal calf serum, 1× MEM nonessential amino acids, 1 mM glutamine, 1.5 mM glucose and 10 mM EDTA. The SOF2 medium contained 5% decomplemented fetal calf serum, MEM nonessential amino acids, MEM essential amino acids, 1 mM glutamine and 1.5 mM glucose. The 8-cell embryos were collected 72 h postfertilization, washed three times in PBS, collected in groups of 10 in small volumes of PBS, frozen and stored at –80°C until RNA extraction.
RNA extraction and standard global mRNA amplification
Total RNA from oocytes and 8-cell embryos was extracted using the PicoPure RNA isolation Kit (Molecular Devices, Sunnyvale, CA, USA) following the manufacturer's protocol and including DNase I treatment to remove genomic DNA. Total RNA integrity and concentration were evaluated using the 2100-Bioanalyzer (Agilent Technologies, Palo Alto, CA, USA) with the RNA PicoLab Chip (Agilent Technologies).
The global mRNA amplifications by T7-IVT were performed using the RiboAmp HS RNA Amplification Kit (Molecular Devices) according to the manufacturer's instructions. In the standard amplification procedure, two amplification rounds of 6 h each were performed. The aRNA output was quantified using the NanoDrop ND-1000 (NanoDrop Technologies, Wilmington, DE, USA).
Determination of first-round amplification kinetics
Total RNA of a pool of 10 GV oocytes and one of 10 eight-cell embryos was extracted as described above, and both samples were subjected to the first amplification round following the manufacturer's instructions. During the IVT incubation, a 1 µl aliquot was removed after 0, 10, 20, 30, 40, 50, 60, 120, 180, 240, 300 and 360 min. The 12 aliquots were then reverse transcribed using the Sensiscript Reverse Transcriptase (Qiagen, Mississauga, ON, Canada) according to the manufacturer's instructions, except that 2.5 µM of random decamers RetroScript (Applied Biosystems/Ambion, Austin, TX, USA) were used instead of an oligo-dT. Real-time PCR (as described below) was performed on each GV oocyte and 8-cell embryo cDNA aliquot using primers targeting a high-abundance gene [actin, gamma 1 (ACTG1)] and a low-abundance candidate [High-mobility group box 1 (HmgB1)].
Determination of second-round amplification kinetics
Total RNA of pools of 10 GV oocytes and 10 eight-cell embryos were amplified by two rounds of amplification with a minor modification. During the second round, 30 µCi of alpha-32P UTP (GE Healthcare, Baie d’Urfé, QC, Canada) was added to the IVT reaction. Two-microliter aliquots were collected after 0, 15, 30, 45, 60, 120, 240 and 360 min of reaction. The eight aliquots were cleaned using the PicoPure RNA isolation Kit (Molecular Devices) and eluted in an 11 µl volume. A 5-µl aliquot of each aRNA eluate was combined with 10 ml of scintillation liquid (Budget solve scintillation cocktail, VWR, Mississauga, ON, Canada). The counts per minute (CPM) were measured using a liquid scintillation counter (Tri-Carb 2100TR, PerkinElmer BioSignal, Inc., Montreal, QC, Canada).
Broken beacon design and validation
The broken beacon is composed of two oligonucleotides (Figure 2) and is designed to target the aRNA coding for the bovine actin, beta (ACTB) gene. The oligonucleotide bearing the complementary sequence to the ACTB aRNA contains a fluorophore (fluo-oligo), whereas the second oligonucleotide contains a quencher (quencher-oligo). The choice of fluorophore was determined according to its suitability with the LightCycler 1.5 instrument (Roche Diagnostics, Laval, QC, Canada). Consequently, the fluo-oligo was synthesized with the 6-FAM on its 5′ extremity (Integrated DNA Technologies, Coralville, IA, USA) while the quencher-oligo was conjugated on its 3′ end with the Dabcyl dark quencher (Integrated DNA Technologies). Since it was observed that, with shorter excitation wavelength fluorophores such as FAM, the adjacent nucleotide can have a strong quenching impact, the design was restricted so the nucleotide adjacent to the fluorophore was a thymidine, which has the lowest quenching capacity (20). Selected sequences for the broken beacon, i.e. the sequences of the fluo-oligo, the quencher-oligo and an antisense single-strand DNA (ssDNA) control, are listed in Supplementary Data Table 1.
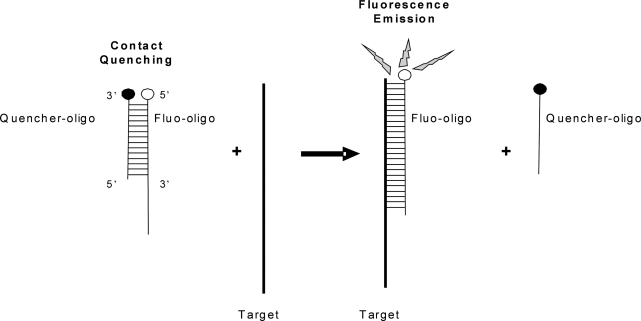
Figure 2.
Schematic representation of the broken beacon. The fluorescent oligonucleotide (fluo-oligo) forms a duplex with the quencher oligonucleotide (quencher-oligo); the proximity of the fluorophore and the quencher prevents fluorescence emission. In the presence of the target sequence, the quencher-oligo is displaced, allowing the fluo-oligo to emit.
