Abstract
Factor H is the primary soluble regulator of activation of the alternative pathway of complement. It prevents activation of complement on host cells and tissues upon association with C3b and surface polyanions such as sialic acids, heparin and other glycosaminoglycans. Here we show that interaction with polyanions causes self-association forming tetramers of the 155,000 Da glycosylated protein. Monomeric human factor H is an extended flexible protein that exhibits an apparent size of 330,000 Da, relative to globular standards, during gel filtration chromatography in the absence of polyanions. In the presence of dextran sulfate (5,000 Da) or heparin an intermediate species of apparent m.w. 700,000 and a limit species of m.w. 1,400,000 were observed by gel filtration. Sedimentation equilibrium analysis by analytical ultracentrifugation indicated a monomer Mr of 163,000 in the absence of polyanions and a Mr of 607,000, corresponding to a tetramer, in the presence of less than a 2-fold molar excess of dextran sulfate. Increasing concentrations of dextran sulfate increased binding of factor H to zymosan-C3b 4.5-fold. This was accompanied by an increase in both the decay accelerating and cofactor activity of factor H on these cells. An expressed fragment encompassing the C-terminal polyanion binding site (complement control protein domains 18–20) also exhibited polyanion-induced self association, suggesting that the C-terminal ends of factor H mediate self-association. The results suggest that recognition of polyanionic markers on host cells and tissues by factor H, and the resulting regulation of complement activation, may involve formation of dimers and tetramers of factor H.
Keywords: Human, Complement, Inflammation, Cell Surface Molecules
Introduction
The alternative pathway of complement activates on surfaces unprotected by complement regulatory proteins, which are responsible for homeostasis of the complement system. Most host cells and tissues are protected by a family of membrane-bound and soluble proteins structurally related to the soluble protein factor H (reviewed in (1–3)). Activation of this arm of the human innate immune system occurs spontaneously (4, 5) on surfaces lacking these protective proteins and this process proceeds without reliance on target-specific recognition by the adaptive immune system. Targets for alternative pathway activation include bacteria, fungi, and most organic and inorganic particles including pollen and dust (6). Adapting to this system, numerous pathogens evade complement by expressing receptors for factor H, the major regulator in plasma, and these proteins are virulence factors for Streptococci, Neisseria, Borrelia, Yersinia, Cryptococci and some parasites (7).
Two diseases have recently been shown to be directly related to defects in regions of factor H that are thought to recognize host markers. Age-related macular degeneration is the leading cause of blindness in the elderly and affects approximately 50 million people worldwide. Age-related macular degeneration is a progressive inflammatory disease that has been strongly linked to the Y402H haplotype of factor H (8–11). Approximately 14% of the human population is homozygous for this haplotype, which is located in the seventh domain of factor H. Atypical hemolytic uremic syndrome (aHUS), an inherited form of HUS, has been shown to be associated with mutations that affect the C-terminal twentieth domain of factor H (7, 12–14). This region has been shown to be critical for interacting with negatively charged molecules on host cells such as sialic acid and sulfated glycosaminoglycans (15–20). In vitro assays have demonstrated that cells normally protected by polyanionic markers on their surface are attacked by the alternative pathway of complement when exposed to human serum containing factor H with mutations affecting its polyanion-binding domains (7, 21). While most human tissues in vivo are adequately protected by the membrane-bound regulators DAF (CD55), CD59, CR1 and MCP (CD46) as well as by binding soluble factor H, the kidney appears to rely heavily on protection by factor H (22, 23). Mutations resulting in functional loss of the C-terminal polyanion binding site of factor H present clinically with acute renal failure (7, 12, 13, 24). Because domain seven also exhibits polyanion binding (25) a similar mechanism is thought to occur in age-related macular degeneration although this interaction is not as well characterized. The existence of two different diseases correlated to different sites on factor H suggests that tissue specific markers are being recognized by different sites on factor H.
Factor H is composed solely of 20 complement control protein (CCP) domains with interdomain spacers of 4 to 7 amino acids (26). While some interdomain linkages appear to be flexible and some more rigid (3, 27–29) the overall structure of this 155,000 Da protein appears in electron micrographs to be sufficiently flexible to fold back on itself (30–33). From NMR and crystal structures of CCP modules from different proteins, these structures appear to be independently folding ellipsoidal modules each composed of approximately 61 amino acids (34–37). Functional sites have been localized along the entire length of factor H and each site occupies from one to four domains (7, 38). The N-terminal 1–4 domains express all of the complement regulatory activities of the protein through their interactions with C3b (39–41). Three polyanion binding sites have been localized in factor H domains 7 (25), 9–14 (42, 43) and 20 (18, 20, 21, 44). Although mutations in the first 18 domains of factor H have been found associated with aHUS, the vast majority of mutations are found in domains 19 or 20, leading to loss of complement control at diverse cell surfaces (21, 44–47). Accordingly, the C-terminus of factor H has been shown to be essential for factor H-mediated recognition and regulation of complement at host cell surfaces (48) and for protecting against an aHUS-like renal pathology in a murine factor H transgenic model (49).
Factor H controls spontaneous activation of the alternative pathway of complement in blood and regulates activation on surfaces in contact with blood (12). Its interaction with host polyanionic markers prevents activation on surfaces bearing such markers and allows activation on almost everything else (17). In this study we report (50) that contact with polyanionic molecules induces specific self-association of the protein resulting in the formation of dimers and tetramers of factor H. The results imply that the interaction with surface polyanions may induce the formation of defined clusters of factor H that would protect broad regions of the surface due to the flexibility and extended shape of factor H.
Materials and Methods
Reagents
Dextran sulfate (average m.w. of base dextran approximately 5,000) and heparin sulfate (average m.w. 12–14,000) were purchased from Sigma Chemical Co. Concentrations of polyanions were determined by weight of dry compounds used to make stock solutions.
Proteins
Complement protein factor H was purified from normal human plasma as previously described (51). Purified factor H was greater than 97% homogenous by polyacrylamide gel electrophoresis with an apparent m.w. on SDS gel electrophoresis of 155,000 in its reduced form. Protein concentration was determined spectrophotometrically using an A280nm (1% solution) of 12.4 for factor H (52). Four different extinction coefficients for factor H have been reported ranging from 9.0 to 19.5 (52–55). The coefficient calculated from the sequence (56) and corrected for the large content of polysaccharide in factor H is 13.2. In spite of these differences, which are typical of widely studied proteins, the value of 12.4 is in the middle of the range and the closest to the calculated value. Regardless of the disagreements in the literature, reporting the coefficient used in this study allows scientists to repeat the work presented here as it was performed. To generate recombinant CCP 18–20, the coding sequence for residues 811–1231 were PCR-amplified from a full length factor H cDNA template (38), using a forward primer (5’-ATA TGC TCT TCA TGC GTG AAT CCG CCC ACA GTA CAA AAT-3’) and a reverse primer (5’-TAT AGC TCT TCA ACA AGT TGG ATA CTC CAG TTT CCC ATC-3’) containing SapI restriction sites (underlined). The SapI-digested PCR product was cloned into pPICZ-α vector (Invitrogen) containing SapI sites added in its multiple cloning site, as previously described (57). Briefly, the original multiple cloning site of pPICZ-α A was replaced by inserting a double stranded oligo containing a SapI site flanked by overhang 5’ XhoI (5’-TCG AGA_AAA GAG AAG ATT GCA GAA GAG CGG CGC GCC GCT CTT CAT GTG CAA AAA GAT GAT-3’) and 3’ XbaI sites (5’-CTA GAT CAT CTT TTT GCA CAT GAA GAG CGG CGC GCC GCT CTT CTG CAA TCT TCT CTT TTC-3’) into the corresponding Xho and XbaI sites located in the vector (57). Following transformation into P. pastoris strain KM71H, protein expression was induced with methanol following manufacturer’s instructions, and the protein was purified from the media by anion exchange chromatography.
Gel Filtration Chromatography
Chromatography was performed on 0.75 × 60 cm TosoHaas TSK-Gel G4000SW or Phenomenex BioSep SEC S4000 columns with guard columns (TosoHass). Gel filtration columns were run at 21°C at a flow rate of 0.5 ml/min in either PBS (10 mM sodium phosphate, 140 mM NaCl, 0.02% sodium azide, pH 7.4) or in half ionic strength PBS (5 mM sodium phosphate, 70 mM NaCl, 0.01% sodium azide, pH 7.4) containing the desired concentrations of polyanions. The following standards (BioRad Laboratories) were used: vitamin B12 (1,350), myoglobin (17,000), ovalbumin (44,000), IgG (158,000), and thyroglobulin (670,000).
