Abstract
The congenital nemaline myopathies are rare hereditary muscle disorders characterized by the presence in the muscle fibers of nemaline bodies consisting of proteins derived from the Z disc and thin filament. In a single large Australian family with an autosomal dominant form of nemaline myopathy, the disease is caused by a mutation in the α-tropomyosin gene TPM3. The typical form of nemaline myopathy is inherited as an autosomal recessive trait, the locus of which we previously assigned to chromosome 2q21.2-q22. We show here that mutations in the nebulin gene located within this region are associated with the disease. The nebulin protein is a giant protein found in the thin filaments of striated muscle. A variety of nebulin isoforms are thought to contribute to the molecular diversity of Z discs. We have studied the 3′ end of the 20.8-kb cDNA encoding the Z disc part of the 800-kDa protein and describe six disease-associated mutations in patients from five families of different ethnic origins. In two families with consanguineous parents, the patients were homozygous for point mutations. In one family with nonconsanguineous parents, the affected siblings were compound heterozygotes for two different mutations, and in two further families with one detected mutation each, haplotypes are compatible with compound heterozygosity. Immunofluorescence studies with antibodies specific to the C-terminal region of nebulin indicate that the mutations may cause protein truncation possibly associated with loss of fiber-type diversity, which may be relevant to disease pathogenesis.
The typical form of congenital nemaline myopathy is characterized by infantile onset of a slowly progressive or nonprogressive weakness of the facial, bulbar, neck flexor, respiratory, and proximal limb muscles, with a later distal involvement (1–4). In more severe forms, in which the course is often fatal, the infants may have arthrogryposis or fractures at birth and no spontaneous movements (1, 2). In the majority of cases, the inheritance is autosomal recessive (4). To date, two loci for nemaline myopathy have been established, the TPM3 locus on chromosome 1 and a locus on chromosome 2q21.2-q22 (5–8). Within the 2q region, the gene encoding nebulin is contained. Nebulin is a skeletal muscle-specific sarcomeric protein that appears to be an integral component of the thin filament (9). Its C-terminal region has been believed to regulate the assembly of Z discs (10, 11) (Fig. 1). This, in conjunction with the linkage data, raised the possibility that mutations in the nebulin gene (NEB) might be causing the typical autosomal recessive form of the disease.
Figure 1.
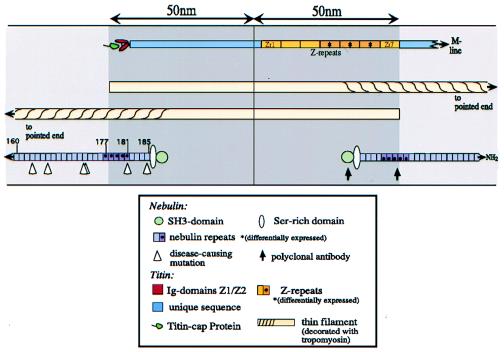
Overview of the layout of actin, tropomyosin, titin, and nebulin filaments in Z lines from vertebrate striated muscles. Titin: The N-terminal region spans the Z line, and, therefore, titin filaments from opposite sarcomeres fully overlap within the Z lines (12). Within the Z line, titin filaments connect to α-actinin Z filaments at multiple sites, which then cross-link the titin and thin filaments (12–15). Nebulin:Nebulin filaments appear to insert about 30 nm away from the center of the Z line (10). Triangles on the left indicate the location of the six identified mutations (Figs. 2 and 3). On the right, arrows indicate the location of the epitopes to which antibodies were raised for immunofluorescence studies (Fig. 4).
To permit screening of nebulin exons and splice sites for mutations, we have partially determined the genomic organization of the gene, by using long-range PCR with primers designed on the basis of the cDNA sequence (GenBank accession no. X83957). The genomic length of the entire gene has been estimated to be about 400 kb (16). As a starting point, sequences encoding the C-terminal 120 kDa of nebulin were determined (bp 17,610–20,881, accession no. X83957), because this part of nebulin is highly conserved in nebulette, a cardiac muscle-specific filamentous protein that is encoded by a distinct gene (10, 11). The conservation suggests that the C termini of both proteins, which are located within the I–Z–I junction (Fig. 2), are important for the assembly and integration of Z discs within the sarcomere (10).