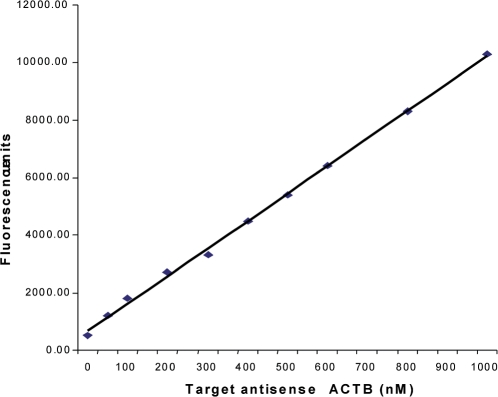
The ratio of fluo-oligo to quencher-oligo was set at 1:2, as this proportion was previously established to be optimal for minimizing background fluorescence (21,22). In order to evaluate the efficiency of the broken beacon to detect the target sequence, various amounts of the ssDNA target control were spiked into a solution mimicking the incubation conditions of the T7-IVT at the temperature used for the global mRNA amplification (42°C). A standard curve using the spiked ssDNA antisense ACTB was performed and fluorescence was monitored using the LightCycler. Each target concentration for the standard curve contained 20 µM of fluo-oligo and 40 µM of quencher-oligo. At this concentration, the fluo-oligo and the quencher-oligo were prehybridized together at room temperature at least 10 min in the dark prior to use. Antisense ssDNA target concentrations of 0, 50, 100, 200, 300, 400, 500, 600, 800 and 1000 nM were tested. The prehybridized fluo-quencher oligos were added to each of the samples spiked with the antisense ssDNA target in the IVT reaction solution (Molecular Devices) in a final volume of 20 µl. Due to the requirement of the T7 RNA polymerase for Mg2+ ions, all IVT samples were put into plastic capillaries (Roche Diagnostics). The size profile of the aRNA products of both rounds was determined using an aliquot of the end products that were run onto a microfluidic Chip read on a 2100-BioAnalyzer system (Agilent).
Real-time monitoring of aRNA production with the broken beacon
Real-time monitoring of aRNA production during the first and second T7-IVT incubations was performed using the broken beacon. For both T7-IVT rounds, the preparation and concentration of each oligo were as described above. All T7-IVT reactions were performed in plastic capillaries (Roche Diagnostics) and incubated at 42°C in a LightCycler apparatus (Roche Diagnostics) where the fluorescence was measured every 5 min in the F1 channel.
Experimental design for the comparative assessment of the impact of the T7-IVT duration
The total RNA of pools of 10 GV oocytes or 8-cell embryos was isolated as described above. Six samples for each developmental stage were extracted. Keeping each developmental stage separated, eluates containing the total RNA samples were pooled together in order to attenuate the biological and technical variations between pools, thus highlighting only the impact of the intrinsic difference in starting material between developmental stages. One-sixth of each pool was subjected to the T7-IVT amplification. The duration of the T7-IVT incubation was determined either to be 6 h as the standard protocol recommends or restricted according to the real-time monitoring of the reactions’ kinetics, the incubations being stopped when one of the samples started showing signs of reduction in aRNA production (roughly 40 and 50 min for the first and second rounds, respectively). The aRNA output was monitored in real-time for all samples. For the microarray hybridizations, a dual-color design using a common reference was used. An RNA reference made by T7-IVT from a pool of oocytes and early embryos was labeled and hybridized on all microarrays to account for hybridization and scanning imprecision. Following the amplification reaction, the aRNA content was quantified using the NanoDrop ND-1000 (NanoDrop Technologies).
Sample labeling and microarray hybridization
To avoid biases induced by lower signal intensities, which are generated by less-concentrated samples, a fixed amount of aRNA (2 µg for developmental samples and 2.5 µg for the common reference) was hybridized to each microarray. To account for the initial proportional differences in sample concentration, a synthetic GFP RNA (23) was spiked in prior to labeling and sampling. The GFP intensity values were latter used to correct the overall intensity values for each of the biological samples. All developmental aRNA samples were spiked with 100 ng of GFP RNA. The labeled aRNA samples spiked with the GFP control and the reference were pooled and purified using the Picopure RNA extraction kit (Molecular Devices) and resuspended in 12 µl of elution buffer before hybridization. Microarray samples (including the aRNA common reference) were labeled indirectly using DY647-ULS for the oocyte/embryo and DY547-ULS for the reference from the ULS aRNA Fluorescent Labelling kit (Kreatec Biotechnology, Amsterdam, The Netherlands) according to the manufacturer's protocol. The labeling efficiency was measured using the NanoDrop ND-1000 (NanoDrop Technologies).
All 12 hybridizations were performed using our custom bovine developmental cDNA microarray (v.1.3) containing 2441 ESTs derived from subtracted libraries (this EST collection represents 1540 Unigenes) (24). Hybridizations were performed in Slidehyb buffer #1 (Ambion, Austin, TX, USA) at 50°C for 18 h in the SlideBooster hybridization station (Advalytix, San Francisco, CA, USA). Slides were then washed twice with 2× SSC-0.5% SDS at 50°C for 15 min and twice with 0.5× SSC-0.5% SDS at 50°C for 15 min. The slides were dipped three times in 1× SSC and three times in H2O. Finally, the slides were dried by centrifugation at room temperature at 1200 g for 5 min.
The slides were scanned using the VersArray ChipReader System (Bio-Rad, Mississauga, ON, Canada) and visualized with the ChipReader software (Media Cybernetics, San Diego, CA, USA). Microarray image processing was performed with the ArrayPro Analyzer software (Media Cybernetics).
Microarray data analysis
A three-step strategy was used to normalize our microarray data. First, in order to remove any spatial effect on the array, an ANOVA analysis was performed on each array, by computing and subtracting any column effect (four columns of subarrays on each array), any row effect (considering only two rows of subarrays, i.e. the top 16 and the bottom 16 subarrays since they are identical replicates) and any column/row interaction. Then, in order to account for hybridization efficiency to normalize data between arrays and account for the array effect, an ANOVA analysis was performed on all the arrays using only the reference samples. An array effect was then estimated and removed from the expression of both the reference and the developmental stage expressions. Finally, since all microarray samples were prepared from a fixed amount of aRNA (2 µg) to avoid any bias introduced by the preponderance of low signal intensities in the less abundant samples, it was essential to use a spiked-in control to normalize and restore the natural divergences found in the initial sample concentrations. As such, the spiked-in GFP control, whose signal is proportional to the volume that was taken from each sample to get 2 µg of aRNA, was normalized to be constant across the different cell stages, which consequently transformed the datasets to account for the natural differences in RNA content that characterize the bovine early developmental stages. The normalized data was then uploaded to the NIA Array Analysis tool for analysis (25). First, background threshold was determined according to the plot of error function [SD (= square root of the error variance) versus expression level (log intensity)]. Clones with a mean log signal intensity above the calculated background threshold (log 2) were considered present. Next, a one factor one-way ANOVA (default parameters, except FDR < 0.05) was performed. Further data processing including correlation coefficient for technical replicates, hierarchical clustering and mean pair-wise comparison between the two biological samples were also performed using the NIA microarray analysis tool (http://lgsun.grc.nia.nih.gov/ANOVA/). The dataset is available on the ArrayExpress website (www.ebi.ac.uk/microarray/as/ae/).