Analytical Ultracentrifugation
Equilibrium sedimentation experiments were carried out in a Beckman XL-A analytical ultracentrifuge. One hundred microliter samples containing factor H at protein concentrations between 0.1 and 0.3 mg/ml in 5 mM sodium phosphate, 70 mM NaCl, 0.01% sodium azide, pH 7.4 were centrifuged at various rotor speeds between 3,000 and 7,000 rpm in two sector cells in an An50Ti rotor at 295°K (22°C) until equilibrium was achieved. The cells were scanned at 280 nm. Data points are the average of five readings at 0.001 cm intervals through the sample column that was approximately 1.4 mm. The endpoint was reached between 24 and 48 hrs when multiple scans did not change over 4 hours. The data was modeled as a single ideal species using the Beckman XL-A data analysis software version 4.0 running in Origin version 4.1 using an estimated solvent density of 1.00 g/cm3. We used a partial specific volume of 0.711 ml/g (53), which was similar to the calculated partial specific volume of 0.717 ml/g based on the predicted amino acid composition (31, 53, 56).
Cell surface factor H binding assays
Varying concentrations of dextran sulfate (0–300 µg/ml) were incubated for 10 min at 22°C with 83 ng of radiolabeled human factor H in GVB in a total volume of 80 µl. Then 20 µl of Zym-C3b (1×109 cells/ml bearing 80,000 to 100,000 C3b/cell) was added and incubated for 20 min. Cell-bound factor H was separated from free protein by layering 80 µl of the mixture on top of 20% sucrose in GVB and centrifuging for 2 min at 10,000 × g as previously described (15). The tube was cut to separate and count the pellet and supernatant.
Cell surface factor H decay acceleration and cofactor assays
Decay accelerating activity expressed by factor H was measured by determining its ability to accelerate the natural release of 125I-labeled Bb from cell bound C3b,Bb. The C3b,Bb complexes were formed by incubating 1 × 107 Zym-C3b, 400 ng 125I-factor B (2 µCi/µg), 250 ng of factor D and 1 mM NiCl2 in 35 µl GVB at 22°C for 3 min as previously described (48). Formation of the C3 convertase was stopped by the addition of 65 µl of GVBE. To generate a standard curve, the C3b,Bb complexes (10 µl) were added immediately to reaction mixtures (40 µl) containing various amounts of factor H (0–8000 ng/ml). After 10 min at 22°C the cells were sedimented rapidly (2 min, 10,000 × g) through 250 µl of 20% sucrose in GVB in a microfuge tube. The bottoms of the tubes were cut off, and the radioactivity in the cell pellet and the supernatant were measured to determine the percent Bb remaining bound. To determine the effect of dextran sulfate on decay acceleration, the concentration of factor H (500 ng/ml) required to release 25% of the 125I-Bb, was incubated in the presence of various concentrations of dextran sulfate (0–150 µg/ml). The increase of decay activity in the presence of dextran sulfate was calculated based on a standard curve using factor H alone. Cofactor activity was determined by measuring the factor H-dependent inactivation of C3b by factor I (20 µg/ml) on Zym-C3b in the presence and absence of dextran sulfate. Following treatment, the amount of functional C3b remaining was measured by quantitating the binding of 125I-factor B in the presence of 0.1 mM NiCl2, 1.3 µg/ml factor D, and excess 125I-factor B.
Results
Size Exclusion Chromatography
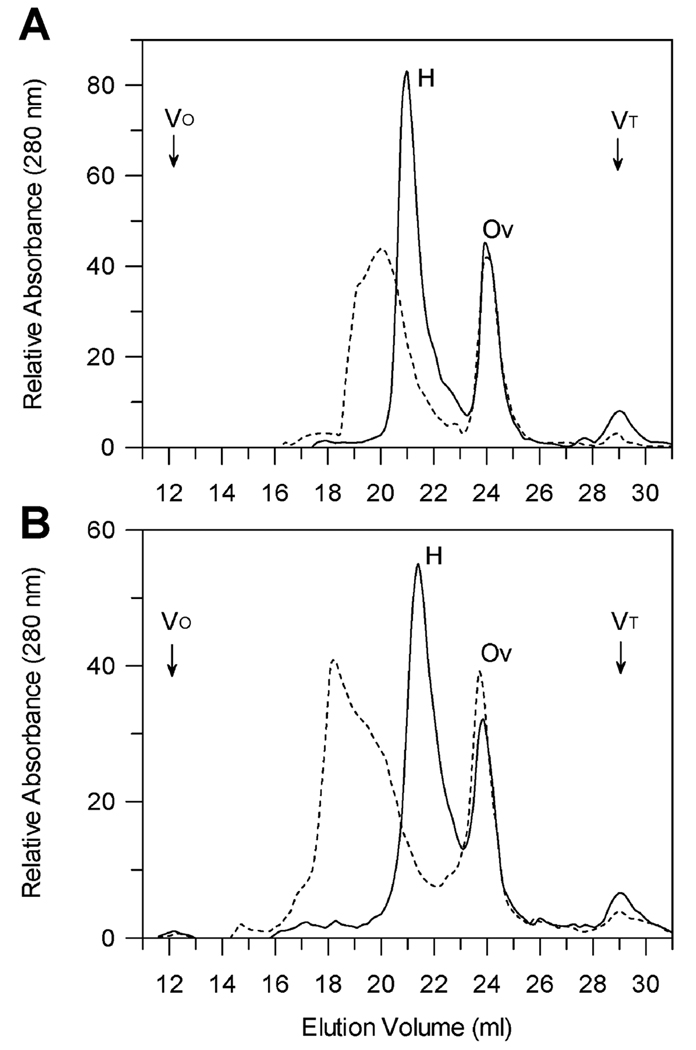
The hydrodynamic properties and oligomeric state of factor H were examined in solution using gel filtration chromatography on the HPLC media TSK-gel G4000SW as indicated in Fig. 1. In normal ionic strength PBS (Fig. 1A) without polyanions factor H migrated as a single species of approximately 330,000 m.w. as previously reported (53, 58). When the sample was preincubated with 1 mg/ml dextran sulfate and the column run in a buffer containing 1 mg/ml dextran sulfate, factor H eluted as a broad peak of higher apparent molecular weight. The void volume (Vo) and the elution volume of vitamin B12 (VT) are indicated in Fig. 1. Chromatography in half ionic strength PBS (Fig. 1B) showed similar profiles except that in the presence of polyanion the majority of factor H eluted at the high m.w. position. Ovalbumin was included as an internal standard. Controls indicated that the elution position of ovalbumin was not affected by ionic strength and its presence did not affect the elution position of factor H with or without polyanions present.
FIGURE 1.
Gel filtration chromatography of purified human factor H in the presence of dextran sulfate. Factor H was chromatographed in normal ionic strength PBS (Panel A) in the presence (dashed line) of 1 mg/ml dextran sulfate or in the absence of dextran sulfate (solid line) using ovalbumin as an internal standard. Samples of 0.3 ml containing 960 µg factor H and 730 µg ovalbumin with or without 1 mg/ml dextran sulfate (average m.w. approx. 5,000 providing an approximately 10-fold molar excess of dextran sulfate over factor H) were preincubated for 15 min at 21°C and loaded onto a 0.75 × 60 cm TosoHaas G4000SW column with a guard column equilibrated with PBS (10 mM sodium phosphate, 140 mM NaCl, 0.02% sodium azide, pH 7.4) (solid line) or PBS + 1 mg/ml dextran sulfate (dashed line). Chromatography was performed at 21°C at a flow rate of 0.5 ml/min. Panel B shows the results of similar chromatography using half ionic strength buffer (5 mM sodium phosphate, 70 mM NaCl, 0.01% sodium azide, pH 7.4). The positions of the column void volume (VO) and salt peak (VT) are indicated.
