Summary
Some human Plasmodium falciparum parasites, but not others, cause malaria in Aotus monkeys. To identify the basis for this variation, we crossed two clones that differ in A. nancymaae virulence and mapped inherited traits of infectivity to erythrocyte invasion by linkage analysis. A major pathway of invasion was linked to polymorphisms in a newly-identified erythrocyte binding protein, PfRH5, found in the apical region of merozoites. Polymorphisms of PfRH5 from the A. nancymaae-virulent parent (GB4) transformed the non-virulent parent (7G8) to a virulent parasite. Conversely, replacements that removed these polymorphisms from PfRH5 converted a virulent progeny clone (LC12) to a non-virulent parasite. A proteolytic fragment of PfRH5 from the infective parasites bound to A. nancymaae erythrocytes. Our results also suggest that PfRH5 is a parasite ligand for human infection, and that amino acid substitutions can cause its binding domain to recognize different human erythrocyte surface receptors.
Introduction
Clinical manifestations of malaria arise from infection of erythrocytes in the human bloodstream and the effects of these parasitized erythrocytes on critical host tissues and organs. Plasmodium falciparum, the greatest killer among the human malaria parasite species, is known for its ability to attack different organs with life-threatening conditions such as cerebral compromise and coma, severe anemia, pulmonary malaria, acidosis, renal failure, algid forms with shock and disseminated intravascular coagulation, and severe pathological effects on the mother and fetus in the malaria of pregnancy. Yet, much remains unclear about the processes by which the parasites enter the host erythrocytes and subsequently interact with host systems to cause these conditions.
P. falciparum parasites do not cause malaria in the majority of non-human primates. Limited episodes of malaria occur in splenectomized chimpanzees and, among monkeys, certain New World species are known to be susceptible to some P. falciparum parasites. Indeed, Aotus, the owl or night monkey, and Saimiri, the squirrel monkey, were frequently used to maintain P. falciparum lines prior to the advent of in vitro culture (Geiman and Meagher, 1967; Trager and Jensen, 1976); today these species are employed in drug and vaccine testing programs (Collins, 1994). However, only some P. falciparum lines infect Aotus and Saimiri whereas other lines do not, and the features of monkey malaria observed with these species can differ substantially from those of human P. falciparum malaria (Young et al., 1975; Heppner et al., 2001).
Malaria parasites can establish disease in the vertebrate host only if they are able to efficiently invade erythrocytes. In molecular terms, invasion requires a complex series of steps initiated by parasite recognition and attachment to the host erythrocyte membrane (Aikawa et al., 1978; Dvorak et al., 1975). Individual species and even strains of a particular species vary in the host receptors they use for these steps. P. falciparum parasites exhibit notable diversity in this regard, as they employ a number of alternative parasite ligand – host receptor combinations to invade human erythrocytes by different invasion pathways (reviewed by Iyer et al., 2007). Such ability to invade by various molecular pathways is likely an adaptation to human erythrocyte polymorphisms that have arisen with malaria in human evolution (Duraisingh et al., 2003; Mitchell et al., 1986; Perkins and Holt, 1988). Some P. falciparum parasites can switch to different pathways of invasion under selection pressure. For example, the Dd2 clone of P. falciparum undergoes an adaptive switch to non-sialic acid dependence when presented with neuraminidase-treated erythrocytes (Dolan et al., 1990); this phenotype change is associated with upregulation of PfRH4, a member of the reticulocyte-binding protein family (Gaur et al., 2006; Stubbs et al., 2005). In other studies, single amino acid changes in two P. falciparum ligands, JESEBL/EBA181 and BAEBL/EBA140, have each been shown to alter host receptor recognition and produce different erythrocyte binding patterns (Mayer et al., 2002; Mayer et al., 2004).
Ligand and receptor differences may also be involved in variation of P. falciparum infectivity to Aotus. One ligand with a possible role in the invasion of Aotus erythrocytes is PfEBA175, a binding protein that recognizes sialic acid residues of glycophorin A on the surface of human erythrocytes (Camus and Hadley, 1985; Sim et al., 1994). Studies have shown that erythrocyte sialic acid residues occur predominantly as N-glycolylneuraminic acid (Neu5Gc) in chimpanzees and Old World monkeys, whereas in humans and New World Aotus monkeys these residues occur only as N-acetylneuraminic acid (Neu5Ac), a precursor of Neu5Gc (Martin et al., 2005). Yet, despite the presence of correct sialic acid residues on Aotus erythrocytes and the ability of P. falciparum to switch to alternative invasion pathways, many P. falciparum lines are unable to infect these New World monkeys (Collins et al., 1997; Kaneko et al., 1999; Young et al., 1975).
In this report, we describe results from a genetic cross between a P. falciparum clone unable to infect Aotus nancymaae and a clone highly virulent to these monkeys. Linkage analysis of the progeny and DNA transfection studies demonstrate a key role of polymorphisms in a newly identified erythrocyte binding protein, PfRH5, for P. falciparum invasion of A. nancymaae erythrocytes. Our evidence also indicates that the PfRH5 protein containing these polymorphisms can bind to alternative receptors on human erythrocytes.
RESULTS
The 7G8×GB4 Cross
The GB4 parent of the cross is a clone from P. falciparum Ghana III (a gift from Bill Collins), a parasite line that produces highly virulent infections in various Aotus species both by mosquito-transmitted sporozoites and by direct inoculation of erythrocyte-stage parasites (Sullivan et al., 2003). Robust and highly reproducible infections were obtained with GB4 in A. nancymaae (Figure 1A). In contrast, the 7G8 clone from the Brazil isolate IMTM 22 (Burkot et al., 1984) did not infect A. nancymaae monkeys on repeated attempts.