Reverse transcription of nonamplified RNA and aRNA from oocytes and 8-cell embryos
Estimation of the impact of the duration of the T7-IVT incubation and validation of the microarray data were performed using real-time PCR measuring transcript abundance for specific candidate genes. Three different sample types were compared: (i) non-amplified, (ii) standard amplification (e.g. two rounds of 6 h) and (iii) restricted amplification using real-time monitoring of aRNA production. In the nonamplified group, total RNA of three pools of 10 oocytes and three pools of 10 eight-cell embryos were reversed transcribed using the Sensiscript Reverse Transcriptase (Qiagen) according to the manufacturer's instructions. With the samples from the standard and restricted T7-IVT, 1 µl of aRNA was diluted in 50 µl of RNase-free water (Sigma), and 1 µl of this dilution was reversed transcribed using the Transcriptor Reverse Transcriptase (Roche Diagnostics) following the manufacturer's protocol, except that 50 mM of random decamers RetroScript (Applied Biosystems/Ambion) and 1 µM of oligo-dT were used in the reaction. This switch in reverse transcriptase enzyme was necessary as the aRNA samples are too concentrated for the SensiScript whereas unamplified samples are too diluted for the Transcriptor.
Real-time PCR
The primers for each candidate were designed using the Primer3 web interface (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi). Sequences, size of amplified product, GenBank accession numbers as well as annealing temperatures are presented in Supplementary Data Table 2. For each target ESTs tested, a standard curve, consisting of PCR products purified with the QIAquick PCR Purification Kit (Qiagen) and quantified with a spectrophotometer (NanoDrop ND-1000, NanoDrop Technologies), was included in the run. The standard curve consisted of five standards of the purified PCR products diluted from 0.10 pg to 0.1 fg. Real-time PCR was performed on a LightCycler apparatus (Roche Diagnostics) using SYBR green incorporation for real-time monitoring of amplicon production. The reaction was performed in glass capillaries (Roche Diagnostics) in a final volume of 20 µl.
For the determination of first-round amplification kinetics, 1.5-µl aliquots of each oocyte and 8-cell cDNA sample were used per PCR reaction. For transcript abundance measurement in the nonamplified, amplified 6 h and restricted amplification samples, 1 µl of cDNA was used. In addition to cDNA template, the reaction mixture consisted of 0.25 mM of each primer, 3 mM of MgCl2 in 1× SYBR green mix containing dNTPs, FastStart DNA polymerase enzyme, and buffer (Roche Diagnostics). The PCR conditions used for all candidates were as follows: denaturation cycle of 10 min at 95°C; 45 PCR cycles (denaturation: 95°C for 5 s; annealing: specific temperature, see Supplementary data Table 2, for 5 s; extension: 72°C for 20 s); followed by a melting cycle. The DNA quantification was performed using LightCycler Software Version 3.5 with reference to the standard curve. The real-time PCR product specificity was confirmed by analysis of the melting curve given by the LightCycler software. The products were then electrophoresed on a 1.5% agarose gel and sequenced to confirm the identity of the amplified product.
DNA sequencing and clone identification
DNA sequencing and clone identification were performed as previously described (26). Briefly, the resulting sequence traces were visualized with the online freeware Chromas 1.45 (http://www.technelysium.com.au/chromas.html) and uploaded to a cDNA Library Manager program (coded by Genome Canada Bioinformatics) that automates sequence analysis and clone identification (27). For clone identification, sequence traces were uploaded to the cDNA Library Manager, trimmed (Phred software) and compared against a locally installed GenBank database (Basic Local Alignment Search Tool (BLAST); http://www.ncbi.nlm.nih.gov/blast/).
RESULTS
Assessing the kinetics of the T7-IVT amplification reaction
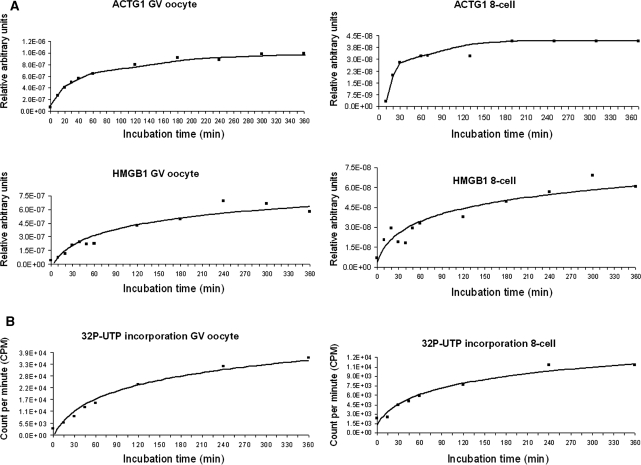
Characterization of the kinetics of aRNA production for both amplification rounds was done using bovine GV oocytes and 8-cell embryos. These tissues were chosen as representative samples for their two very different intrinsic RNA contents. Monitoring of the first amplification round was performed by real-time PCR. Aliquots were taken at different time points during IVT incubation and reverse transcribed before a real-time PCR was performed using ACTG1, a highly abundant mRNA, and HMGB1, a low-abundance transcript. The relative transcript abundance was plotted as a function of the incubation time. As shown in Figure 3A, the kinetics of aRNA production was linear for the first 40 min. Then the slope dramatically changed and aRNA production slowly and gradually reached a stationary-like phase. This phenomenon was observed irrespectively of the sample or candidate gene studied.
Figure 3.
Characterization of the kinetics of each T7-IVT amplification round. Two tissues were tested, i.e. germinal-stage oocytes and 8-cell embryos. (A) The aRNA production during the first round was followed using real-time PCR targeting the actin gamma 1 (ACTG1) and high-mobility group box 1 (HmgB1) transcripts. (B) The aRNA production during the second round was followed by measuring the incorporation of radiolabeled UTP.