Figure 2.
Genomic organization of the 3′ end of NEB, corresponding to the repeat domains M163–M185, the serine-rich domain with potential RS-phosphorylation motifs, and the SH3-domain of the protein. The exons are shown as boxes numbered from 153a to 187, and the introns are shown as lines. Exons 177b, 177c, 177d, and exon 178b are exons that are not present in the published nebulin sequence (accession no. X83957) but identified in cDNA from one patient with nemaline myopathy. The C-terminal protein structure from the end of the super-repeat region (S21R1–S22R7) to the SH3 domain is shown below the exon/intron organization. The mutation sites are indicated by arrows. The alternatively spliced region of nebulin is shown as gray exons and protein domains.
The nebulin exons, including their intron boundaries, were screened for mutations by single-strand conformation polymorphism (SSCP) analysis, and aberrant bands were sequenced. In addition, we analyzed the region spanning exons 163–172 (bp 17,699–18,807, accession no. X83957) by reverse transcription (RT)–PCR and sequencing, by using RNA extracted from lymphoblastoid cultures of 10 unrelated patients. Altogether, 22 families with affected children of healthy parents from seven different countries were included in the study; mutations were identified in five families.
MATERIALS AND METHODS
Patients and Samples.
Altogether, 22 families (11 Finnish, 6 British, 1 Danish, 1 Dutch, 1 French, 1 German, and 1 Australian) were included in the study. They had the typical form (1) of congenital autosomal recessive nemaline myopathy and linkage results compatible with linkage to the 2q region. Genomic DNA was extracted from blood or lymphoblastoid cell lines from all 28 patients and their families according to standard procedures. RNA was isolated from lymphoblastoid cell lines of 10 unrelated patients by the Trizol method (GIBCO/BRL). DNA samples from 121 unrelated individuals with no known neuromuscular disorder were used as controls.
Haplotype Analysis.
Haplotypes for the following polymorphic microsatellite markers were constructed: D2S2324, D2S2277, D2S2275, D2S2236, and D2S2299 (17). PCR conditions were as described (7). The PCR products were run on 5–6% polyacrylamide gels, and the alleles were visualized by silver staining (18). The alleles were numbered consecutively, “1” being the largest.
Long-Range PCR and Sequencing.
Long-range PCR was performed with the GeneAmp XL PCR kit (Perkin–Elmer) as described by the manufacturer. The primers were designed on the basis of the nebulin cDNA sequence (accession no. X83957). Sequencing of the PCR fragments was done with either ABI PRISM dye terminator or ABI PRISM dRhodamine cycle sequencing kits (Perkin–Elmer) according to the manufacturer’s instructions, and reactions were run on an ABI 373 A or 377 sequencer (Perkin–Elmer), respectively.
RT-PCR.
Total RNA (0.8 μg) was reverse-transcribed with Moloney murine leukemia virus reverse transcriptase (Promega) and random hexamers as primers in a total volume of 20 μl. RT-PCR was performed on 5 μl of cDNA with the following primer pairs: RT1F (ACAGGCCAGTGAGGTGGAGTACAGAG) and RT1R (TTGGCAGTGATGTACGTTGGTGTTTC), 519-bp product (bp 17,699–18,217 in the cDNA); RT2F (GAAACACCAACGTACATCACTGCCAA) and RT2R (TGAAGTTCTTTTGGTTCTCCCGGACT), 592-bp product (bp 18,192–18,783); RT3F (GGAAGTCAAAGGAAGAGGCCTGAATG) and RT3R (GGTCACACTGGTATTGGAGAAGGCTG), 317-bp product (bp 18,491–18,807). The abnormally spliced mRNA in family 4 was detected with the following primer pair: RT4F (TATGGAAGTGGCCAAGAAGCAAAGTG) and RT4R (TGGATTGTATTTTGGTGTCTTGCCCT), 293-bp product (bp 17,582–17,874).
The annealing temperature for the primers was 68°C, and the cycling conditions were as follows. Cycles: 1, 95°C, 5 min; 2, 95°C, 1 min; 3, 68°C, 1 min; 4, 72°C, 1 min; 5, go to 2. 39 times; 6, 95°C, 1 min; 7, 68°C, 1 min; 8, 72°C, 5 min; 9, 4°C. DynaZyme DNA polymerase (Finnzymes, Espoo, Finland) was used for amplification.