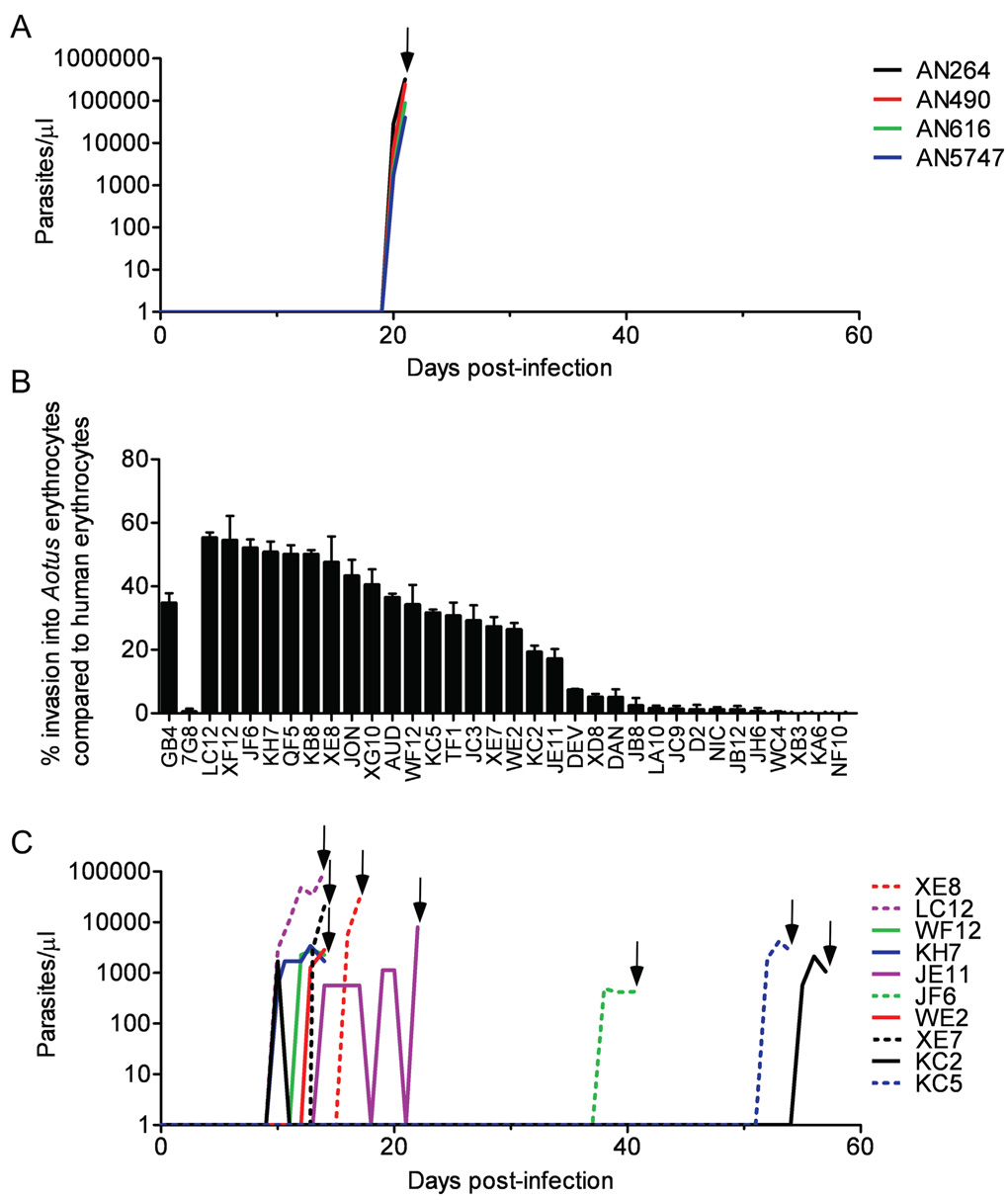
FIGURE 1. P. falciparum parasite infectivities to Aotus monkeys and to their erythrocytes in vitro.
A) Parasitemias obtained in four A. nancymaae monkeys after intravenous inoculation of 1×107 GB4-parasitized erythrocytes. All infections were cured with 25 mg/kg mefloquine (arrow). B) A. nancymaae erythrocyte invasion rates by the parents and 32 independent recombinant progeny from the P. falciparum 7G8×GB4 cross. Percentage values are relative to rates of invasion into human control erythrocytes ± SEM. C) Infectivity of 10 7G8×GB4 progeny clones to A. nancymaae monkeys. All infections were cured with 25 mg/kg mefloquine (arrows).
To cross 7G8 and GB4, we induced gametocytes from each parent in vitro, combined these sexual stages in equal numbers and fed them to about 1000 Anopheles freeborni and Anopheles gambiae mosquitoes. Tests of individual oocysts from the mosquito midguts on day 11 confirmed genetic recombination (data not shown). On day 15, approximately 300 infectious mosquitoes were fed on a splenectomized chimpanzee. Samples of parasitemic blood were collected from the chimpanzee on days 13, 16, 18, 19, 20 and 21 after mosquito feeding. Of more than 200 individual parasites cloned from these samples and typed by microsatellite analysis, we identified 28 progeny clones from 7G8 self-fertilization events, a single clone from GB4 selfing, and 32 unique haploid recombinant progeny clones (independent recombinants) for phenotype assessment and genetic linkage analysis.
Erythrocyte Invasion and A. nancymaae Infectivity by 7G8×GB4 Progeny Clones
In vitro, the 7G8 parent showed no invasion of A. nancymaae erythrocytes, consistent with its inability to infect the monkeys, whereas invasion by the virulent GB4 parent proved to be robust, 35% that of human erythrocytes (Figure 1B). The 32 independent recombinant progeny from the 7G8×GB4 cross varied in their ability to invade A. nancymaae erythrocytes. Some progeny were similar to the 7G8 parent, exhibiting little or no invasion of A. nancymaae erythrocytes. Other progeny invaded A. nancymaae erythrocytes at rates ranging from approximately 0.5× to 1.6× that of the GB4 parent (17% to 55% of the control rates in human erythrocytes; Figure 1B).
We also investigated the ability of 14 of the 32 progeny clones to infect splenectomized A. nancymaae monkeys. JB8, a progeny clone that had inherited the 7G8 parent’s poor ability to invade A. nancymaae erythrocytes in vitro, failed to produce an observable infection with an inoculum of 1×107 parasites. Among the 13 remaining progeny, all of which were able to invade A. nancymaae erythrocytes in vitro, 10 clones produced patent monkey infections with marked differences in pre-patent periods: some progeny blood stages were detected within 10 days while others required 30–60 days to reach detectable parasitemia (Figure 1C). Clone KC2 gave an early peak that spontaneously remitted and was followed by a later peak before treatment. Clones KB8, JC3 and XF12 failed to produce detectable infections despite their ability to invade A. nancymaae erythrocytes in vitro (KB8 and JC3 were each tried in two animals and XF12 in one animal). This variability among the progeny was in contrast to the consistent appearance of parasitemia from the GB4 parent 19–20 days after inoculation in four different A. nancymaae (Figure 1A), suggesting that multiple inherited factors in the 7G8×GB4 progeny affect both virulence and time to parasitemia in A. nancymaae.