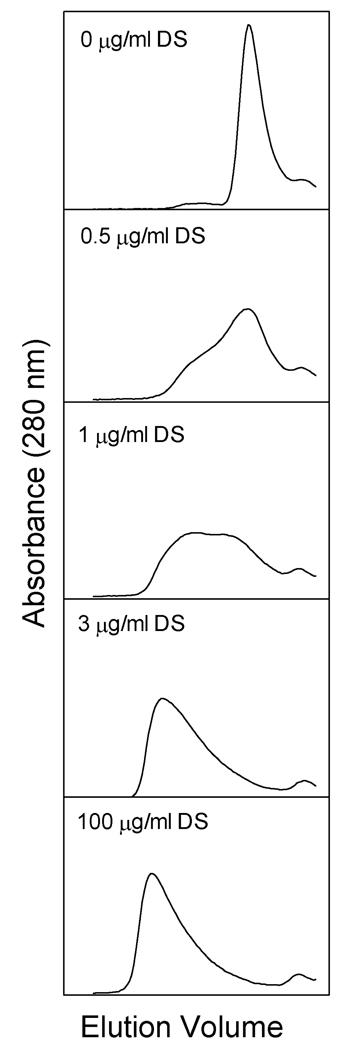
Fig. 2 demonstrates the dependence of the elution position of factor H on the concentration of dextran sulfate present in the buffer used for chromatography. An identical concentration of dextran sulfate was preincubated with the sample for 15 min prior to loading. Comparison of the panels in Fig. 2 shows a concentration dependent decrease in the elution volume of factor H with increasing dextran sulfate concentration. The sharp frontal boundary of the peak in the presence of 100 µg/ml dextran sulfate indicates that a stable maximum size has been reached which is considerably smaller than the void volume of the column (Fig. 1B). The trailing edge of the factor H peak was greatly extended (Fig. 1B) possibly indicating a complex in dynamic equilibrium with the lower molecular weight forms and with a dissociation half-life similar to the time factor H spent subjected to the HPLC gel filtration process (approximately 45 min).
FIGURE 2.
Gel filtration chromatography of purified human factor H in the presence of increasing concentrations of dextran sulfate. Samples containing 75 µg factor H in the indicated concentrations of dextran sulfate (average m.w. ~ 5,000) in 0.5 ml were preincubated for 15 min at 21°C and loaded onto a 0.75 × 60 cm TosoHaas G4000SW column with a guard column equilibrated with the indicated concentration of dextran sulfate in 5 mM sodium phosphate, 70 mM NaCl, 0.01% sodium azide, pH 7.4. Chromatography was performed at 21°C at a flow rate of 0.5 ml/min. Proper alignment was verified by including ovalbumin as an internal standard (peak is not shown). The region of the chromatograms shown here corresponds to elution volumes from 15 ml to 23 ml (see Fig. 1).
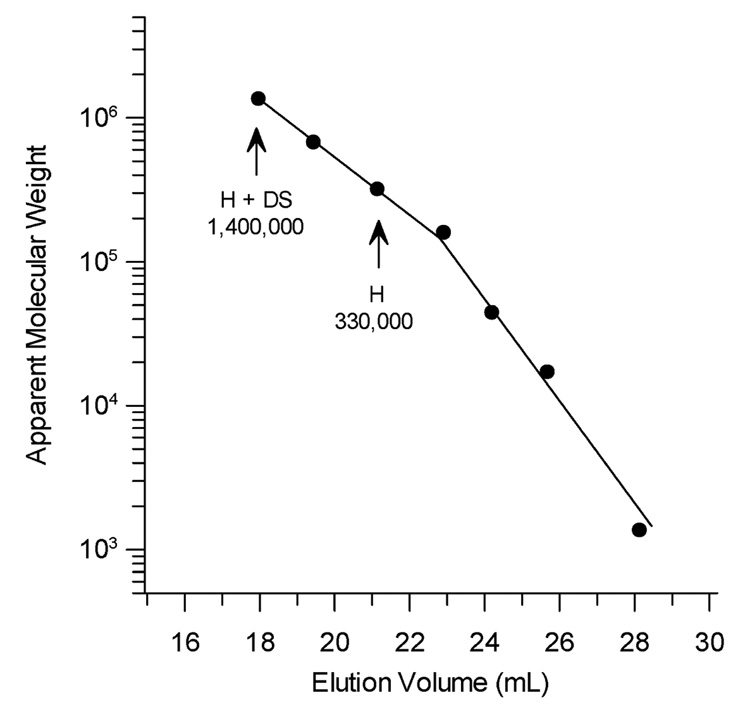
The apparent size of the complex formed upon exposure of factor H to dextran sulfate was estimated from the elution positions of factor H in buffer alone and in the presence of the polyanion relative to standard proteins (Fig. 3). Factor H has been shown to have an extended shape in solution and to exhibit an apparent molecular mass of approximately 300,000 Da upon gel filtration through agarose gel filtration media (31, 53, 58). Using TosoHass G4000SW media an apparent size of 330,000 Da was observed for factor H in agreement with previous studies. In the presence of 100 µg/ml of dextran sulfate the apparent molecular mass, compared to standards, was 1,400,000 Da. This is almost four times the molecular weight found for factor H in the absence of polyanions (Fig. 3). Although this result suggests that a homo-tetramer of factor H was induced by interaction with the polyanion, size determination of any molecule other than globular proteins by gel filtration is an approximation due to the hydrodynamic behavior of highly asymmetrical molecules compared to globular proteins. As shown below, analysis of sedimentation equilibrium profiles provide a more reliable measure of the molecular weight of a complex because such analyses are not sensitive to molecular shape, only to mass.
FIGURE 3.
Apparent size of factor H in the presence and absence of dextran sulfate compared to globular standards. Gel filtration chromatography was performed on a 0.75 × 60 cm TosoHaas G4000SW column with a guard column equilibrated with 10 mM sodium phosphate, 500 mM NaCl, 0.02% sodium azide, pH 7.4 using the following standards (Mr): vitamin B12 (1,350), myoglobin (17,000), ovalbumin (44,000), IgG (158,000), IgG dimers (316,000), thyroglobulin (670,000), thyroglobulin dimers (1,340,000). The positions and estimated Mr of factor H and the high molecular weight complex are indicated. All chromatography was run in the high salt buffer (0.5 M NaCl) except factor H plus dextran sulfate (H + DS) that was run in 5 mM sodium phosphate, 70 mM NaCl, 0.01% sodium azide, pH 7.4.
The pattern of factor H polymerization observed during gel filtration in the presence of the polyanion heparin (not shown) was similar to that observed with dextran sulfate. In the presence of increasing concentrations of heparin (12–14,000 Da) the majority of factor H eluted with an apparent size of 640,000 Da similar to the pattern shown with dextran sulfate in Fig. 1A. In half ionic strength PBS at the highest heparin concentration tested (1000 µg heparin/ml) only 10% of the factor H emerged in the 1,400,000 m.w. position and the predominant species was the apparent dimer form of factor H at a m.w. of ~640,000. Taken together these data suggest that higher molecular weight complexes were formed with heparin, but that these were less stable than those formed with dextran sulfate even at half physiological ionic strength.
Sedimentation Equilibrium Analysis
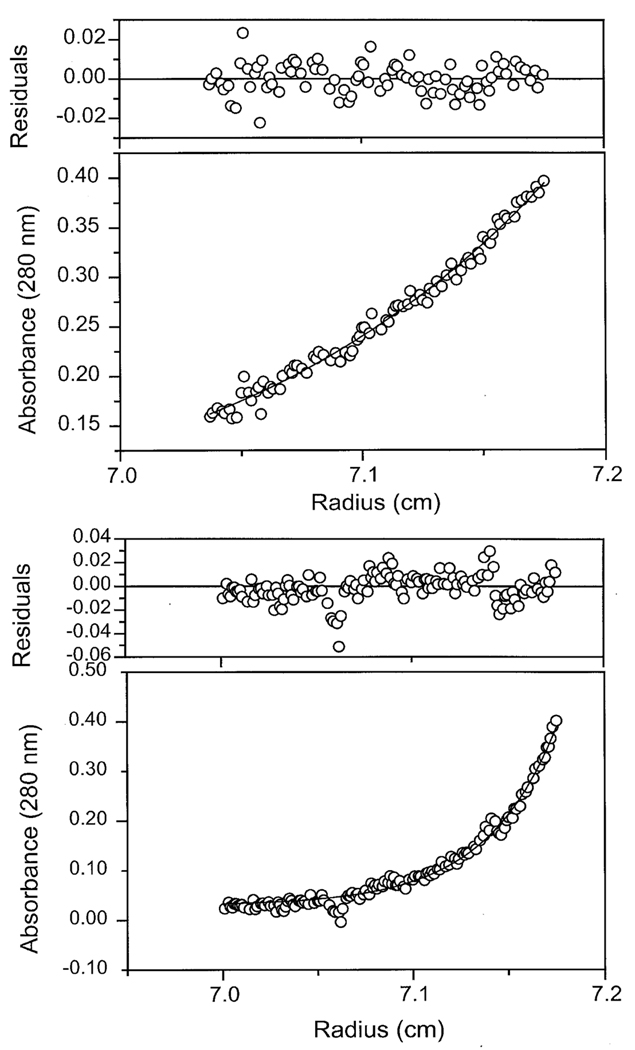
Because of the asymmetrical structure of factor H no reliable information regarding the number of factor H molecules in either the low or high molecular weight species can be derived from gel filtration measurements. Therefore, the subunit structure of the complexes was analyzed by sedimentation equilibrium using analytical ultracentrifugation. Fig. 4 shows the A280nm scans from sedimentation equilibrium analyses of factor H in the absence of polyanions (Fig. 4, upper panel) and in the presence of 100 µg/ml of dextran sulfate (Fig 4, lower panel). The molecular mass of factor H determined in half ionic strength PBS was 163,264 Da when the data in Fig. 4 (upper panel) were fit to the equation for a single species. The monomer molecular mass varied from 158,183 to 165,238 Da with a mean of 162,133 for four separate experiments at two speeds. Previous similar analyses (31) in normal ionic strength PBS found 153,000 Da at similar factor H concentrations and 159,000 Da at higher concentrations (1.2 mg/ml). These observations suggest that there may be some propensity for self-association and that this may be enhanced by high concentrations of factor H or by the low ionic strength used in the experiments shown in Fig. 4. In the presence of 100 µg/ml dextran sulfate a four-fold increase in mass was observed. The data in Fig 4 (lower panel) were best fit to an average molecular mass of 607,244 Da using the equation for a single species.