The additional exons in the cDNA from a muscle biopsy of the Australian patient were detected with the following primer pairs: 23F (CCACTACCACACCATACCCGATA) and 24R (CTATGGTTTTTCTGTGTCAATGTGAT), bp 18,068–20,572, and 31F (GAATCCCAACACCTATCACTC) and 32R (TTCTTTATTCTGATAGTTTCCG), bp 19,201–19,660.
SSCP.
MDE gel solution (FMC) was used for SSCP analysis of exons according to the protocol of the manufacturer. Genomic DNA was used as the template for PCR with primers constructed for each exon (primer sequences are available on request) and the 150- to 250-bp PCR products were run on 0.6–0.9× MDE gels. The gels were run at room temperature for 16–19 h at 5 W, fixed in 10% citric acid for 30 min, stained for 30 min with a 0.1% silver nitrate solution supplemented with 0.15% formaldehyde, and developed in a 3% sodium carbonate solution supplemented with 0.15% formaldehyde and 0.0002% sodium thiosulfate.
Immunocytochemistry.
Unfixed cryostat sections (5 μm) were immunolabeled for 30 min with rabbit polyclonal antisera raised against the SH3 domain (diluted 1:10) and the M176–M181-repeat region (diluted 1:40) (10). Antibodies were visualized with biotinylated anti-rabbit IgG (Amersham 1:200, 30 min) and then by streptavidin conjugated to Texas Red (Amersham 1:200, 15 min). Sections were viewed with a Leica (Deerfield, IL) microscope fitted with epifluorescence.
RESULTS
Genomic Structure of the 3′ Region of the Nebulin Gene.
The 3′ end of the 20.8-kb nebulin cDNA, bp 17,610–20,881 (accession no. X83957), consists of at least 30 exons and 29 introns, which span about 50 kb of genomic DNA. These exons encode the nebulin C terminus-specific repeat domains M163–M185, the serine-rich domain with RS-phosphorylation motifs, and the SH3 domain (10, 16, 19). The sizes of the exons vary from 93 to 184 bp, and the introns vary from 99 bp to 4–5 kb. We numbered the exons to correspond with the protein repeat domains described previously (16, 19) (Fig. 2).
Four segments were identified in genomic DNA that have 83–96% sequence homology to the C-terminal portion of nebulin but are not present in the published cDNA sequence (accession no. X83957). They presumably correspond to exons and were named 177b, 177c, 177d, and 178b. Their sequences were amplified from genomic DNA of healthy controls. On the transcription level, their transcripts could not be amplified from healthy controls, which is in agreement with their absence from the sequenced human nebulin (accession no. X83957). However, M177b-, M177c-, M177d-, and M178b-positive transcripts were amplified from a muscle biopsy tissue sample from an Australian patient with nemaline myopathy in whom mutational analysis has not yet been completed.
For all sequenced nebulin exons, the exon/intron boundaries are located within the SDXXYK consensus sequence, in the middle of the protein repeat domains M162–M184 (Fig. 2). Therefore, the regular modular architecture of the nebulin protein is also reflected in the genomic organization of the gene.
In summary, based on the genomic organization, the previously described multiple nebulin isoforms differing in the M176–M181 region (10) are generated by exon-skipping events involving exons 176–181 in the alternatively spliced region (Fig. 2). Expression of the exons M177b, M177c, M177d, and M178b is predicted to produce a novel type of nebulin isoform, whose tissue or developmental stage expression is currently not known.
Mutation Analysis.
For SSCP analysis, exon-flanking primer pairs were designed on the basis of the genomic sequence encoding exons 162–187. RT-PCR analysis was performed on RNA extracted from lymphoblastoid cultures.
Family 1.
A 1-bp (G) deletion in exon 165, inferred to cause a truncation of the protein at the repeat domain M166 (Table 1; Fig. 2), was first detected by SSCP. Two affected British children and their unaffected father were heterozygous for the deletion, which was also confirmed by mRNA sequencing. Based on the family’s haplotypes in this region, we expect that the children’s maternal allele harbors an as yet unknown NEB mutation (Fig. 3).