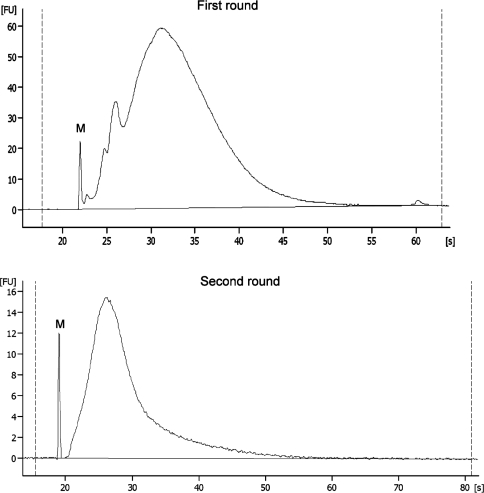
To study the kinetics of the second amplification round, a different methodological approach was required since aRNA fragments were then much smaller (Figure 4) due to the necessary use of random primers to initiate reverse transcription. This shorter product size prevented proper real-time PCR amplification (data not shown), most probably because this high fragmentation also occurs between both priming sites and results in fragments bearing only one primer sequence, thus preventing exponential amplification.
Figure 4.
Total RNA micro-electrophoretic profiles of first-round and second-round T7-IVT aRNA outputs. S = seconds and FU = Fluorescent units. M = spiked in marker.
We thus used radiolabeled UTP incorporation to follow aRNA production during the second IVT round. The CPMs were plotted as a function of reaction time (Figure 3B). For both sample types, the second amplification round was truly linear during the first 50 min, then the incorporation rate began to slowly decline. These aRNA production profiles indicated that the true linear phases of both T7-IVT rounds are relatively short.
Broken beacon design and validation
The design of the broken beacon was inspired by Blair and colleagues (22), who used this type of probe to monitor RNA polymerase activity. As shown in Figure 2, the broken beacon consists of two complementary oligonucleotide probes of different lengths. The longer sense probe is labeled with a fluorophore at the 5′ end (fluo-oligo) and the shorter antisense probe harbours a nonfluorescent quencher on its 3′ end (quencher-oligo). When hybridized together, the complex is nonfluorescent due to the proximity of the fluorophore to the quencher. In the presence of the aRNA target, the fluorescent sense probe forms a more stable duplex with the target and displaces the quencher strand, allowing the fluorophore of the sense probe to emit. The designed broken beacon specifically targets the bovine ACTB sequence.
The potential of the broken beacon to form a duplex with its target sequence under the T7-IVT reaction conditions (especially at the low temperature and high Mg2+ concentration used), as well as its ability to do so quantitatively, was investigated by using various amounts of a specific antisense ssDNA template that was added to the T7-IVT reaction master mix without the RNA polymerase. The calculated fluorescence intensity was plotted as a function of DNA target concentration (Figure 5). The fluorescence concentration curve indicates that the broken beacon can efficiently hybridize with the ssDNA target and its fluorescence is proportional to the target concentration. Moreover, when the broken beacon was incubated without template, the emitted fluorescence was weak and near the background threshold.
Figure 5.
Standard curve of fluorescence emitted by the broken beacon versus concentration of the single-stranded antisense actin beta (ACTB) target.
Monitoring aRNA production in real time
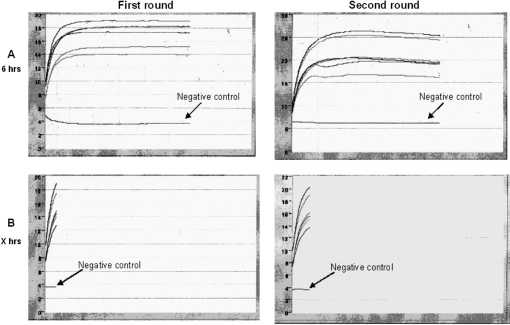
The two amplification rounds of the T7-IVT reaction were monitored in real time with the broken beacon. In Figure 6A, the amplification reaction lasted 6 h, in accordance with the manufacturer's protocol. As observed previously (Figure 3) for the first round, the reduction in aRNA production was initiated only after a 40-min incubation, while it was initiated after 50 min for the second round. This kinetics was observed for both sample types. This T7-IVT amplification was repeated with aliquots of the same samples from either oocytes or 8-cell embryos, but this time the reaction was stopped during the linear phase (Figure 6B), as determined by a change in the slope of the fluorescence profiles. The aRNA yields obtained after these standard or restricted amplifications are listed in Table 1. The aRNA yields for the 8-cell samples following the first round in the restricted context were lower than the absorbance detection limit of the spectrometer, resulting in the absence of aRNA measurements for these samples. Overall, the aRNA output from restricted incubations were much lower than the ones obtained following the standard 6-h incubation protocol. In the full 6-h incubation protocol, a difference of about 3.3-fold between oocyte and 8-cell samples was observed following the first round, whereas little difference was observed following the second round. By contrast, in the restricted samples, the proportional difference in aRNA output following the second round between the two tissue types was about 4.5-fold. This lack of proportional output difference across tissues when performing the full 6-h incubation clearly indicates the introduction of an intersample methodological bias. To study the extent of the impact of this skew, a microarray-based comparative approach was used.
Figure 6.
(A and B) Fluorescence history charts of real-time monitoring of the amplification reaction using the broken beacon. Both rounds were monitored in both sample types, i.e. GV oocytes and 8-cell embryos.
Table 1.
Total aRNA yield for the first and second rounds of the IVT reaction for the standard (6 h) and restricted (X h) amplification for both the GV oocyte and 8-cell embryo samples
| Samples | First-round final aRNA yield (µg) after 6 h | Second-round final aRNA yield (µg) after 6 h | First-round final aRNA yield (µg) after X h | Second-round aRNA yield (µg) after X h |
|---|---|---|---|---|
| Oocytes_pool 1 | 0.71 | 59.03 | 0.21 | 20.44 |
| Oocytes_pool 2 | 0.78 | 61.58 | 0.18 | 15.36 |
| Oocytes_pool 3 | 0.85 | 60.03 | 0.21 | 18.12 |
| 8-cell_pool 1 | 0.20 | 49.32 | NA | 4.39 |
| 8-cell_pool 2 | 0.23 | 52.04 | NA | 4.66 |
| 8-cell_pool 3 | 0.27 | 49.12 | NA | 4.48 |
NA, not applicable: the aRNA concentration measured was below the spectrophotometer detection limit.