FIGURE 4.
Sedimentation equilibrium scans of factor H alone (upper panel) and factor H in the presence of 100 µg/ml dextran sulfate (lower panel). Representative equilibrium scans of factor H alone and of factor H in the presence of 100 µg/ml dextran sulfate at 295 °K. Both samples were centrifuged at 7,000 rpm for comparative purposes. Samples contained 273 µg/ml (1.8 µM) factor H and either no dextran sulfate (upper panel) or 100 µg/ml dextran sulfate (lower panel) in 5 mM sodium phosphate, 70 mM NaCl, 0.01% sodium azide, pH 7.4.
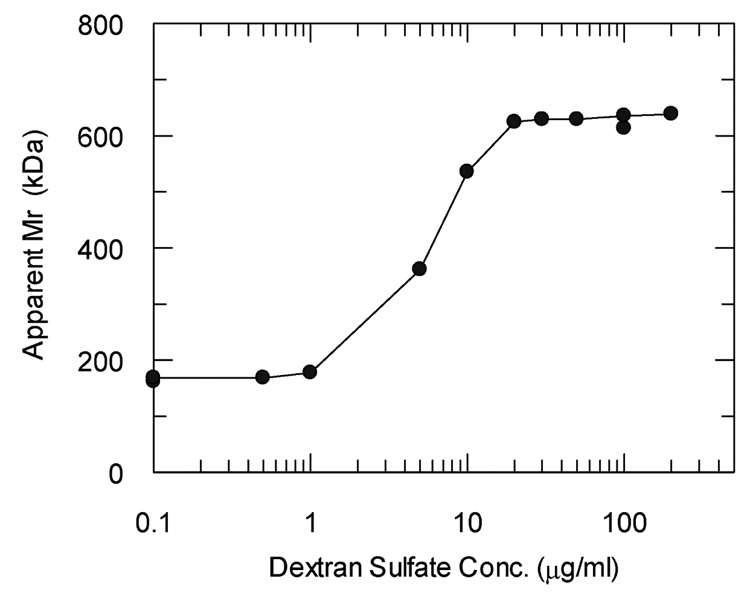
Similarly, Fig. 5 shows the effect of polyanion concentration on the average molecular mass of the factor H complexes. Analytical ultracentrifugation was performed in the presence of the indicated dextran sulfate concentrations and each of the protein gradients observed was fit to an equation for a single species. Little or no association was found until the polyanion concentration was above 1 µg/ml. A maximum of 638,000 Da was observed at 200 µg/ml and a plateau was observed between 20 and 200 µg/ml suggesting that there is a defined size to the complexes and that the assembly is self-limiting. The mean molecular mass in the presence of dextran sulfate concentrations ranging from 20 µg/ml to 500 µg/ml was 619,355 Da for nine separate determinations at two different speeds. This indicates that in the presence of a saturating concentration of dextran sulfate the high molecular weight complex contains 4 molecules of factor H. Furthermore, the plateau suggests that saturation of the polyanion site on factor H was achieved at 20 µg/ml. At a concentration of dextran sulfate of 7 µg/ml, polymerization was half maximal and the molar ratio of dextran sulfate to factor H was approximately 1.4 to 1, clearly indicating that the increase in molecular mass was not due to binding of multiple dextran sulfate molecules of only 5,000 Da. Sedimentation equilibrium analysis of the effect of heparin on factor H indicated similar polymerization was caused by comparable concentrations of heparin although there were indications of the presence of larger complexes of factor H (700,000 to 900,000 Da). The heparin molecules used were larger than the dextran sulfate and it is possible that the multiple heparin binding sites on factor H bound additional heparin or that the longer heparin polyanions provided nucleation sites for binding of additional factor H molecules.
FIGURE 5.
Average apparent molecular weights of factor H in the presence of increasing concentrations of dextran sulfate. Sedimentation equilibrium analyses of factor H at 273 µg/ml (1.8 µM) in the presence of increasing concentrations of dextran sulfate were performed at 3,000 rpm and 7,000 rpm in 5 mM sodium phosphate, 70 mM NaCl, 0.01% sodium azide, pH 7.4 at 295 °K (22 °C) as described in the Materials and Methods. Note that the concentrations of factor H and dextran sulfate were approximately equimolar at 7 µg/ml dextran sulfate.
Analysis of recombinant CCP factor H domains by size exclusion chromatography
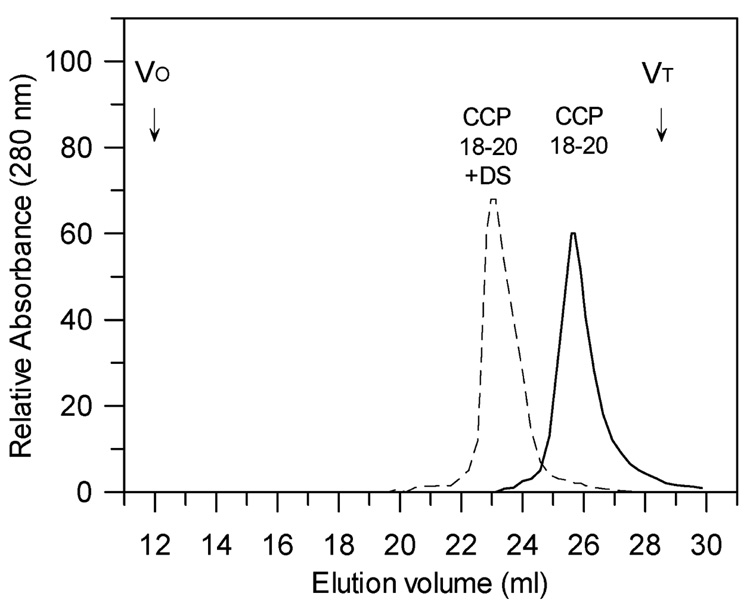
It has been recently described that CCP domains 16–20 and whole factor H have a weak ability to self associate in solution to form dimers when analyzed by X-ray scattering and analytical ultracentrifugation (59, 60). In addition, the crystal structure of CCP 19–20 indicates that monomers of this region form a tetramer composed of two tightly packed antiparallel dimers (29). In order to determine if this ability of the C-terminal domains to self associate is polyanion-enhanced, we examined a three domain recombinant protein composed of CCP 18–20 from the C-terminal region of factor H. In normal ionic strength buffer (PBS) in the absence of polyanions, factor H CCP 18–20 (22,000 Da) migrated as a single low molecular weight species (Fig. 6) during gel filtration. However, in the presence of dextran sulfate CCP 18–20 eluted as a sharp single peak of apparent molecular weight 90,000 Da (Fig. 6). Other three-domain regions of factor H (CCP 1–3 and CCP 11–13) were used as controls and no difference in their mobility was observed in the presence or absence of dextran sulfate (data not shown). Taken together, our results indicate that the C-terminal region of factor H can self associate at physiological pH and ionic strength and that this association is dependent on the presence of polyanions.
FIGURE 6.
Gel filtration chromatography of human factor H recombinant domains 18–20 in the presence of dextran sulfate. Recombinant CCP 18–20 of factor H was chromatographed in normal ionic strength PBS in the presence (dashed line) of 1 mg/ml dextran sulfate or in the absence of dextran sulfate (solid line). A 0.5 ml sample of 210 µg recombinant CCP 18–20 in 1 mg/ml dextran sulfate (10-fold molar excess of dextran sulfate over CCP 18–20) was preincubated for 15 min at 21°C and loaded onto a 0.75 × 60 cm Phenomenex BioSep SEC S4000 column with a guard 3.5 × 7.8 cm Phenomenex BioSep SEC 3000 column equilibrated with PBS or PBS + 1 mg/ml dextran sulfate. The chromatogram was performed at 21°C at a flow rate of 0.5 ml/min. The position of the column void (VO) and salt peak (VT) are indicated.