Table 1.
Nebulin mutations in patients with nemaline myopathy
| Family no. | Mutation | Location, exon | Codon | Predicted effect | Control chromosomes |
|---|---|---|---|---|---|
| 1 | 1-bp deletion | 165 | 5,846 | Frameshift, stop at codon 5,850 | 0/242 |
| 2 | 4-bp insertion | 172 | 6,099 | Frameshift, stop at codon 6,123 | 0/216 |
| 3 | 2-bp deletion | 172 | 6,108 | Frameshift, stop at codon 6,123 | 0/216 |
| 2-bp deletion | 181 | 6,373 | Frameshift, stop at codon 6,391 | 0/226 | |
| 4 | G → C | 163 | 5,792 | Abnormal splicing of exon 163 | 0/236 |
| 5 | G → T | 185 | 6,636 | Nonsense, stop at codon 6,636 | 0/238 |
Figure 3.
Haplotypes of families 1 and 2 in the 4-centimorgan region on chromosome 2q21.2-q22 harboring the nebulin gene (7). Black symbols indicate affected persons and white symbols indicate unaffected persons.
Family 2.
In a Finnish family, we found a 4-bp (GTTT) duplication/insertion in exon 172 encoding part of the repeat domains M172 and M173 (Fig. 2). The only affected child inherited the mutation from her unaffected mother. The duplication should cause a frameshift leading to a stop codon in the beginning of exon 173 and truncation of the protein from domain M173 (Table 1). Haplotype analysis suggests that the child is likely to have inherited a different, as yet unidentified, mutation from her unaffected father (Fig. 3).
Family 3.
The two affected children of a French family were found to be compound heterozygotes for two different frameshift mutations. From their unaffected father they inherited a 2-bp (AG) deletion in exon 172, and from their unaffected mother they inherited a 2-bp (GA) deletion in exon 181. The first mutation would lead to a stop codon in the beginning of exon 173 and a truncation of the protein from domain M173. The second mutation would lead to a stop codon in exon 181 and truncation of the protein from domain M182 (Table 1). Exon 181 is within the alternatively spliced region of NEB (Fig. 2), and the maternal mutation may therefore affect only some of the nebulin isoforms.
Family 4.
A homozygous G to C transversion in the last codon of exon 163 was detected in the only affected child of a German family (Table 1; Fig. 2). The German consanguineous parents of Turkish descent are heterozygous, and two healthy siblings are heterozygous for the mutation and homozygous for the normal allele, respectively. The mutation interferes with the 5′ consensus splice site of intron 163 and is predicted to cause in-frame skipping of exon 163 or partial retention of intron 163, because of activation of a cryptic splice site 100 bp downstream in intron 163. The latter mRNA has a STOP codon 36 bp downstream of exon 163. The abnormally spliced mRNA is seen as two distinct RT-PCR products different from the control mRNA (Fig. 4).
Figure 4.
RT-PCR analysis of the region spanning nebulin exons 162, 163, and 164 in the patient from family 4. The center lane shows that mRNA from the patient gave two abnormally spliced products, about 190 and 400 bp, respectively. Sequencing of the fragments revealed absence of exon 163 in the smaller fragment, and the larger fragment contained exon 163, as well as 100 bp of intron 163. Right lane, the expected 293-bp product from control mRNA; left lane, size marker φX174/HaeIII.
Family 5.
A homozygous G to T transversion in the ninth codon of exon 185 was detected in one affected British child of consanguineous parents. The mutation changes a glutamic acid (GAG) to a stop codon (TAG) and should result in loss of the last 134 amino acids, i.e., from the beginning of the serine-rich domain in the C-terminal region of nebulin (Table 1; Fig. 2).
None of the six mutations were detected by SSCP in control DNA from more than 100 healthy individuals (Table 1).
Histological Analysis of Muscle Biopsy Specimens.