A Key Locus of A. nancymaae Erythrocyte Invasion Maps to P. falciparum Chromosome 4
We identified 285 polymorphic microsatellite markers that distinguish the 7G8 and GB4 parents across all 14 P. falciparum nuclear chromosomes and determined the inheritance of these markers in the 32 independent recombinant progeny. Recombination among these markers in the progeny indicated an average linkage separation of 2.2 cM and a map coverage of 628 cM over 14 linkage groups (corresponding to an average map unit distance of 36 kb/cM). Most genetic markers exhibited an approximately equal distribution of parental alleles in accord with random Mendelian inheritance (Figure 2A). However, highly skewed ratios of 7G8 and GB4 inheritance were evident in some regions, particularly at one end of chromosome 7, where every independent recombinant carried markers of this chromosome region from the 7G8 parent (Figure 2A, Table S1).
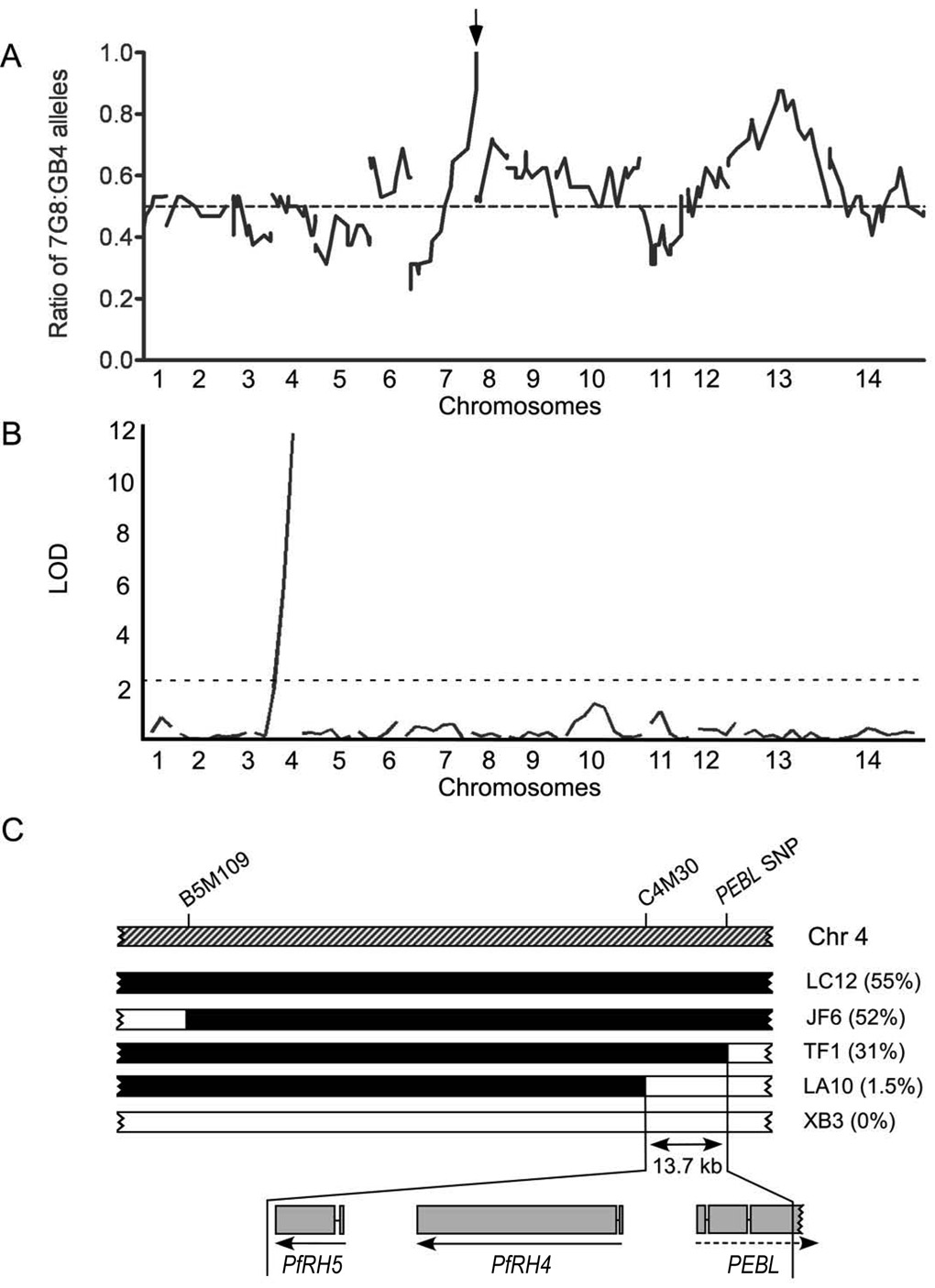
FIGURE 2. Identification of candidate genes for the A. nancymaae invasion phenotype.
A) Genome-wide plot of the ratios of 7G8:GB4 markers among the 32 independent recombinants. A peak containing PfEBA175 (arrow) identifies a region of chromosome 7 that is severely skewed in favor of the 7G8 parent. A dashed line indicates the 0.5 ratio expected from random assortment. B) Logarithm of odds (LOD) scores from the primary scan for quantitative trait loci (QTL) associated with A. nancymaae erythrocyte invasion. The peak with LOD of 11.8 maps to the sub-telomeric region of chromosome 4. A dashed line represents the significance threshold (0.05). C) Map of the chromosome region linked to A. nancymaae invasion phenotype. A. nancymaae erythrocyte invasion phenotypes (%’s in brackets) are shown for five progeny clones carrying different chromosome segments from each of the parents in the mapped region (chr 4, chromosome 4; solid black segments, GB4; open segments, 7G8). The Aotus invasion locus maps to a 13.7 kb sequence defined by the microsatellite marker C4M30 and a single nucleotide polymorphism (SNP) in the pseudogene PEBL. The exons and transcription directions of the PfRH4 and PfRH5 genes are indicated by connected gray bars and arrows, respectively, while the reading frame direction of the pseudogene PEBL is shown with a dashed arrow. Comparisons of the genomic and cDNA sequences of the 7G8 PfRH5 gene, including 5'- and 3'-rapid amplification of cDNA ends (RACE) sequencing, confirmed the two exon structure documented in PlasmoDB (www.plasmodb.org).
Associations between the invasion rates shown in Figure 1B and the markers from complete microsatellite maps of the 32 progeny were assessed by quantitative trait loci (QTL) analysis. Results from a primary scan identified a major locus for A. nancymaae erythrocyte invasion near the telomere of chromosome 4 with a log of odds (LOD) score of 11.8 (Figure 2B). A secondary scan to search for residual effects did not identify additional peaks with statistical significance (Figure S1).