Effect of polyanions on the binding of factor H to cell-bound C3b and on its complement regulatory functions
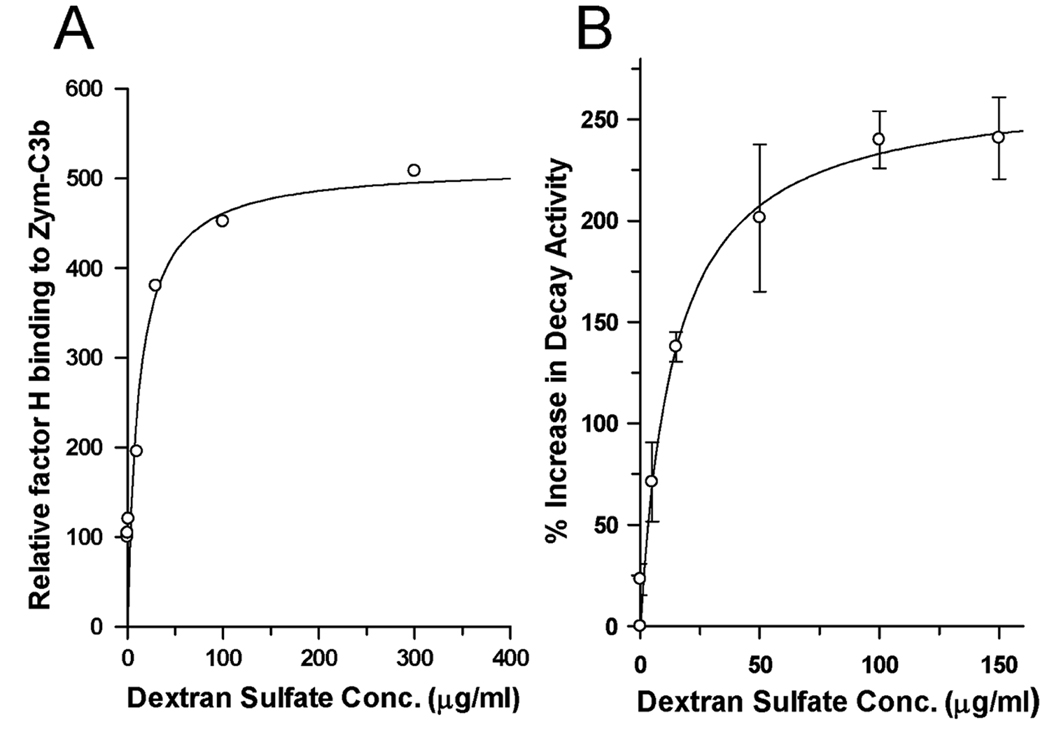
Current understanding of the interactions of factor H with polyanions is that host cells and tissues bear various polyanionic structures which bind the C-terminus of factor H thus enhancing its ability to control complement activation on host surfaces (17). If the interactions with polyanions produce dimers and tetramers these complexes should exhibit greater affinity for C3b on surfaces lacking natural polyanions (zymosan). Thus, we examined whether soluble dextran sulfate increases the binding of factor H to Zym-C3b and whether it enhances the complement regulatory activities of factor H at the cell surface. Fig. 7A shows that soluble dextran sulfate increases the binding of factor H to Zym-C3b by 4.5-fold. Polyanion-rich host cells such as human erythrocytes have been shown to exhibit a maximum of 10-fold enhanced binding compared to Zym-C3b suggesting that the observed effect represents a significant increase. We also determined the effect of dextran sulfate on the ability of factor H to increase the rate of dissociation of the two subunits of the alternative pathway C3/C5 convertase (C3b,Bb) in a decay acceleration assay. The release of radiolabeled Bb from surface-bound C3b was measured in the presence and absence of various concentrations of dextran sulfate (Fig 7B). The results showed that dextran sulfate increased the decay accelerating activity of factor H ~2.5-fold. A similar (>2-fold) polyanion-dependent increase in factor H cofactor activity using Zym-C3b and factor I was found (data not shown). The effective concentrations of dextran sulfate for binding and for each functional assay were similar to the concentration required for oligomer formation.
FIGURE 7.
Dextran sulfate increases factor H binding and factor H-mediated decay acceleration on the surface of Zym-C3b. (A) Binding of factor H to Zym-C3b was measured by incubating 125I-labeled factor H (83 ng, 50,000 cpm) with various concentrations of dextran sulfate and Zym-C3b for 10 min at 37°C, in 100 µl of GVB. Binding was normalized to the level of factor H binding observed in the control without dextran sulfate. (B) Decay acceleration assays measured the ability of human factor H to release 125I-Bb from cell-bound C3b,Bb in the presence of various concentrations of dextran sulfate. The C3b, 125I-Bb complexes were formed on Zym-C3b as described in Materials and Methods. The 125I-Bb-loaded cells were added to reaction mixtures containing factor H (500 ng/ml) and the indicated concentrations of dextran sulfate. The relative increase of decay accelerating activity (n=3) was calculated by comparison to the decay by factor H without dextran sulfate.
Discussion
Inactivation of clusters of complement enzymes on host or foreign cell surfaces requires interaction with regulatory proteins such as factor H. The concentration of factor H is approximately 500 µg/ml in plasma, but its affinity for a single C3b is low. The regulatory effectiveness of the protein toward surface-bound clusters is enhanced by the presence of three C3b binding sites in each factor H molecule (38, 61). On host and host-like surfaces bearing polyanionic markers a ten-fold greater effectiveness has been measured due to the interactions between the polyanion binding sites on factor H and surface polyanions (12, 15–18, 21, 44, 62). The present data suggests that all or part of this increased effectiveness could be due to polyanion-induced self-association of factor H.
Factor H is an extended protein (approximately 35 × 380 to 730 Å) that exhibits great flexibility and even appears to fold back on itself in electron micrographs (30–33). Before this asymmetry was recognized, its gel filtration properties were thought to indicate that it was a dimer of the 150,000 Da subunit seen on SDS gels (53, 58). A later report detected a tendency to dimerize at high concentrations in solution (32) and as a result factor H was depicted as a dimer in a review on complement in Scientific American in 1991 (63). Although later studies (30) questioned these findings, a very recent report (60) documented a weak tendency for dimer formation. The present results demonstrate that polyanions greatly enhance this weak interaction and promote formation of stable dimers as well as tetramers. Recently published structures for the C-terminal polyanion binding site derived from NMR (64) and from X-ray crystallography (29) further support our conclusions. The NMR study identified the amino acid residues forming the polyanion binding site in domain 20 (64). The X-ray structure of domains 19–20 yielded the interesting result that the unit cell was a D2 tetramer composed of two tightly packed antiparallel dimers (29). This observation confirms that a tendency exists in the C-terminal polyanion binding domains of factor H toward dimer and tetramer formation. Our data suggest that under physiological conditions in solution, defined oligomer formation is initiated by the interaction of the C-terminus of factor H with polyanions.
High performance gel filtration chromatography yields mostly qualitative information about asymmetrical proteins such as factor H. Fig. 1, Fig. 2 and Fig. 3 show that high m.w. species of factor H were formed in a polyanion-dependent manner. The peaks of the largest species exhibited sharp leading edges (Fig. 1 and Fig. 2, bottom panels) indicating that a defined maximum size exists. Comparison of the position of this peak (Fig. 1) with the void volume and salt peaks shows that the sharp leading edge is not due to oligomers that exceed the exclusion limit of the resin used, which was over 10,000,000 Da. The chromatograms also show that an intermediate species predominates at some concentrations of polyanion (Fig. 2), but whether this is due to a preferred complex or to the dynamics of association and dissociation cannot be determined. Nevertheless, higher concentrations of polyanion converted most of the factor H to the maximum size. The trailing edge of this peak (Fig. 2, 100 µg/ml dextran sulfate) indicates that this species is not completely stable during the approximate 25 to 30 min elution time, but the sharp leading edge shows that the majority of the complexes spent most of their time in the highest molecular weight form during the run. Thus, although gel filtration results are qualitative, they do provide information about the stability of these complexes. The complexes formed with heparin were never observed to migrate as a distinct peak at the higher molecular weight position. Instead, the main protein peak migrated in the position of the intermediate peak observed with dextran sulfate. It trailed off on the leading edge in high concentrations of heparin and on the trailing edge in low concentrations of heparin. Because high molecular weight species were observed in the analytical ultracentrifuge with heparin, the gel filtration results can be interpreted as suggesting that the high molecular weight complexes formed with heparin were less stable than those formed with dextran sulfate. The stability of complexes in vivo may be higher because the specific polyanionic receptors for factor H on human cells have yet to be identified and thus it is possible that we have not used the optimal ligands in this study.