Microarray data normalization
In addition to the duration of the amplification procedure, the relative RNA contents will be affected by microarray data normalization. The choice of an appropriate data normalization method is paramount for subsequent analysis and will vary depending on the tissues or cells studied. In the context of samples with considerable differences in RNA content and overall RNA abundance that might be significantly different, the largely used Lowess (or Loess) normalization for dual-labeled microarrays is inappropriate. The main assumption of the Lowess normalization method is that most genes are not differentially expressed between studied samples (28). This assumption clearly does not match the present context of oocytes and 8-cell embryos. The normalization must then rely on spikes and exogenous standards. Herein, the microarray samples were prepared from a fixed amount of aRNA (2 µg) to avoid introducing a bias originating from the low signal intensities that would be more prevalent in the low-abundance tissue. Indeed, comparison of signal intensities generated from hybridization samples prepared using different aRNA input would suffer from the increased variance found in the low-intensity signals. In order to account for the initial differences in RNA contents between sample types, a normalization strategy based on the concentration of the samples in aRNA was utilized. Since the concentration of aRNA varies between samples, the volume taken from each sample to obtain 2 µg of aRNA differed proportionally. In order to take into account such a difference, an equal amount of an in vitro produced RNA coding for the GFP protein was spiked into every sample following amplification. The signal strength generated by the GFP spike was therefore representative of the volume taken, and thus of aRNA abundance, and was used to normalize the microarray data. This method maintains the differences in relative abundance between these two developmental stages.
Microarray profile of standard and restricted amplifications
The RNA abundance data analysis of oocytes and 8-cell embryos from standard and restricted amplifications was performed with the NIA Array Analysis tool, and candidates with mean log signal intensities above the established cutoff value threshold (log-intensity 2) were considered to be expressed, whereas candidates with signal intensities below the threshold were designated as absent.
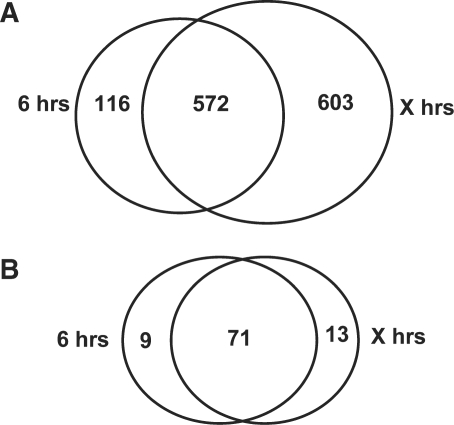
A pair-wise comparison of means for RNA abundance was done with the oocyte and 8-cell embryo data sets. To increase the level of confidence, a combination of both false discovery rate control (FDR) of 0.05 and fold change criteria of 2 were used to identify the over- and underexpressed. Such stringent sorting was used to highlight the change in gene lists resulting from the impact of the amplification strategies. As shown in Figure 7A, a total of 688 and 1175 ESTs were found to be more abundant in the oocyte compared to the 8-cell embryo with hybridization samples prepared from the standard 6-h protocol and the restricted protocol, respectively. Among these, 572 ESTs were common to both lists, whereas 116 were found only in the 6-h list and 603 were specifically identified with the restricted protocol. In contrast, the majority of the underexpressed candidates, that is 71, were mostly common for both protocols, whereas only nine candidates were found only with the standard incubation and 13 with the restricted protocol (Figure 7B).
Figure 7.
Venn diagrams highlighting the proportional overlaps and divergences in gene lists of ESTs with significant abundance difference between both amplification protocols [standard (6 h) versus restricted (X h)]. In both cases, the candidates selected were those that were more prevalent in (A) the GV oocyte or (B) the 8-cell embryo.
Comparative RNA abundance of candidates by real-time PCR
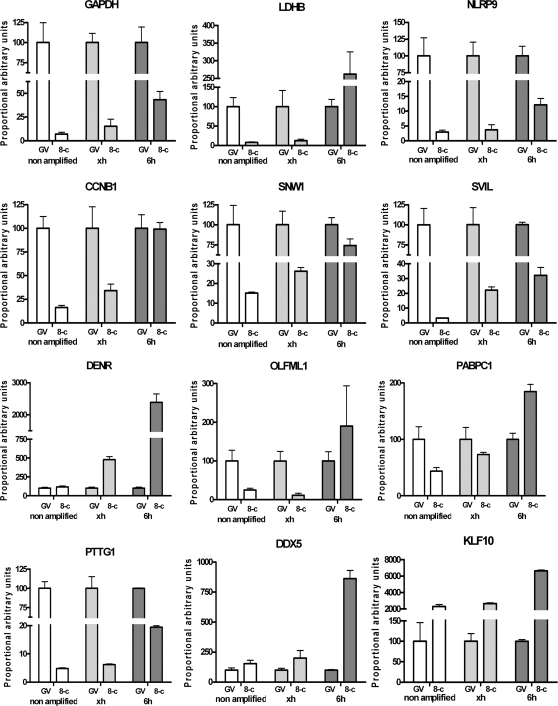
To validate the microarray results and to assess the extent of the relative distortion, the abundance in RNA content of candidate genes was measured using real-time PCR. For both tissues, i.e. GV oocytes and 8-cell embryos, the abundance in mRNA of specific candidates was measured from the outputs of the two amplification protocols and proportionally compared to unamplified samples (Figure 8). As a basis for comparison, the mean relative abundance found in the oocyte samples was set at 100% for every condition. This allowed for the comparison of the relative proportion between the oocyte and the 8-cell embryo for both of the amplification procedures. These proportions can also be compared to the unamplified samples considered as the gold standard bearing the true proportional difference between these developmental stages.
Figure 8.