We defined the boundaries of the principal QTL invasion locus more precisely with progeny clones carrying recombinant crossovers in the region of the peak from chromosome 4. One of these progeny clones, LA10, showed little or no invasion of A. nancymaae erythrocytes, indicating a position for the locus distal (right) of marker C4M30 on chromosome 4 (Figure 2C). Another clone, TF1, showed an A. nancymaae erythrocyte invasion rate comparable to that of the GB4 parent, indicating a locus position internal to a SNP (single nucleotide polymorphism) in the pseudogene PEBL (Figure 2C) (Triglia et al., 2001). Together, these results mapped the locus within a 13.7 kb region of chromosome 4. This region contains two predicted genes, PfRH4 (PFD1150c) and PfRH5 (PFD1145c), both of which are members of the reticulocyte-binding protein homolog (RH) family (Cowman and Crabb, 2006).
Distinct Coding Polymorphisms Occur in the Aotus-virulence Locus of GB4 Parasites
To evaluate potential roles of PfRH4 and PfRH5 in A. nancymaae erythrocyte invasion, we searched for an association of mRNA transcription levels or sequence polymorphisms with invasion by P. falciparum parasites from geographically distant regions with distinct genetic backgrounds (Wootton et al., 2002). No significant differences in the transcript levels of PfRH4 or PfRH5 were detected in synchronized 7G8 and GB4 schizont-stage parasites by microarray analysis (Figure S2). Further, subclones of the Dd2 parasite that express both PfRH4 and PEBL at upregulated levels (Dd2/Nm) (Gaur et al., 2006) showed no change from Dd2’s poor ability to invade A. nancymaae erythrocytes in vitro (data not shown).
Comparison of the 7G8 and GB4 sequences identified two unique non-synonymous polymorphisms in the GB4 PfRH5 open reading frame (Table 1), whereas a single codon polymorphism in PfRH4 (encoding Q438 in GB4 vs. K438 in 7G8) was found in other parasites whether or not they were invasive to A. nancymaae erythrocytes (data not shown). In light of these observations, we decided to focus on PfRH5 as a possible determinant of A. nancymaae infectivity in the 7G8×GB4 cross.
TABLE 1.
Position of amino acid polymorphisms in PfRH5 from parasites with various A. nancymaae erythrocyte invasion phenotypes.
| Parasite | 48 | 197 | 203 | 204 | 347 | 358 | 362 | 407 | 410 | 429 |
A. nancymaae invasion (% ± SEM)a |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3D7 | E | S | C | I | N | Y | E | I | I | K | 0 ± 0.3 |
| Tm284 | E | S | C | I | N | Y | E | I | M | K | 0 ± 0.7 |
| 7G8 | E | S | Y | I | N | Y | E | I | I | K | 0.4 ± 0.2 |
| Haiti | E | S | Y | I | N | Y | E | I | I | K | 0.4 ± 1.0 |
| Mont S1b | E | Y | Y | I | N | Y | E | I | I | K | 1.9 ± 0.0 |
| JAV | E | S | Y | I | N | Y | E | I | I | K | 2.4 ± 2.0 |
| Ecu110 | E | S | Y | I | N | Y | E | I | I | K | 3.0 ± 1.6 |
| ECP | E | S | Y | I | N | Y | E | I | I | K | 3.7 ± 2.1 |
| Dd2 | E | S | C | I | N | Y | E | I | M | K | 3.4 ± 3.4 |
| Liberia II | E | S | Y | I | N | Y | E | I | I | K | 3.8 ± 2.6 |
| D10 | E | S | Y | I | N | Y | E | I | I | K | 4.7 ± 2.2 |
| HB3 | E | S | Y | I | N | Y | E | I | I | K | 4.8 ± 0.2 |
| Mal Camp | K | S | Y | I | Y | Y | E | I | I | N | 22.6 ± 5.9 |
| Genevec | E | Y | Y | I | N | F | E | I | I | K | 26.4 ± 0.8 |
| GB4d,e | E | S | Y | K | N | Y | E | V | I | K | 34.7 ± 3.2 |
| Santa Lucia | E | S | Y | I | D | Y | E | I | I | K | 46.5 ± 8.0 |
| Palo Alto | K | S | Y | R | N | Y | E | I | I | N | 46.8 ± 3.3 |
| FVO | E | Y | Y | I | N | F | D | I | I | K | 58.5 ± 1.8 |
Merozoite invasion rates of A. nancymaae erythrocytes are shown as a percent of the invasion into human erythrocytes ± SEM.
Infects A. lemurinus griseimembra (Collins et al., 1997).
Grows in Saimiri sciureus (Fajfar-Whetstone et al., 1987).
Previously shown to infect A. lemurinus griseimembra, A. nancymaae and A. vociferans (Sullivan et al., 2003).
In addition to open reading frame polymorphisms, PfRH5 in GB4 contains a microsatellite TA-repeat in its intron that is 17 nucleotides shorter than in 7G8.
Allelic modifications of PfRH5 alter the ability of P. falciparum to invade A. ancymaae erythrocytes
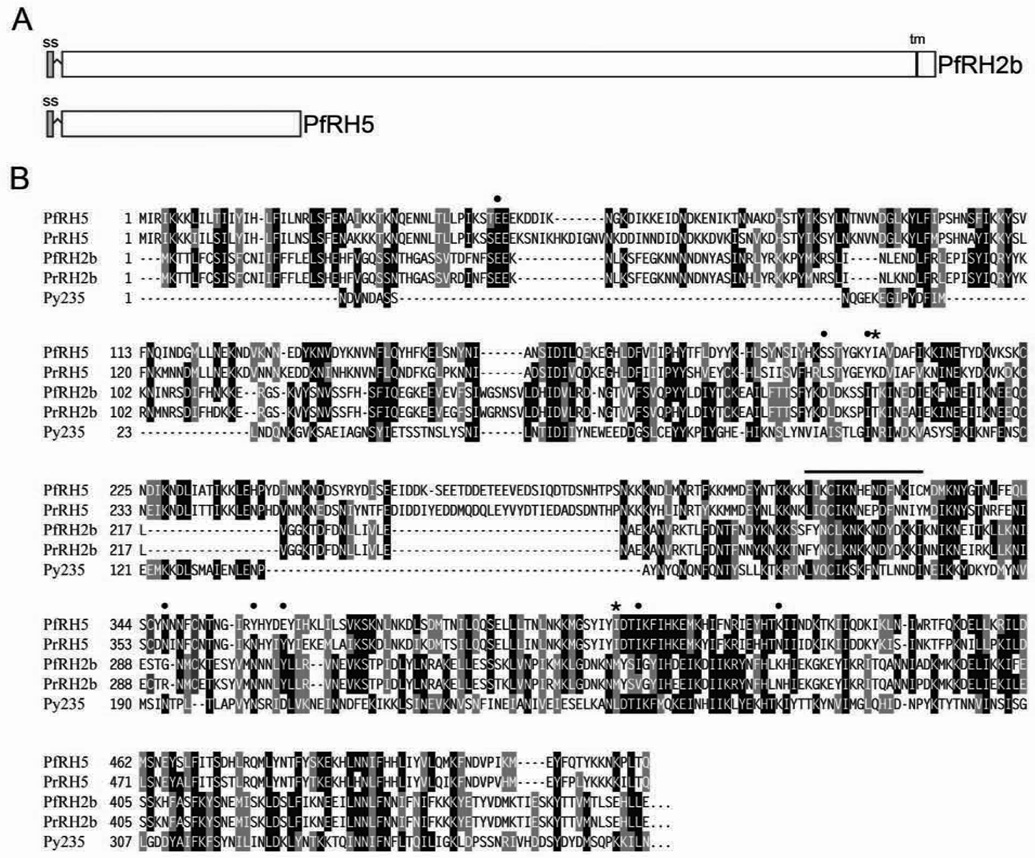
The PfRH5 gene occurs in a sub-telomeric region of P. falciparum chromosome 4 that lacks synteny with any of the rodent malaria parasite chromosomes and contains species-specific genes likely to be involved in host-parasite interactions (Kooij et al., 2005). Our genome database searches identified an ortholog of PfRH5 only in the P. falciparum-like chimpanzee parasite P. reichenowi; we detected no orthologs in the human P. vivax parasite, the monkey parasite P. knowlesi, or the rodent parasites P. berghei and P. yoelii. PfRH5 is nevertheless conserved among P. falciparum lines and, as a member of the RH superfamily, it aligns with high similarity to the amino terminal regions of PfRH2a and PfRH2b (much larger 370 kDa and 383 kDa members of the RH family, respectively), the N-terminal regions of the P. reichenowi PrRH2b and the P. yoelii 235 proteins (Ogun and Holder, 1996; Rayner et al., 2000; 2004) (Figure 3).