Fig. 3 shows the apparent size of factor H during gel filtration relative to more globular standards. The apparent molecular mass found here in low ionic strength buffer (330,000 Da) agrees with the 300,000 Da found by others in physiological buffers (30, 53, 58). The apparent size of the dextran sulfate-induced high molecular weight species was 1,400,000 Da, which is very close to four times that of the monomer. While this may be fortuitous, it is consistent with the analytical ultracentrifuge results and the gel filtration results suggesting the existence of tetramers of significant stability as discussed in the preceding paragraph. The intermediate species (Fig. 1A and Fig. 2 at 1 µg/ml) exhibited an apparent size of 590,000 Da which is approximately twice that of the monomer. This peak may indicate the formation of dimers, consistent with the previous reports (32) and with the tightly packed dimer structures observed in the X-ray crystallography study of domains 19–20 (29). Alternatively, owing to the low concentration of polyanion, this intermediate peak may be due to the presence of a tetramer/monomer equilibrium that migrates at this position because of a relatively rapid dynamic equilibrium between the monomers and tetramers.
In contrast to gel filtration, sedimentation equilibrium allows accurate determination of molecular weights independent of the shape of a molecule or the shape of a complex of molecules. Fig. 4 and Fig. 5 show that polyanions induced the formation of high molecular weight species of factor H with Mr of 607,000 to 638,000. The polyanion is unseen in these scans because detection was at 280 nm, but it is important to note that 50% conversion to the tetramer form required only 7 µg/ml dextran sulfate, which is a 1.4-fold molar excess of polyanion over factor H. Complete conversion to the tetramer form was achieved with 20 µg/ml dextran sulfate. This represents less than a four-fold molar excess of polyanion (5000 Mr) over factor H (165,000 Mr) and thus the dextran sulfate could not significantly contribute to the apparent size of the factor H without inducing self-assembly of the protein. The presence of a maximum size of 638,000 that remains even in the presence of 10-fold higher concentrations of dextran sulfate suggests a defined, self-limiting structure has formed. This is in agreement with the high performance gel filtration results where a sharp front on the peak at high polyanion concentrations suggests a defined maximum size (1,400,000 Da), four times that of the monomer (330,000 Da), as measured by gel filtration. Interestingly, the C-terminal region of factor H (CCP 18–20), elutes at high molecular weight in the presence of dextran sulfate (Fig. 6) suggesting that this region may be responsible, at least in part, for conferring the oligomerization properties of full-length factor H when it is in contact with polyanions. These results are consistent with the tetramer structure seen in crystals of domains CCP 19–20 (29) and the weak self association seen with CCP 16–20 (59, 60) suggesting that the C-terminal domain of factor H, may form a contact point for tetramer formation. In addition, since the vast majority of aHUS-associated mutations in factor H are found in CCP 19–20 (7, 12, 13, 65), it is possible that these mutations may affect the capacity of factor H to oligomerize on polyanion-containing cell surfaces, with possible consequences in disease pathogenesis.
One previous observation of factor H behavior that has been difficult to explain may have its basis in polyanion-induced tetramer formation. It was observed (17, 66) that while the affinity of factor H for C3b on host-like cells was high, its affinity for C3b on cells lacking polyanions was as much as 10-fold lower. However, in the presence of a variety of soluble polyanions the affinity of soluble factor H for C3b, on particles that activate the alternative pathway of complement, increased to within 2-fold of the affinity for C3b on host cells. Figure 7A shows that fluid phase dextran sulfate induces a 4.5-fold increase in factor H binding to C3b on a cell lacking polyanions (zymosan). Dimerization of factor H by monoclonal antibodies produced a similar, but only 2-fold, increase in binding (67).
In addition to demonstrating that fluid phase dextran sulfate induced increased binding of factor H to an alternative pathway activator (Fig. 7A), we have also demonstrated that the cell surface functional activity of factor H was increased more than 2-fold in the presence of polyanion (Fig. 7B). Although this effect seems small it was necessary to measure both the decay acceleration and cofactor activities at approximately 100-fold lower concentrations of factor H than used in the ultracentrifuge and gel filtration experiments. Such low concentrations of factor H must shift the equilibrium toward the dissociated forms and reduce the concentration of dimers and tetramers in these experiments. Thus, the increase in functional activities would be expected to be greater at serum concentrations of factor H (i.e., 300-fold higher) which would enhance self-association in the presence of host polyanions.
It is known that human cells and tissues possess surface-bound polyanionic structures including heparan, other glycosaminoglycans, and sialic acids all of which are known to interact with factor H. The alternative pathway of complement spontaneously deposits C3b on all surfaces, host and foreign alike (4, 5). Effective control of amplification of the initial C3b must occur on host cells or tissue damage will occur. We have recently shown that the C-terminus of factor H is essential for factor H-mediated recognition and regulation of complement at cell surfaces (48). Formation of multimers of factor H bound to surface polyanions would be consistent with this previous observation and with our current data. Although our work indicates that the C-terminus may be the site of polyanion-induced self-association, these findings do not rule out participation of other polyanion-binding regions of factor H, such as CCP 6–8 (60, 68). Efforts are currently underway to examine the participation of other sites on factor H in this phenomenon as well as to determine whether factor H tetramers are, in fact, formed on host cell surfaces.
Acknowledgments
We express our appreciation to Kerry L. Wadey-Pangburn, Connie Elliott, Nicole S. Narlo and Gerald Arnett for their excellent technical assistance.
Footnotes
This research was supported by Grants DK-35081 (to M.K.P) and HL-073804 (to N.R.) from the National Institutes of Health and by Grants 0265178Y (to N.R.) and 0735101N (to V.P.F.) from the American Heart Association.
The abbreviations used are: aHUS atypical hemolytic uremic syndrome; CCP, complement control protein domain; VBS, Veronal-buffered saline; GVB, VBS containing 0.1% gelatin; GVBE, GVB containing 10 mM EDTA; NHS, normal human serum.
Disclosures
Drs. M.K. Pangburn and N. Rawal are officers of and have a financial interest in Complement Technology, Inc. (www.ComplementTech.com) a supplier of complement reagents.