Relative abundance of candidate gene transcripts in unamplified or amplified samples following the restricted amplification protocol (identified on the charts by ‘xh’) or following the standard procedures (two rounds of 6 h) identified by ‘6h’. For each group, the mean abundance value of the GV oocyte was set as standard relative basis. CCNB1, cyclin B1; DDX5, DEAD box polypeptie 5; DENR, density-regulated protein; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; KLF10, Kruppel-like factor 10; LDHB, lactate dehydrogenase B; NLRP9, NLR family, pyrin domain containing 9; OLFML1, olfactomedin-like 1; PABPC1, poly(A)-binding protein cytoplasmic 1; PTTG1, pituitary tumor-transforming 1; SNW1, SNW domain containing 1; SVIL, supervillin.
For all candidates, a distorted proportion was found for the samples prepared using the standard 6-h protocol when compared to the proportion of the unamplified sample used as reference. The skew was clearly caused by an over-amplification of the 8-cell samples. In some cases, such as the ones presented for cyclin B1 (CCNB1), lactate dehydrogenase B (LDHB), olfactomedin-like 1 (OLFML1) and poly(A)-binding protein cytoplasmic 1 (PABPC1), conclusions drawn from the relative RNA abundance measurements using the samples amplified under standard conditions would be erroneous. By contrast, the relative abundances from the restricted amplification were highly similar to the unamplified control (Figure 8).
DISCUSSION
When working with scarce tissues, the success of global RNA abundance profiling using microarrays largely depends on the efficiency of the amplification step to preserve the relative abundance of transcripts in the resulting amplified sample. Concerns over sequence-specific alterations in the amplification kinetics that would result in the introduction of a skew in the relative abundance between candidate gene mRNAs within sample (Figure 1, columns A–C) have been addressed previously (5–7,29). The aim of the present study was rather to determine the presence of a different type of distortion that could impact the relative RNA contents across samples of different total contents (as represented in Figure 1, columns D–F). This is of particularly high relevance when considering the fact that cell types or cells under different physiological states can exhibit major variations in their overall gene expression activity. Such a situation is becoming more prevalent with our growing ability to compare samples of known cell contents, including single-cell transcriptome profiling. Our initial premise that the amplification step was introducing an important bias came from observations using early mammalian embryos, in which we observed that aRNA outputs were similar regardless of the embryonic stage and did not reflect the physiology of the developing embryo. In the present study, characterization of the kinetics of aRNA production, followed using different approaches, highlighted a production that quickly slowed to gradually reach a plateau-like phase (Figures 3 and 6). These observations confirm the presence of a nonlinear amplification phase that introduces a skew in the amplified end products by homogenizing the outputs and therefore eliminating the initial relative proportions in RNA contents. The observed drastic reduction in aRNA production that occurs early on is not a plateau phase in its strict sense, as we observed constant increases in the yield, indicating the presence of residual productivity. This observation is in contrast with a previous publication showing a reduction in aRNA yield due to degradation when incubations are extended for over 4 h (30). We have not noticed such RNA decay in our assays. Indeed, the kinetics of the reaction did not plummet, even after the recommended 6-h incubation.
Another point to consider when performing T7-IVT amplification for microarray sample preparation is the final output of the reaction. This final output is of critical importance since standard microarray sample preparation requires a minimal 2 µg of input material. This minimal requirement is plentifully fulfilled when using the standard 6-h incubation, which generates yields in the range of 50–60 µg following a two-round reaction. To be implementable, the proposed restricted amplification protocol must also meet this minimal output requirement. In the present study, the current output from the restricted amplification protocol, using the least abundant tissue (8-cell embryo) was found to be sufficient for the preparation of two microarray hybridization samples, proving it to be suitable for technical replications. Moreover, in spite of this shorter amplification duration, aRNA yields were still higher than PCR-based amplification outputs (such as the SuperSMART), whose final content in double-stranded cDNA is in the range of 1–2 µg (Clontech's user manual).
The use of a fluorescent probe targeting a candidate sequence to monitor the aRNA production in real time provides the required mean to ensure that the amplification step performs following a true linear kinetic. Furthermore, the use of the broken beacon may also provide a basis to study and better control the efficiency divergences observed between platforms. These divergences were shown to arise from the different T7 promoter sequences used to recruit the RNA polymerase (29).
In addition to highlighting the presence of an amplification distortion caused by homogenization of the yields, we assessed its impact on microarray data interpretation. In the case of scarce samples, such as early embryos or micro-dissections, two amplification rounds are inevitable; we have thus not assessed the presence of a distortion following the first amplification round. In fact, due to insufficient output following the first restricted amplification, it was not possible to measure differences in the output between both sample types. However, when monitoring the aRNA production during the recommended full 6-h incubations, both rounds showed flattening kinetics (Figures 3 and 6), suggesting an impact on relative RNA abundance during both rounds. Nonetheless, we cannot rule out that nonlinear kinetics could have a less profound impact when performing only one amplification round. It was noticed that under the standard 6-h incubation protocol, the difference between sample types was about 3.3-fold and 1.2-fold following the first and second rounds, respectively. By comparison, the difference between the GV oocyte and the 8-cell samples was 4.5-fold after two rounds of controlled amplifications. Given that the restricted protocol was confirmed to preserve the natural proportional difference between samples, this result suggests that the distortion introduced during the second amplification round is more important. Further validations to assess the extent of the distortion introduced during the first round would be required for an in-depth analysis of its impact.