FIGURE 3. Schematic representation and sequence alignment of PfRH5 with members of the RH superfamily.
A) Organization of PfRH5 and PfRH2b. ss, putative signal sequence (gray box); tm, transmembrane region (black box). PfRH5 has 526 predicted amino acids and an estimated molecular weight of 63 kDa. B) Alignments of predicted RH5 and homologous N-terminal RH sequences from P. falciparum, P. reichenowi and P. yoelii. Conserved residues are shaded black and semi-conserved regions shaded grey. PfRH5 carries cysteine residues (purple highlight) at four of the five positions where there are conserved or semi-conserved cysteines in the other sequences (185, 224, 317, 345, 351). Amino acids 1–24 of PfRH5 constitute a possible signal peptide (SignalP 3.0 program; Bendtsen et al., 2004). Asterisks mark positions 204 and 407 where polymorphisms occur between the 7G8 and GB4 clones; positions of differences in other isolates (Table 1) are marked with closed circles. Lysine occurs in position 204 of PfRH5 in GB4, but not in other P. falciparum lines of our survey (Table 1); this residue is also encoded at the equivalent codon position in the P. reichenowi PrRH5 sequence and may represent a persisting ancestral codon. Amino acids of the synthetic MAP4 peptide are overlined. Sequences: PfRH5, (7G8 clone, accession number EU433391), PfRH2b (amino acids 1–375, AF312917), PrRH2b (1–473, Y166677), PrRH5 (EF142858), Py235 (partial sequence, 1–464, XM_719883); Pf, P. falciparum; Pr, P. reichenowi, Py, P. yoelii. Sequence homology searches were performed using tblastn and alignments were generated with T-Coffee software (Notredame et al., 2000).
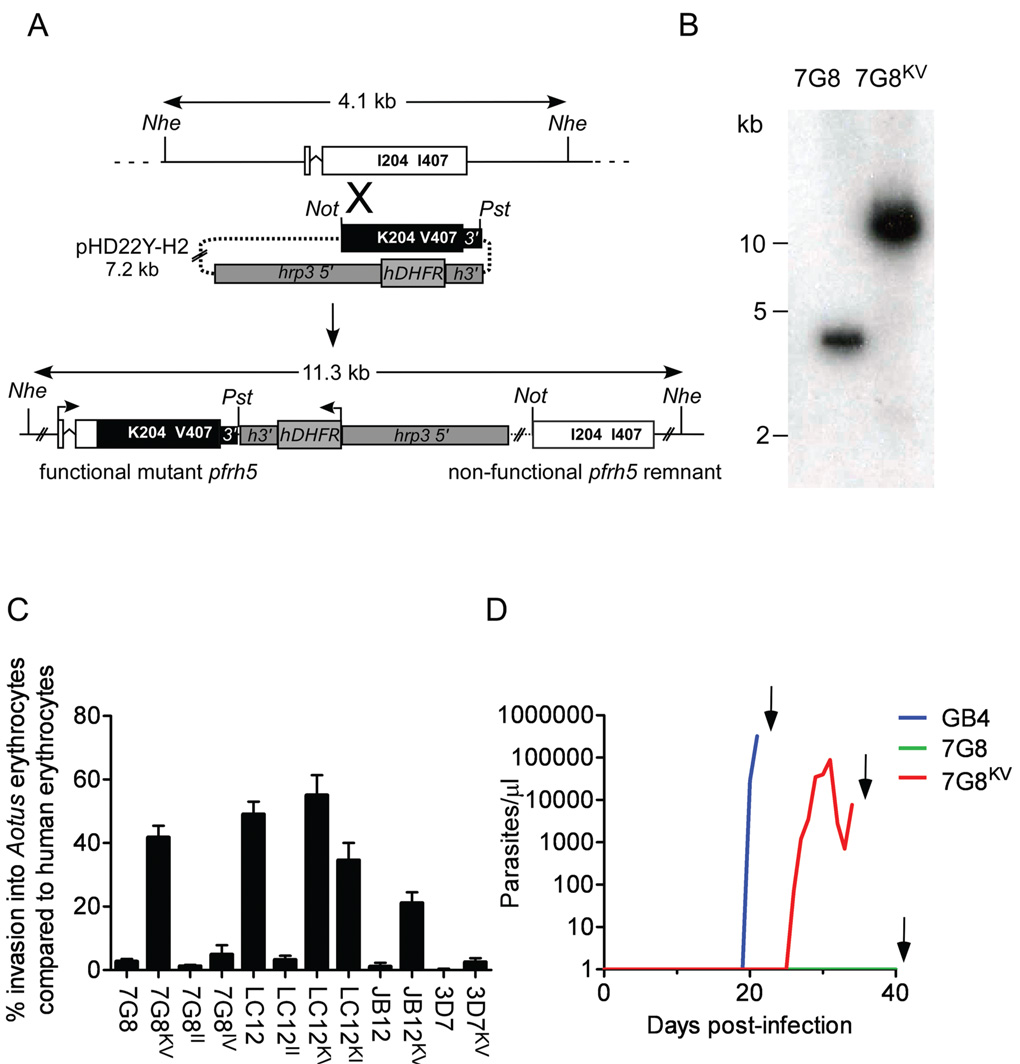
To directly evaluate the role of amino acid substitutions in PfRH5 in A. nancymaae erythrocyte invasion, we modified the 7G8 PfRH5 sequence to contain the I204K and I407V codons of the GB4 parasite. Following single site crossover and homologous integration of a transfected episome carrying the GB4 sequence, modified 7G8 parasites were obtained that carry a full-length version of GB4 PfRH5 upstream of the drug selection cassette and a 3’ non-translated remnant of the original chromosomal 7G8 PfRH5 sequence (Figure 4A). The appropriate allelic exchange in this line, 7G8KV, was confirmed by DNA restriction and Southern blotting (Figure 4B).