To test whether the mutations lead to truncation beyond the respective mutations, we used an antiserum to the C-terminal SH3 domain and an antibody directed against nebulin simple repeats M176–M181 (10) (Fig. 1) for staining muscle sections of nine cases of nemaline myopathy, including the patients from families 1–5. In controls, striations were visible with both antibodies, and the antibody to M176–M181 labeled slow (type 1) fibers more intensely than fast (type 2) fibers. All cases of nemaline myopathy showed immunoreactivity with the M176–M181 antibody and cross striations were indistinguishable from those of the controls (Figs. 5 and 6). All cases except that from family 5 showed labeling with the SH3 antibody (Figs. 5 and 6). The absence of labeling in this case is consistent with the finding of the homozygous nonsense mutation in exon 185. The cases from families 1 and 5 also showed more intense labeling of slow fibers than of fast fibers with the M176–M181 antibody. In the patients, particularly patients 3 and 4, it was not possible to define the type of all fibers, because many fibers coexpressed slow, fast, and/or fetal isoforms of myosin. However, the case from family 3 showed differences in labeling intensity between fibers with the SH3 antibody and the case from family 4 showed differences with the M176–M181 antibody (Fig. 5). It is noteworthy that the patient studied in family 3 had a population of fibers that did not label with the SH3 antibody, in contrast to the uniform labeling in other cases. In the case from family 5, the fast fibers appeared to be less intensely labeled with the M176–M181 antibody, rather than negative, and some patchiness and uneven labeling was seen (Fig. 6). This was not apparent with other antibodies to myofibrillar proteins and may, therefore, represent specific loss of one form of nebulin.
Figure 5.
Cryostat sections from a control (a and b), patient 4 (c and d), and patient 3 (e and f) immunolabeled with antibodies to the SH3 (a, c, and e) and M176–M181 (b, d, and f) domains of nebulin. Note the striation pattern with both antibodies in the control (a and b), and the fiber-typing effect (enhanced in slow fibers) with the M176–M181 antibody in the control and patient 4 (b and d). The difference in fiber types with the SH3 antibody in patient 3 (e) does not relate to a specific fiber type, but note that some fibers appear to be negative. (Bars = 50 μm.)
Figure 6.
Immunolabeling of muscle from patient 5 showing absence of labeling with the SH3 antibody in longitudinal and transverse orientation (a) but positive labeling with the M176–M181 antibody (b). Labeling is enhanced in the small fibers expressing slow myosin (c), but the larger fibers are not completely negative and show uneven labeling (arrow). (Bar = 50 μm.)
To summarize, in one patient, immunohistochemical analysis failed to detect the C-terminal nebulin SH3 domain, as predicted by the mutation in M185. In six other patients, despite frame shifts in the region M162–M181, the C-terminal SH3 epitope located further toward the 3′ end was, however, detected, although in one of these patients a population of fibers was SH3 negative.
DISCUSSION
In the nebulin gene segment studied, we found six different mutations associated with autosomal recessive nemaline myopathy. Two patients were homozygous for their mutations; one was shown to be a compound heterozygote. Two patients with one detected mutation each were likely, in view of their haplotypes, also to be compound heterozygotes (Fig. 3). In summary, all mutations identified to date are located in the segment M162–M185.
The frame shifts/stop codons caused by the six mutations are predicted to cause the loss of 25–80 kDa from the C-terminal end of the nebulin filament system. In agreement with the gene sequence data, the histochemical analysis of one patient biopsy suggests loss of the C-terminal SH3 domain epitope. However, at present, it is unclear why, in the other six patients analyzed, the SH3 domain is detectable.
Possibly, it is relevant that, for the SH3-positive cases, the five observed mutations are located within the M176–M181 segment or in the 5′ vicinity of this segment, a region in which extensive differential splicing occurs. Therefore, we tested whether the splicing pattern is altered by some of the mutations. In one case, a misspliced nebulin transcript was detected in which an intron is retained (Fig. 4). Because in this patient the C-terminal SH3 domain epitope is detectable (Fig. 4), despite the presence of stop codons in the amplified aberrant transcript (Fig. 2), we speculate that mutant transcripts leading to abnormal splice intermediates may be further processed by splicing out the mutated sequences. Thus, the mature transcripts may encode the extreme C-terminal epitopes but would be internally truncated within the M176–M181 region. Such a model may suggest that the aberrant nebulins in nemaline myopathy would lack the normal physiological isoform diversity, which appears to be linked to the fiber-type and Z line diversity in different muscle tissue types (10). In another, an Australian patient, transcripts including the exons M177b, M177c, M177d, and M178b were observed, which are not expressed in the nebulins from healthy controls sequenced to date.