This outcome of the aRNA output homogenization on microarray data interpretation was assessed in the specific physiological context of the early 8-cell bovine embryo, which corresponds to the latest developmental stage of the pre-EGA window. In 8-cell bovine embryo, total RNA content represent residual maternal mRNA and as expected, in the absence of transcriptional activity, which characterizes this developmental window, nearly all of the transcripts were found to be more abundant in the GV oocyte than in the 8-cell embryo (Figure 7). The few candidates found to be more abundant in the 8-cell embryo (Figure 7B) could represent false positives or genes involved in the early onset of embryonic genome activation either through de novo RNA synthesis or re-adenylation of maternal transcripts that were stored in a de-adenylated state in the GV stage oocyte. Maternal RNAs stored in the cytoplasm of the oocyte have been shown to be stabilized partly by the removal of their poly(A) tail and enclosed within ribonucleoprotein complexes (31). The recruitment mechanisms by which these dormant RNA are targeted for either translation or decay are still largely uncharacterized. The current model involves lengthening of the poly(A) tail, which triggers binding of the poly(A)-binding protein, followed by the formation of a loop bringing the 3′ end in proximity to the 5′ cap structure which facilitates binding of translation initiation factors (32). It is thus generally accepted that de-adenylated mRNAs are part of the stored maternal RNA pools. During the initial step of the T7-IVT procedure, reverse transcription of mRNA using an oligo-dT bearing a T7 RNA polymerase promoter sequence is selecting mRNA bearing long poly(A) tail and could therefore explain the apparent preponderance of some maternal transcripts in the 8-cell embryo compared to the GV oocyte.
To validate the microarray data and further confirm the introduction of a relative skew by the standard T7-IVT amplification protocol, real-time PCR abundance measurements of candidate gene transcripts were used as a reference. This method showed an overall close concordance between the restricted amplification protocol and the unamplified reference (Figure 8). It also clearly highlighted that the recommended T7-IVT protocol systematically overrepresented the abundance of transcripts found in the 8-cell embryo, thus confirming the distortion in the relative proportions of all ESTs across these two developmental stages. Furthermore, when comparing candidate genes lists between the standard and restricted protocols, 116 candidates, or 10% (116/1175), of candidates identified by the standard procedure appear to be false positives. In addition to the distortion of relative RNA abundance, these false positives could be the result of the sequence driven bias introduced by extended incubations. Although these sequence driven biases have been reported to be marginally frequent (5,7), it has been recently reported that this sequence related skew can affect up to 16% of microarray reporters (6). Thus, the overall loss of natural representation due to the T7-IVT kinetics is about five times more important (603/1175) than the combined false-positive rate. In other words, by using the recommended procedure, more than half of the truly differently abundant transcripts would not have been detected. The candidates common to both techniques were found to be those with the highest fold-change difference, indicating that more subtle differences were lost with aRNA output normalization.
CONCLUSION
Monitoring aRNA production in real time as provided a novel perspective for the study of gene expression in situations where the natural RNA contents are divergent between studied samples. Aside from situations involving management of stored RNA pools such as the one presented herein, situations where the biological samples are bearing different RNA contents are most certainly prevalent in numerous physiological states through fluctuation in overall transcriptional activity. Within this context, the use of housekeeping candidates for sample normalization becomes problematic and the comparison of fixed RNA contents may provide irrelevant information. The comparison of samples of known cell numbers offers a novel discovery potential for comparative gene expression. To take advantage of this opportunity, the amplification step must preserve the initial proportional difference in RNA contents. The loss in relative proportions across samples during microarray sample preparation had not been considered nor assessed. We have shown here for the first time that ignoring the amplification reaction kinetics significantly reduces the quality of microarray data. The resulting high incidence of undetected differences and skewed proportions translate in a significantly lower level of discovery potential and may lead to irrelevant physiological interpretation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Fonds québécois de la recherche sur la nature et les technologies (107924 and 121560). Funding for open access charge: The Fonds québécois de la recherche sur la nature et les technologies (121560).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Isabelle Laflamme for her technical assistance during the embryo production in addition to Dr Marc-André Laniel and Dr Julie Nieminen for critical review of the manuscript.
REFERENCES
- 1.Kurn N, Chen P, Heath JD, Kopf-Sill A, Stephens KM, Wang S. Novel isothermal, linear nucleic acid amplification systems for highly multiplexed applications. Clin. Chem. 2005;51:1973–1981. doi: 10.1373/clinchem.2005.053694. [DOI] [PubMed] [Google Scholar]
- 2.Wellenreuther R, Schupp I, Poustka A, Wiemann S. SMART amplification combined with cDNA size fractionation in order to obtain large full-length clones. BMC Genomics. 2004;5:36. doi: 10.1186/1471-2164-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Gelder RN, von Zastrow ME, Yool A, Dement WC, Barchas JD, Eberwine JH. Amplified RNA synthesized from limited quantities of heterogeneous cDNA. Proc. Natl Acad. Sci. USA. 1990;87:1663–1667. doi: 10.1073/pnas.87.5.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puskas LG, Zvara A, Hackler L, Jr, Van Hummelen P. RNA amplification results in reproducible microarray data with slight ratio bias. Biotechniques. 2002;32:1330–1334. doi: 10.2144/02326mt04. 1336, 1338, 1340. [DOI] [PubMed] [Google Scholar]