FIGURE 4. Allelic modification of PfRH5 and confirmation of its role in erythrocyte invasion.
A) Schematic of the strategy to generate a 7G8KV parasite by homologous recombination. A portion of the GB4 PfRH5 gene containing the K204 and V407 codons along with 0.2 kb of 3’ flanking sequence replaces its counterpart in the expressed endogenous allele. The hDHFR (human dihydrofolate reductase) drug selection marker is located downstream of the modified PfRH5 allele. Abbreviations: 3’, 3’ UTR of PfRH5; hrp3 5’, promoter region from P. falciparum histidine-rich protein 3; h3’, 3’ UTR of P. falciparum histidine-rich protein 2; Nhe, Nhe I; Not, Not I; Pst, Pst I. B) Southern blot verification of the 11.3 kb Nhe I band resulting from homologous integration in the PfRH5 locus. Genomic DNAs were isolated from untransformed 7G8 parasites and from transformed 7G8KV parasites containing the GB4 PfRH5 allele. C) Invasion rates of untransformed and transformed P. falciparum lines into A. nancymaae erythrocytes ± SEM. D) Results in three naïve A. nancymaae monkeys after intravenous inoculation of bloodstream forms of GB4 (blue line), 7G8 (green line) or PfRH5-modified 7G8KV parasites (red line). All animals were splenectomized prior to infection and cured with 25 mg/kg mefloquine (arrows).
In assays with A. nancymaae erythrocytes, 7G8KV parasites proved to have converted from the non-invasion 7G8 phenotype to invasion rates comparable to or better than the GB4 parent (42% of the control rates in human erythrocytes, Figure 4C; compare Figure 1B). Independent transformation of 7G8 parasites with the GB4 allele on a second occasion yielded similar results (data not shown). Control experiments, in which the transfection plasmid containing the native 7G8 PfRH5 sequence was inserted by single site crossover into the chromosomal copy of 7G8 parasites (7G8II control), showed no significant change from the non-invasion phenotype (Figure 4C).
We performed further experiments to transform and test two progeny clones: LC12 that carries the GB4 PfRH5 allele and invades A. nancymaae erythrocytes with high efficiency; and JB12 that carries the 7G8 PfRH5 allele and does not invade A. nancymaae erythrocytes (Figure 1B). As expected, insertion of the transfection plasmid containing the GB4 PfRH5 allele into the progeny clone LC12 (generating control LC12KV parasites) produced no significant change in A. nancymaae red cell invasion, whereas replacement of the LC12 PfRH5 gene with the 7G8 allele resulted in a dramatic decrease of invasion efficiency (LC12II transformants; Figure 4C). In experiments with the non-invasive progeny clone JB12, we modified the PfRH5 allele inherited from the 7G8 parent so that it contained the GB4 codons K204 and V407. The resulting parasites, JB12KV, showed conversion to an invasion phenotype, which we measured as an increase from zero to about two-thirds of the ability of transformed 7G8KV parasites to enter A. nancymaae erythrocytes (Figure 4C).
In these allelic exchange experiments, we also found that allelic crossovers occurred between the codons at positions 204 and 407 so that alternative combinations of parental PfRH5 polymorphisms were obtained. 7G8 transformants lacking a change to codon K204 but containing codon V407 (7G8IV) showed little change from the baseline non-invasion phenotype, suggesting a critical role for the K204 residue in A. nancymaae invasion by 7G8KV parasites (Figure 4C). LC12 transformants containing codon K204 but lacking codon V407 (LC12KI) retained substantial invasion rates, again consistent with an important effect of the charged K204 residue on A. nancymaae invasion relative to much less influence from the conservative I→V substitution at position 407 (Figure 4C).
We performed parallel experiments to replace the endogenous PfRH5 allele in a P. falciparum clone, 3D7, which is unrelated to parasites of the 7G8×GB4 cross. In three separate transformation experiments, recovered parasites carrying the GB4 allele (3D7KV) showed little change from their original non-invasion phenotype (Figure 4C). A possible explanation for this result is that, while PfRH5 is a major determinant of A. nancymaae virulence in the 7G8×GB4 cross, different P. falciparum parasites may require manipulation of other determinants for A. nancymaae invasion.
Table 1 shows the distribution of non-synonymous polymorphisms in 7G8, GB4 and 16 additional parasite isolates with different infectivities to New World monkeys and their erythrocytes. Among the various polymorphisms, amino acid class differences stand out among the parasites able to invade A. nancymaae erythrocytes with efficiencies > 20%. These include the I204K/I407V substitutions in the GB4 parasite, the S197Y/Y358F substitutions of the Geneve parasite, the E48K/N347Y/K429N substitutions of Malayan Camp, the E48K/I204R/K429N substitutions in Palo Alto, the S197Y/Y358F/E362D substitutions in FVO and the single N347D substitution of the Santa Lucia parasite (Table 1). Possible involvement of these different changes in monkey erythrocyte invasion by the PfRH5-mediated pathway remains to be examined.
Transformed 7G8KV parasites are virulent to A. nancymaae
The ability of 7G8KV transformants to efficiently invade A. nancymaae erythrocytes in vitro suggested they could also infect the monkeys in vivo. To test this possibility, we challenged each of six A. nancymaae monkeys with an inoculum of 1 × 107 7G8KV parasites. A successful infection was detected in one animal on day 26, and the parasitemia was monitored until day 34 when it was drug cured (Figure 4D). Parasite samples recovered from this infection all expressed the I204K and I407V substitutions of the introduced GB4 allele, despite the absence of WR99210 drug pressure for >30 days (data not shown). The remaining five animals remained parasite negative until the 90 day limit of the experiment, at which time they were treated with mefloquine and retired from the study.