Currently, we are investigating whether changes in splice isoforms and the aberrations in fiber-typing patterns generally seen in nemaline myopathies are linked to each other and thus might be causally related to the disease.
Within the determined nebulin gene sequence, exon/intron boundaries are invariably located within the central SDXXYK motif, which is characteristic of the nebulin module family. As observed previously, the segment M176–M181 gives rise to multiple different-sized isoform variants (10). It is now clear, based on the exon/intron structures, that these isoforms are generated by differential exon-skipping events of the exons encoding M176–M181 (Fig. 2). If the positions of the exon/intron boundaries are conserved throughout the complete nebulin gene, it will become possible in the near future to develop primer pairs for the amplification of all, more than 185, nebulin repeats.
Interestingly, tissue culture studies have previously shown that overexpression of α-actinin also results in the formation of nemaline bodies (20, 21). On the genetic level, mutations in TPM3 (5) and NEB (this study) both lead to nemaline myopathy. Therefore, genetic as well as cell biology approaches suggest that alterations in nebulin, and other thin filament proteins, and their connecting partners such as α-actinin, are the molecular basis for nemaline body formation. According to this model, perturbation of their physiological interactions in Z disc assembly leads to nemaline body formation. The mechanisms of the tight coordination of the coassembly of titin, nebulin, α-actinin, and actin are not understood. Possibly, a study of the Z disc structure and Z disc proteins present in the patient with the M185 stop codon, leading to lack of the C-terminal regulatory 25-kDa sequences of nebulin, will lead to further insights into the mechanisms controlling this assembly.
For future studies, a complete panel of mAbs specific to single exons of the M162–M181 repeat region, to the unique serine-rich domain, and to the SH3 domain is needed to determine which parts of nebulin are lacking in nemaline myopathies. Thereby, possibly, a better understanding of the relationship between disease severity and the nature of the nebulin mutation present in patients with nemaline myopathy may emerge and provide a basis for genetic counseling.
Acknowledgments
We thank Dr. Carsten Bönnemann for his kind help regarding the German patient and Dr. El Hadi Hammouda of the Assocation Française contre les Myopathies–Myobank and Marie Bourgeois regarding the French patient. Dr. Lauri Aaltonen is gratefully acknowledged for providing the control samples and Dr. Sue Brown for immunocytochemical work. We are grateful to the European Neuromuscular Centre and its associate members for organizational support. K.P., P.H., and K.D. were supported by grants to C.W.-P. and A.d.l.C. from the Association Française contre les Myopathies, the Swedish Cultural Foundation, the Finska Läkaresällskapet, and the Medicinska understödsföreningen Liv och Hälsa. A.d.l.C. was supported by Grants CA67941 and CA16058 from the National Institutes of Health. P.A.A., N.G.L., and S.D.W. were supported by Australian National Health and Medical Research Council Project Grant 970104. T.C. and S.L. were supported by the Human Frontier Science Program and the Deutsche Forschungsgemeinschaft (La668/3-3). D.W. and A.H.B. were supported by National Institutes of Health Grants AR44345 and AR02026 and by the Muscular Dystrophy Association of the United States.