References
- 1.Atkinson JP, Oglesby TJ, White D, Adams EA, Liszewski MK. Separation of self from non-self in the complement system: a role for membrane cofactor protein and decay accelerating factor. Clin. Exp. Immunol. 1991;86:27–30. doi: 10.1111/j.1365-2249.1991.tb06203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liszewski MK, Farries TC, Lublin DM, Rooney IA, Atkinson JP. Control of the complement system. Adv. Immunol. 1996;61:201–283. doi: 10.1016/s0065-2776(08)60868-8. [DOI] [PubMed] [Google Scholar]
- 3.Kirkitadze MD, Barlow PN. Structure and flexibility of the multiple domain proteins that regulate complement activation. Immunol. Rev. 2001;180:146–161. doi: 10.1034/j.1600-065x.2001.1800113.x. [DOI] [PubMed] [Google Scholar]
- 4.Pangburn MK. The alternative pathway: activation and regulation. In: Rother K, Till GO, editors. The Complement System. New York: Springer-Verlag; 1998. pp. 93–115. [Google Scholar]
- 5.Pangburn MK, Schreiber RD, Müller-Eberhard HJ. Formation of the initial C3 convertase of the alternative complement pathway. Acquisition of C3b-like activities by spontaneous hydrolysis of the putative thioester. J. Exp. Med. 1981;154:856–867. doi: 10.1084/jem.154.3.856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dervaderics M, Hidvegi T, Schmidt B, Fust G, Varga L. Ragweed allergy: correlation between skin reactivity and in vitro complement activation. Immunol. Lett. 1998;64:119–123. doi: 10.1016/s0165-2478(98)00101-1. [DOI] [PubMed] [Google Scholar]
- 7.Rodriguez de Cordoba S, Esparza-Gordillo J, Goicoechea de Jorge E, Lopez-Trascasa M, Sanchez-Corral P. The human complement factor H: functional roles, genetic variations and disease associations. Mol. Immunol. 2004;41:355–367. doi: 10.1016/j.molimm.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 8.Hageman GS, Anderson DH, Johnson LV, Hancox LS, Taiber AJ, Hardisty LI, Hageman JL, Stockman HA, Borchardt JD, Gehrs KM, Smith RJH, Silvestri G, Russell SR, Klaver CCW, Barbazetto I, Chang S, Yannuzzi LA, Barile GR, Merriam JC, Smith RT, Olsh AK, Bergeron J, Zernant J, Merriam JE, Gold B, Dean M, Allikmets R. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc. Natl. Acad. Sci. USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edwards AO, Ritter RI, Abel KJ, Manning A, Panhuysen C, Farrer LA. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 10.Klein RJ, Zeiss C, Chew EY, Tsai J-Y, Sackler RS, Haynes C, Henning AK, SanGiovanni JP, Mane SM, Mayne ST, Bracken MB, Ferris FL, Ott J, Barnstable C, Hoh J. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haines JL, Hauser MA, Schmidt S, Scott WK, Olson LM, Gallins P, Spencer KL, Kwan SY, Noureddine M, Gilbert JR, Schnetz-Boutaud N, Agarwal A, Postel EA, Pericak-Vance MA. Complement factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 12.Pangburn MK. Host recognition and target differentiation by factor H, a regulator of the alternative pathway of complement. Immunopharmacology. 2000;49:149–157. doi: 10.1016/s0162-3109(00)80300-8. [DOI] [PubMed] [Google Scholar]
- 13.Zipfel PF, Hellwage J, Friese MA, Hegasy G, Jokiranta TS, Meri S. Factor H and disease: a complement regulator affects vital body functions. Mol. Immunol. 1999;36:241–248. doi: 10.1016/s0161-5890(99)00038-3. [DOI] [PubMed] [Google Scholar]
- 14.Kavanagh D, Goodship TH, Richards A. Atypical haemolytic uraemic syndrome. Br. Med. Bull. 2006;77–78:5–22. doi: 10.1093/bmb/ldl004. [DOI] [PubMed] [Google Scholar]
- 15.Pangburn MK, Müller-Eberhard HJ. Complement C3 convertase: cell surface restriction of βIH control and generation of restriction on neuraminidase treated cells. Proc. Natl. Acad. Sci. USA. 1978;75:2416–2420. doi: 10.1073/pnas.75.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fearon DT. Regulation by membrane sialic acid of βIH-dependent decay-dissociation of amplification C3 convertase of the alternative complement pathway. Proc. Natl. Acad. Sci. USA. 1978;75:1971–1975. doi: 10.1073/pnas.75.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meri S, Pangburn MK. Discrimination between activators and nonactivators of the alternative pathway of complement: regulation via a sialic acid/polyanion binding site on factor H. Proc. Natl. Acad. Sci. USA. 1990;87:3982–3986. doi: 10.1073/pnas.87.10.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ram S, Sharma AK, Simpson SD, Gulati S, McQuillen DP, Pangburn MK, Rice PA. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J. Exp. Med. 1998;187:743–752. doi: 10.1084/jem.187.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kazatchkine MD, Fearon DT, Austen KF. Human alternative complement pathway: membrane-associated sialic acid regulates the competition between B and βIH for cell-bound C3b. J. Immunol. 1979;122:75–81. [PubMed] [Google Scholar]
- 20.Blackmore TK, Hellwage J, Sadlon TA, Higgs N, Zipfel PF, Ward HM, Gordon DL. Identification of the second heparin-binding domain in human complement factor H. J. Immunol. 1998;160:3342–3348. [PubMed] [Google Scholar]
- 21.Pangburn MK. Cutting edge: Localization of the host recognition functions of complement factor H at the carboxyl-terminal: implications for hemolytic uremic syndrome. J. Immunol. 2002;169:4702–4706. doi: 10.4049/jimmunol.169.9.4702. [DOI] [PubMed] [Google Scholar]
- 22.Hogasen K, Jansen JH, Mollnes TE, Hovdenes J, Harboe M. Hereditary porcine membranoproliferative glomerulonephritis type II is caused by factor H deficiency. J. Clin. Invest. 1995;95:1054–1061. doi: 10.1172/JCI117751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pickering M, Cook H, Warren J, Bygrave A, Moss J, Walport MJ, Botto M. Uncontrolled C3 activation causes membranoproliferative glomerulonephritis in mice deficient in complement factor H. Nat. Genet. 2002;31:424–428. doi: 10.1038/ng912. [DOI] [PubMed] [Google Scholar]
- 24.Kavanagh D, Richards A, Atkinson J. Complement regulatory genes and hemolytic uremic syndromes. Annu. Rev. Med. 2008;59:293–309. doi: 10.1146/annurev.med.59.060106.185110. [DOI] [PubMed] [Google Scholar]
- 25.Blackmore TK, Sadlon TA, Ward HM, Lublin DM, Gordon DL. Identification of a heparin binding domain in the seventh short consensus repeat of complement factor H. J. Immunol. 1996;157:5422–5427. [PubMed] [Google Scholar]
- 26.Ripoche J, Day AJ, Harris TJR, Sim RB. The complete amino acid sequence of human complement factor H. Biochem. J. 1988;249:593–602. doi: 10.1042/bj2490593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirkitadze MD, Henderson C, Price NC, Kelly SM, Mullin NP, Parkinson J, Dryden DT, Barlow PN. Central modules of the vaccinia virus complement control protein are not in extensive contact. Biochem. J. 1999;344:167–175. [PMC free article] [PubMed] [Google Scholar]
- 28.Kirkitadze MD, Krych M, Uhrin D, Dryden DT, Smith BO, Cooper A, Wang X, Hauhart RE, Atkinson JP, Barlow PN. Independently melting modules and highly structured intermodular junctions within complement receptor type 1. Biochemistry. 1999;38:7019–7031. doi: 10.1021/bi982453a. [DOI] [PubMed] [Google Scholar]
- 29.Jokiranta TS, Jaakola V-P, Lehtinen MJ, Parepalo M, Goldman A. Structure of complement factor H carboxyl-terminus reveals molecular basis of atypical haemolytic uremic syndrome. EMBO J. 2006;25:1784–1794. doi: 10.1038/sj.emboj.7601052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Discipio RG. Ultrastructures and interactions of complement factors H and I. J. Immunol. 1992;149:2592–2599. [PubMed] [Google Scholar]
- 31.Aslam M, Perkins SJ. Folded-back solution structure of monomeric factor H of human complement by synchroton X-ray and neutron scattering, analytical ultracentrifugation and constrained molecular modelling. J. Mol. Biol. 2001;309:1117–1138. doi: 10.1006/jmbi.2001.4720. [DOI] [PubMed] [Google Scholar]
- 32.Perkins SJ, Nealis AS, Sim RB. Oligomeric domain structure of human complement factor H by X-ray and neutron solution scattering. Biochemistry. 1991;30:2847–2857. doi: 10.1021/bi00225a017. [DOI] [PubMed] [Google Scholar]
- 33.Sim RB, Perkins SJ. Molecular modelling of C3 and its ligands. Curr. Top. Microbiol. 1989;153:209–222. doi: 10.1007/978-3-642-74977-3_11. [DOI] [PubMed] [Google Scholar]
- 34.Barlow PN, Steinkasserer A, Norman DG, Kieffer B, Sim RB, Campbell ID. Solution structure of a pair of complement modules by nuclear magnetic resonance. J. Mol. Biol. 1993;232:268–284. doi: 10.1006/jmbi.1993.1381. [DOI] [PubMed] [Google Scholar]
- 35.Moore MD, Discipio RG, Cooper NR, Nemerow GR. Hydrodynamic, electron microscopic, and ligand-binding analysis of the Epstein-Barr virus/C3dg receptor (CR2) J. Biol. Chem. 1989;264:20576–20582. [PubMed] [Google Scholar]
- 36.Weisman HF, Bartow T, Leppo MK, Marsh HC, Carson GR, Concino MF, Boyle MP, Roux KH, Weisfeldt ML, Fearon DT. Soluble human complement receptor type 1: in vivo inhibitor of complement suppressing post-ischemic myocardial inflammation and necrosis. Science. 1990;249:146–151. doi: 10.1126/science.2371562. [DOI] [PubMed] [Google Scholar]