Substitution I204K directs PfRH5 binding to neuraminidase-sensitive receptors on Aotus and human erythrocytes
To investigate the erythrocyte binding and cellular localization of PfRH5 protein, we generated and validated rabbit antisera against a peptide from the PfRH5 sequence (predicted amino acids 314–329; overlined in Figure 3B). These antisera specifically detected single bands from Escherichia coli expressing transfected subsequences of PfRH5, but not from control E. coli cells (Mr 16K or 28K bands, Figure 5A). When tested against extracts of parasitized erythrocytes, the antisera detected a protein band with a Mr of 67K (Figure 5B), in the range expected from the predicted molecular weight of PfRH5 (~63 kDa). This band was present in preparations of erythrocytes parasitized by schizonts but not by rings or early trophozoites, in agreement with published PfRH5 expression data (Le Roch et al., 2004).
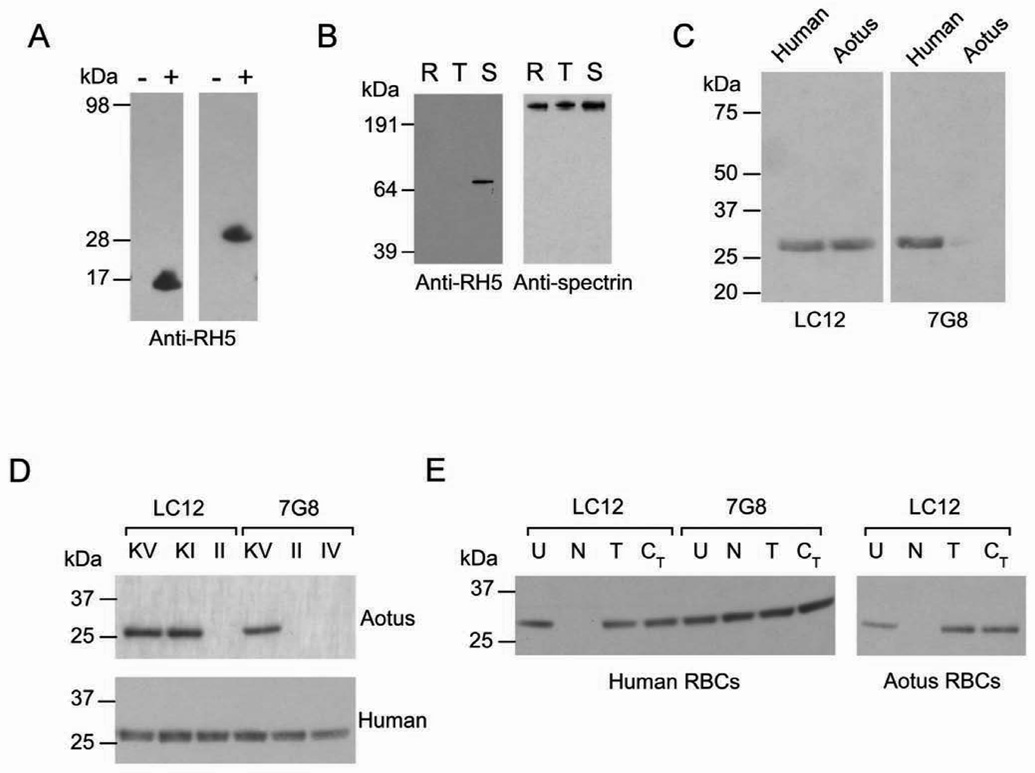
FIGURE 5. Immunoblot detection of PfRH5 and binding of a Mr 28K fragment to human and A. nancymaae erythrocytes.
A) Rabbit antisera raised against a 16 amino acid peptide recognize 16 and 28 kDa fragments of PfRH5 in Escherichia coli. Polypeptides of the appropriate size are present in induced (+) but not uninduced (−) cells. B) Immunoblot analysis of 7G8 parasite extracts from synchronized ring stages (R), early trophozoites (T) and mature schizonts (S). A Mr 67K band is evident in the 7G8 schizont preparation probed by anti-PfRH5 peptide antisera. The signals from repeat probing of the blot with antibodies against human spectrin, an abundant red cell protein present in parasite extracts, verify that protein from comparable numbers of cells was present in each lane. C) The Mr 28K PfRH5 fragment of LC12 parasites binds to A. nancymaae as well as human erythrocytes, consistent with the ability of LC12 to infect both types of cells, while the PfRH5 fragment from 7G8 parent parasites only binds human erythrocytes. D) The Mr 28K PfRH5 fragment from parasites transformed to express K204 (LC12KV, LC12KI, 7G8KV), but not I204 (LC12II, 7G8II, 7G8IV) binds to A. nancymaae erythrocytes. Human erythrocytes bind the PfRH5 fragment from all parasites. E) The human erythrocyte receptor for the PfRH5 fragment of 7G8 parasites is neuraminidase-, trypsin-and chymotrypsin-resistant, while both the human and A. nancymaae erythrocyte receptor for the PfRH5 fragment of LC12 parasites is trypsin- and chymotrypsin-resistant, but neuraminidase-sensitive. U, untreated; N, neuraminidase; T, trypsin; CT, chymotrypsin.
We tested for associations of parasite infectivity with PfRH5 attachment to erythrocytes by standard binding assays with parasite culture supernatants (Camus and Hadley, 1985; Gaur et al., 2007). Salt-wash eluates from human erythrocytes that had been incubated with 7G8 or LC12 culture supernatants contained a strong Mr 28K band recognized by anti-PfRH5 sera (Figure 5C). The mobility of this band relative to the Mr 67K band from merozoites suggested it is a proteolytic fragment of the full length PfRH5 protein.
In assays with A. nancymaae erythrocytes, binding of the Mr 28K fragment was detected only for the parasite lines that were able to invade these cells. Anti-PfRH5 sera did not detect bound protein from culture supernatants of the 7G8 parasites that are unable to invade A. nancymaae erythrocytes, whereas the Mr 28K fragment was readily detected in the eluates after incubation with the LC12 supernatant (Figure 5C). Experiments with allelically-modified transfectants provided further evidence for a strict association between the PfRH5 fragment binding and PfRH5 polymorphisms as well as their ability to invade A. nancymaae erythrocytes. Among the 7G8 transformants, binding of the Mr 28K fragment was detected with the A. nancymaae-invasive 7G8KV parasite culture supernatants but not with the non-invasive 7G8II or 7G8IV parasites (Figure 5D). Among the LC12 transformants, binding was detected with the invasive LC12KV and LC12KI parasites but not with the non-invasive LC12II parasites (Figure 5D), again indicating a critical role for the position 204 I→K substitution in A. nancymaae receptor binding and invasion.