ABBREVIATIONS
- SSCP
single-strand conformation polymorphism
- RT
reverse transcription
Footnotes
References
- 1.Wallgren-Pettersson C. Neuromusc Disord. 1998;8:401–404. doi: 10.1016/s0960-8966(98)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.North K N, Laing N G, Wallgren-Pettersson C the ENMC International Consortium on Nemaline Myopathy. J Med Genet. 1997;34:705–713. doi: 10.1136/jmg.34.9.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dubowitz V. Muscle Disorders in Childhood. 2nd Ed. London: Saunders; 1995. pp. 147–155. [Google Scholar]
- 4.Wallgren-Pettersson C, Clarke A. In: Emery and Rimoin’s Principles and Practice of Medical Genetics. 3rd Ed. Rimoin D L, Connor J M, Pyeritz R E, Emery A E H, editors. New York: Churchill Livingstone; 1996. pp. 2367–2386. [Google Scholar]
- 5.Laing N G, Wilton S D, Akkari P A, Dorosz S, Boundy K, Kneebone C, Blumbergs P, White S, Watkins H, Love D R, et al. Nat Genet. 1995;9:75–79. doi: 10.1038/ng0195-75. [DOI] [PubMed] [Google Scholar]
- 6.Wallgren-Pettersson C, Avela K, Marchand S, Kolehmainen J, Tahvanainen E, Juul Hansen F, Muntoni F, Dubowitz V, de Visser M, van Langen I M, et al. Neuromusc Disord. 1995;5:441–443. doi: 10.1016/0960-8966(95)00022-f. [DOI] [PubMed] [Google Scholar]
- 7.Pelin K, Ridanpää M, Donner K, Wilton S, Krishnarajah J, Laing N, Kolmerer B, Labeit S, de la Chapelle A, Wallgren-Pettersson C. Eur J Hum Genet. 1997;5:229–234. [PubMed] [Google Scholar]
- 8.Wallgren-Pettersson C, Beggs A H, Laing N. Neuromusc Disord. 1998;8:53–56. [PubMed] [Google Scholar]
- 9.Pfuhl M, Winder S, Pastore A. EMBO J. 1994;13:1782–1789. doi: 10.1002/j.1460-2075.1994.tb06446.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Millevoi S, Trombitas K, Kolmerer B, Kostin S, Schaper J, Pelin K, Granzier H, Labeit S. J Mol Biol. 1998;282:111–123. doi: 10.1006/jmbi.1998.1999. [DOI] [PubMed] [Google Scholar]
- 11.Moncman C L, Wang K. Cell Motil Cytoskeleton. 1995;32:205–225. doi: 10.1002/cm.970320305. [DOI] [PubMed] [Google Scholar]
- 12.Gregorio C C, Trombitás K, Centner T, Kolmerer B, Stier G, Kunke K, Suzuki K, Obermayr F, Herrmann B, Granzier H, et al. J Cell Biol. 1998;143:1013–1027. doi: 10.1083/jcb.143.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ohtsuka H, Yajima H, Kimura S, Maruyama K. FEBS Lett. 1997;401:65–67. doi: 10.1016/s0014-5793(96)01432-9. [DOI] [PubMed] [Google Scholar]
- 14.Sorimachi H, Freiburg A, Kolmerer B, Ishiura S, Stier G, Gregorio C C, Labeit D, Linke W A, Suzuki S, Labeit S. J Mol Biol. 1997;270:688–695. doi: 10.1006/jmbi.1997.1145. [DOI] [PubMed] [Google Scholar]
- 15.Young P, Ferguson C, Bañuelos S, Gautel M. EMBO J. 1998;17:1614–1624. doi: 10.1093/emboj/17.6.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labeit S, Kolmerer B. J Mol Biol. 1995;248:308–315. doi: 10.1016/s0022-2836(95)80052-2. [DOI] [PubMed] [Google Scholar]
- 17.Dib C, Fauré S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, et al. Nature (London) 1996;14:152–154. doi: 10.1038/380152a0. [DOI] [PubMed] [Google Scholar]
- 18.Bassam B J, Caetano-Anolles G, Gresshoff P M. Anal Biochem. 1991;196:80–83. doi: 10.1016/0003-2697(91)90120-i. [DOI] [PubMed] [Google Scholar]
- 19.Pfuhl M, Winder S J, Castiglione Morelli M A, Labeit S, Pastore A. J Mol Biol. 1996;257:367–384. doi: 10.1006/jmbi.1996.0169. [DOI] [PubMed] [Google Scholar]
- 20.Schultheiss T, Choi J, Lin Z X, DiLullo C, Cohen-Gould L, Fischman D, Holtzer H. Proc Natl Acad Sci USA. 1992;89:9282–9286. doi: 10.1073/pnas.89.19.9282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Z, Hijikata T, Zhang Z, Choi J, Holtzer S, Sweeney H L, Holtzer H. Dev Biol. 1998;199:291–308. doi: 10.1006/dbio.1998.8920. [DOI] [PubMed] [Google Scholar]