- 37.Gaboriaud C, Rossi V, Bally I, Arlaud GJ, Fontecilla-Camps JC. Crystal structure of the catalytic domain of human complement C1s: a serine protease with a handle. EMBO J. 2000;19:1755–1765. doi: 10.1093/emboj/19.8.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sharma AK, Pangburn MK. Identification of three physically and functionally distinct binding sites for C3b in human complement factor H by deletion mutagenesis. Proc. Natl. Acad. Sci. USA. 1996;93:10996–11001. doi: 10.1073/pnas.93.20.10996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alsenz J, Lambris JD, Schulz TF, Dierich MP. Localization of the complement component C3b binding site and the cofactor activity for factor I in the 38 kDa tryptic fragment of factor H. Biochem. J. 1984;224:389–398. doi: 10.1042/bj2240389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kühn S, Skerka C, Zipfel PF. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H. J. Immunol. 1995;155:5663–5670. [PubMed] [Google Scholar]
- 41.Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin DM. Identification of complement regulatory domains in human factor H. J. Immunol. 1995;155:348–356. [PubMed] [Google Scholar]
- 42.Pangburn MK, Atkinson MAL, Meri S. Localization of the heparin-binding site on complement factor H. J. Biol. Chem. 1991;266:16847–16853. [PubMed] [Google Scholar]
- 43.Ormsby RJ, Jokiranta TS, Duthy TG, Griggs KM, Sadlon TA, Giannakis E, Gordon DL. Localization of the third heparin-binding site in the human complement regulator factor H. Mol. Immunol. 2006;43:1624–1632. doi: 10.1016/j.molimm.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 44.Richards A, Buddles MRH, Donne RL, Kaplan BS, Kirk E, Venning MC, Tielemans CL, Goodship JA, Goodship TH. Factor H mutations in hemolytic uremic syndrome cluster in exons 18–20, a domain important for host cell recognition. Am. J. Hum. Genet. 2001;68:485–490. doi: 10.1086/318203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bettinaglio CJ, Zipfel PF, Amadei B, Daina E, Gamba S, Skerka C, Marziliano N, Remuzzi G, Noris M. The molecular basis of familial hemolytic uremic syndrome: mutation analysis of factor H gene reveals a hot spot in short consensus repeat 20. J. Am. Soc. Nephrol. 2001;12:297–307. doi: 10.1681/ASN.V122297. [DOI] [PubMed] [Google Scholar]
- 46.Sanchez-Corral P, Perez-Caballero D, Huarte O, Simckes AM, Goicoechea E, Lopez-Trascasa M, Rodriguez de Cordoba S. Structural and functional characterization of factor H mutations associated with atypical hemolytic uremic syndrome. Am. J. Hum. Genet. 2002;71:1285–1295. doi: 10.1086/344515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perez-Caballero D, Gonzalez-Rubio C, Gallardo ME, Vera M, Lopez-Trascasa M, Rodriguez de Cordoba S, Sanchez-Corral P. Clustering of missense mutations in the C-terminal region of factor H in atypical hemolytic uremic syndrome. Am. J. Hum. Genet. 2001;68:478–484. doi: 10.1086/318201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ferreira VP, Herbert AP, Hocking HG, Barlow PN, Pangburn MK. Critical role of the C-terminal domains of factor H in regulating complement activation at cell surfaces. J. Immunol. 2006;177:6308–6316. doi: 10.4049/jimmunol.177.9.6308. [DOI] [PubMed] [Google Scholar]
- 49.Pickering MC, de Jorge EG, Martinez-Barricarte R, Recalde S, Garcia-Layana A, Rose KL, Moss J, Walport MJ, Cook HT, de C, Sr, Botto M. Spontaneous hemolytic uremic syndrome triggered by complement factor H lacking surface recognition domains. J. Exp. Med. 2007;204:1249–1256. doi: 10.1084/jem.20070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pangburn MK, Rawal N, Ferreira VP, Atkinson MAL. Polyanion-induced self-association of complement factor H: possible mechanism of host protection from an innate immune system. J. Immunol. 2007;178 doi: 10.4049/jimmunol.182.2.1061. 53.11 (abstract) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pangburn MK, Schreiber RD, Müller-Eberhard HJ. Human complement C3b inactivator: Isolation, characterization, and demonstration of an absolute requirement for the serum protein βIH for cleavage of C3b and C4b in solution. J. Exp. Med. 1977;146:257–270. doi: 10.1084/jem.146.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pangburn MK, Müller-Eberhard HJ. Kinetic and thermodynamic analysis of the control of C3b by the complement regulatory proteins factors H and I. Biochemistry. 1983;22:178–185. doi: 10.1021/bi00270a026. [DOI] [PubMed] [Google Scholar]
- 53.Sim RB, Discipio RG. Purification and structural studies on the complement system control protein βIH (factor H) Biochem. J. 1982;205:285–293. doi: 10.1042/bj2050285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nagaki K, Iida K, Okubo M, Inai S. Reaction mechanims of β1H globulin. Int. Arch. Allergy Appl. Immunol. 1978;57:221–232. doi: 10.1159/000232106. [DOI] [PubMed] [Google Scholar]
- 55.Hakobyan S, Harris CL, Tortajada A, Goicochea de JE, Garcia-Layana A, Fernandez-Robredo P, Rodriguez de CS, Morgan BP. Measurement of factor H variants in plasma using variant-specific monoclonal antibodies: application to assessing risk of age-related macular degeneration. Invest Ophthalmol. Vis. Sci. 2008;49:1983–1990. doi: 10.1167/iovs.07-1523. [DOI] [PubMed] [Google Scholar]
- 56.Perkins SJ. Protein volumes and hydration effects: the calculation of partial specific volumes, neutron scattering matchpoints and 280 nm absorption coefficients for proteins and glycoproteins from amino acid sequences. Eur. J. Biochem. 1986;157:169–180. doi: 10.1111/j.1432-1033.1986.tb09653.x. [DOI] [PubMed] [Google Scholar]
- 57.Alam MN, Haque A, Sreedhar M, Pangburn MK. A novel vector for the expression of SCR domains in insect cells. J. Immunol. Methods. 2004;293:107–113. doi: 10.1016/j.jim.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 58.Whaley K, Ruddy S. Modulation of the alternative complement pathway by βIH globulin. J. Exp. Med. 1976;144:1147–1163. doi: 10.1084/jem.144.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Okemefuna AI, Gilbert HE, Griggs KM, Ormsby RJ, Gordon DL, Perkins SJ. The regulatory SCR-1/5 and cell surface-binding SCR-16/20 fragments of factor H reveal partially folded-back solution structures and different self-associative properties. J. Mol. Biol. 2008;375:80–101. doi: 10.1016/j.jmb.2007.09.026. [DOI] [PubMed] [Google Scholar]
- 60.Nan R, Gor J, Perkins SJ. Implications of the progressive self-association of wild-type human factor H for complement regulation and disease. J. Mol. Biol. 2008;375:891–900. doi: 10.1016/j.jmb.2007.11.015. [DOI] [PubMed] [Google Scholar]
- 61.Jokiranta TS, Hellwage J, Koistinen V, Zipfel PF, Meri S. Each of the three binding sites on factor H interacts with a distinct site on C3b. J. Biol. Chem. 2000;275:27657–27662. doi: 10.1074/jbc.M002903200. [DOI] [PubMed] [Google Scholar]
- 62.Pangburn MK, Pangburn KLW, Koistinen V, Meri S, Sharma AK. Molecular mechanisms of target recognition in an innate immune system: interactions among factor H, C3b and target in the alternative pathway of human complement. J. Immunol. 2000;164:4742–4751. doi: 10.4049/jimmunol.164.9.4742. [DOI] [PubMed] [Google Scholar]
- 63.Fischetti VA. Streptococcal M protein. Scientific American. 1991;264:58–65. doi: 10.1038/scientificamerican0691-58. [DOI] [PubMed] [Google Scholar]
- 64.Herbert AP, Uhrin D, Lyon M, Pangburn MK, Barlow PN. Disease-associated sequence variations congregate in a polyanion-recognition patch on human factor H revealed in 3D structure. J. Biol. Chem. 2006;281:16512–16520. doi: 10.1074/jbc.M513611200. [DOI] [PubMed] [Google Scholar]
- 65.Pichette V, Querin S, Schurch W, Brun G, Lehner-Netsch G, Delage JM. Familial hemolytic-uremic syndrome and homozygous factor H deficiency. Am. J. Kidney Disease. 1994;24:936–943. doi: 10.1016/s0272-6386(12)81065-1. [DOI] [PubMed] [Google Scholar]
- 66.Meri S, Pangburn MK. Regulation of alternative pathway complement activation by glycosaminoglycans: specificity of the polyanion binding site on factor H. Biochem. Biophys. Res. Commun. 1994;198:52–59. doi: 10.1006/bbrc.1994.1008. [DOI] [PubMed] [Google Scholar]
- 67.Jokiranta TS, Zipfel PF, Hakulinen J, Kuhn S, Pangburn MK, Tamerius JD, Meri S. Analysis of the recognition mechanism of the alternative pathway of complement by monoclonal anti-factor H antibodies: evidence for multiple interactions between H and surface bound C3b. FEBS Lett. 1996;393:297–302. doi: 10.1016/0014-5793(96)00905-2. [DOI] [PubMed] [Google Scholar]
- 68.Fernando AN, Furtado PB, Clark SJ, Gilbert HE, Day AJ, Sim RB, Perkins SJ. Associative and structural properties of the region of complement factor H encompassing the Tyr402His disease-related polymorphism and its interactions with heparin. J. Mol. Biol. 2007;368:564–581. doi: 10.1016/j.jmb.2007.02.038. [DOI] [PubMed] [Google Scholar]