- 5.van Haaften RI, Schroen B, Janssen BJ, van Erk A, Debets JJ, Smeets HJ, Smits JF, van den Wijngaard A, Pinto YM, Evelo CT. Biologically relevant effects of mRNA amplification on gene expression profiles. BMC Bioinformatics. 2006;7:200. doi: 10.1186/1471-2105-7-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Degrelle SA, Hennequet-Antier C, Chiapello H, Piot-Kaminski K, Piumi F, Robin S, Renard JP, Hue I. Amplification biases: possible differences among deviating gene expressions. BMC Genomics. 2008;9:46. doi: 10.1186/1471-2164-9-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duftner N, Larkins-Ford J, Legendre M, Hofmann HA. Efficacy of RNA amplification is dependent on sequence characteristics: implications for gene expression profiling using a cDNA microarray. Genomics. 2008;91:108–117. doi: 10.1016/j.ygeno.2007.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat. Genet. 1999;21:20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 9.Mahadevappa M, Warrington JA. A high-density probe array sample preparation method using 10- to 100-fold fewer cells. Nat. Biotechnol. 1999;17:1134–1136. doi: 10.1038/15124. [DOI] [PubMed] [Google Scholar]
- 10.Robert C, McGraw S, Massicotte L, Pravetoni M, Gandolfi F, Sirard MA. Quantification of housekeeping transcript levels during the development of bovine preimplantation embryos. Biol. Reprod. 2002;67:1465–1472. doi: 10.1095/biolreprod.102.006320. [DOI] [PubMed] [Google Scholar]
- 11.Waxman S, Wurmbach E. De-regulation of common housekeeping genes in hepatocellular carcinoma. BMC Genomics. 2007;8:243. doi: 10.1186/1471-2164-8-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Derks NM, Muller M, Gaszner B, Tilburg-Ouwens DT, Roubos EW, Kozicz LT. Housekeeping genes revisited: different expressions depending on gender, brain area and stressor. Neuroscience. 2008;156:305–309. doi: 10.1016/j.neuroscience.2008.07.047. [DOI] [PubMed] [Google Scholar]
- 13.Minner F, Poumay Y. Candidate housekeeping genes require evaluation before their selection for studies of human epidermal keratinocytes. J. Invest. Dermatol. 2009;129:770–773. doi: 10.1038/jid.2008.247. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, He F, Song S, Wang J, Yu J. How many human genes can be defined as housekeeping with current expression data? BMC Genomics. 2008;9:172. doi: 10.1186/1471-2164-9-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathupala SP, Rempel A, Pedersen PL. Aberrant glycolytic metabolism of cancer cells: a remarkable coordination of genetic, transcriptional, post-translational, and mutational events that lead to a critical role for type II hexokinase. J. Bioenerg. Biomembr. 1997;29:339–343. doi: 10.1023/a:1022494613613. [DOI] [PubMed] [Google Scholar]
- 16.Barnes FL, First NL. Embryonic transcription in in vitro cultured bovine embryos. Mol. Reprod. Dev. 1991;29:117–123. doi: 10.1002/mrd.1080290205. [DOI] [PubMed] [Google Scholar]
- 17.Memili E, Dominko T, First NL. Onset of transcription in bovine oocytes and preimplantation embryos. Mol. Reprod. Dev. 1998;51:36–41. doi: 10.1002/(SICI)1098-2795(199809)51:1<36::AID-MRD4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 18.Gandhi AP, Lane M, Gardner DK, Krisher RL. A single medium supports development of bovine embryos throughout maturation, fertilization and culture. Hum. Reprod. 2000;15:395–401. doi: 10.1093/humrep/15.2.395. [DOI] [PubMed] [Google Scholar]
- 19.Ali A, Sirard MA. Effect of the absence or presence of various protein supplements on further development of bovine oocytes during in vitro maturation. Biol. Reprod. 2002;66:901–905. doi: 10.1095/biolreprod66.4.901. [DOI] [PubMed] [Google Scholar]
- 20.Marras SA. Selection of fluorophore and quencher pairs for fluorescent nucleic acid hybridization probes. Methods Mol. Biol. 2006;335:3–16. doi: 10.1385/1-59745-069-3:3. [DOI] [PubMed] [Google Scholar]
- 21.Li Q, Luan G, Guo Q, Liang J. A new class of homogeneous nucleic acid probes based on specific displacement hybridization. Nucleic Acids Res. 2002;30:E5. doi: 10.1093/nar/30.2.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blair RH, Rosenblum ES, Dawson ED, Kuchta RD, Kuck LR, Rowlen KL. Real-time quantification of RNA polymerase activity using a ‘broken beacon’. Anal. Biochem. 2007;362:213–220. doi: 10.1016/j.ab.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vigneault C, McGraw S, Massicotte L, Sirard MA. Transcription factor expression patterns in bovine in vitro-derived embryos prior to maternal-zygotic transition. Biol. Reprod. 2004;70:1701–1709. doi: 10.1095/biolreprod.103.022970. [DOI] [PubMed] [Google Scholar]
- 24.Sirard MA, Dufort I, Vallee M, Massicotte L, Gravel C, Reghenas H, Watson AJ, King WA, Robert C. Potential and limitations of bovine-specific arrays for the analysis of mRNA levels in early development: preliminary analysis using a bovine embryonic array. Reprod. Fertil. Dev. 2005;17:47–57. doi: 10.1071/rd04113. [DOI] [PubMed] [Google Scholar]
- 25.Sharov AA, Dudekula DB, Ko MS. A web-based tool for principal component and significance analysis of microarray data. Bioinformatics. 2005;21:2548–2549. doi: 10.1093/bioinformatics/bti343. [DOI] [PubMed] [Google Scholar]
- 26.Sirard MA, Dufort I, Vallee M, Massicotte L, Gravel C, Reghenas H, Watson AJ, King WA, Robert C. Potential and limitations of bovine-specific arrays for the analysis of mRNA levels in early development: preliminary analysis using a bovine embryonic array. Reprod. Fertil. Dev. 2005;17:47–57. doi: 10.1071/rd04113. [DOI] [PubMed] [Google Scholar]
- 27.Vallee M, Gravel C, Palin MF, Reghenas H, Stothard P, Wishart DS, Sirard MA. Identification of novel and known oocyte-specific genes using complementary DNA subtraction and microarray analysis in three different species. Biol. Reprod. 2005;73:63–71. doi: 10.1095/biolreprod.104.037069. [DOI] [PubMed] [Google Scholar]
- 28.Do JH, Choi DK. Normalization of microarray data: single-labeled and dual-labeled arrays. Mol. Cell. 2006;22:254–261. [PubMed] [Google Scholar]
- 29.Kerkhoven RM, Sie D, Nieuwland M, Heimerikx M, De Ronde J, Brugman W, Velds A. The T7-primer is a source of experimental bias and introduces variability between microarray platforms. PLoS ONE. 2008;3:e1980. doi: 10.1371/journal.pone.0001980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiess AN, Mueller N, Ivell R. Amplified RNA degradation in T7-amplification methods results in biased microarray hybridizations. BMC Genomics. 2003;4:44. doi: 10.1186/1471-2164-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paynton BV, Bachvarova R. Polyadenylation and deadenylation of maternal mRNAs during oocyte growth and maturation in the mouse. Mol. Reprod. Dev. 1994;37:172–180. doi: 10.1002/mrd.1080370208. [DOI] [PubMed] [Google Scholar]
- 32.Groisman I, Jung MY, Sarkissian M, Cao Q, Richter JD. Translational control of the embryonic cell cycle. Cell. 2002;109:473–483. doi: 10.1016/s0092-8674(02)00733-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.