We also examined PfRH5 binding to human erythrocytes before and after treatment with neuraminidase to remove sialic acid residues from cell surface receptor molecules (Breuer et al., 1983). Whereas untreated human erythrocytes bound the Mr 28K fragment from the culture supernatants of all parasites in our experiments: 7G8, LC12 and the allelically-transformed versions of these clones (Figure 5D), neuraminidase-treated human erythrocytes bound the Mr 28K fragment from 7G8 but not from LC12 (Figure 5E).
The inability of the LC12 PfRH5 fragment to bind neuraminidase-treated human erythrocytes suggested that binding might also be lost with neuraminidase treatment of A. nancymaae erythrocytes. Indeed, neuraminidase-treated A. nancymaae erythrocytes showed no binding of the Mr 28K fragment from culture supernatants of LC12 parasites (Figure 5E). To see if the sialic acid-mediated binding of the Mr 28K fragment was associated with invasion of A. nancymaae erythrocytes, we tested the ability of LC12 to invade neuraminidase-treated A. nancymaae erythrocytes. Results confirmed that sialic acid-deficient A. nancymaae erythrocytes were fully refractory to invasion (Figure S3). LC12 and 7G8 parasites remained invasive to neuraminidase-treated human erythrocytes (Figure S3), in agreement with previous observations that P. falciparum parasites can use various ligand-receptor combinations in different pathways to enter human erythrocytes (Iyer et al., 2007).
PfRH5 localizes to the apical end of merozoites
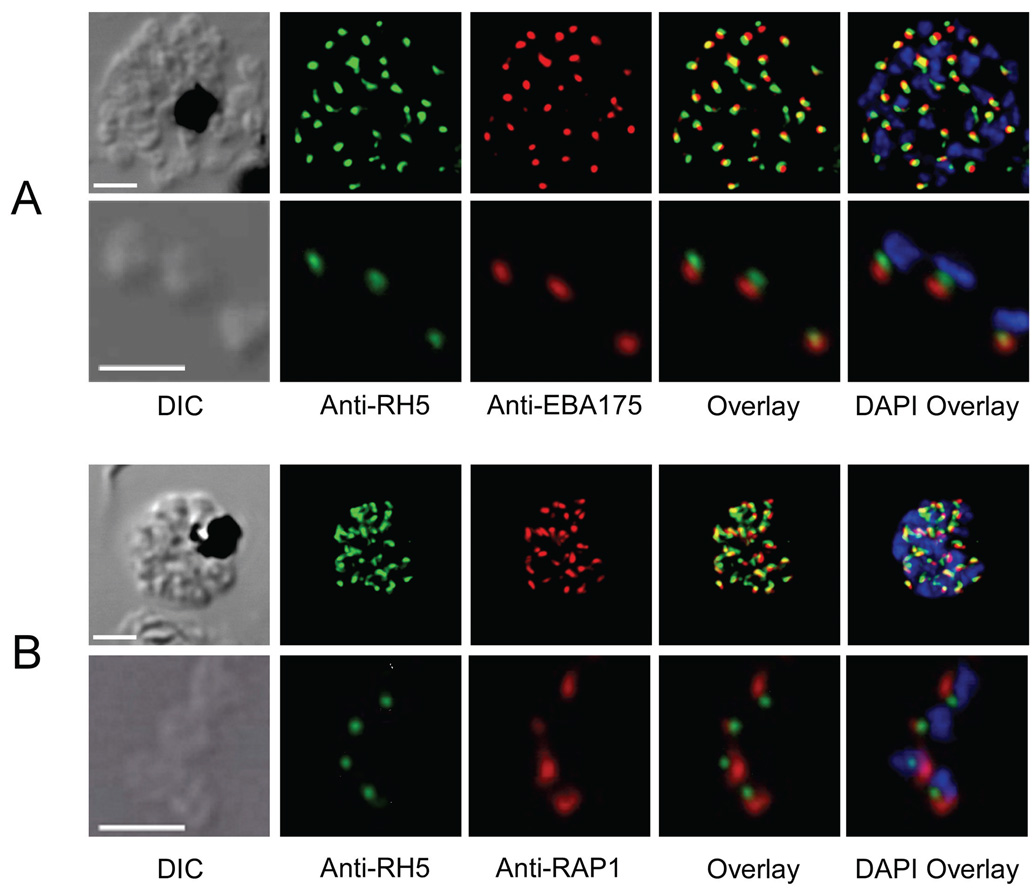
Immunofluorescence (IF) images of P. falciparum parasites probed with anti-PfRH5 sera showed signals from the apical region in developing merozoites of late schizonts and in mature individual merozoites released from parasitized erythrocytes (Figure 6A, B). PfRH5 signals were only faintly observed in the cytoplasm of late trophozoites (data not shown), and they were not detected at all in early trophozoite or ring stage parasites. We used dual-labeled indirect IF confocal laser scanning microscopy to determine the location of PfRH5 signals relative to those of other proteins known to reside in various organelles of the merozoite apical complex. PfRH5 signals at the apical end of merozoites were consistently observed near, but not overlapping with, signals from PfEBA175, a marker of merozoite micronemes (Adams et al., 1990) (Figure 6A). PfRH5 also localized near signals from PfRAP1, a protein of merozoite rhoptry organelles (Stowers et al., 1996) (Figure 6B). PfRH5 signals were identical in wild-type and allelically transformed parasites (data not shown).
Figure 6. PfRH5 localization in schizonts and merozoites.
A) Immunoconfocal microscopy showing the localization of PfRH5 relative to PfEBA175, a microneme marker. Signals from mature segmented schizonts are shown in the top panes; signals from free merozoites are shown in the bottom panes. B) Immunoconfocal microscopy showing the localization of PfRH5 relative to PfRAP1, a rhoptry marker. Signals from mature schizonts are shown in the top panes; signals from free merozoites are shown in the bottom panes. Scale bar, 2 µm.
Discussion
Our findings link polymorphisms in PfRH5, a newly identified erythrocyte binding protein, to switches of host receptor recognition by P. falciparum malaria parasites. In a genetic cross of two parasites that differ dramatically in their ability to infect A. nancymaae monkeys (7G8, GB4), inheritance of the PfRH5 allele from one parent or the other in the haploid progeny proved to be the dominant determinant of infectivity. Transfection and allelic exchange results show that non-synonymous changes in just two codons were sufficient to convert the non-invasive parent (7G8) to a parasite (7G8KV, carrying the equivalent of the GB4 parent allele) able to invade A. nancymaae erythrocytes and infect a monkey in vivo. Of these two polymorphisms, codon change I204K accounted for nearly all of the conversion in invasion phenotype. Furthermore, the invasion of A. nancymaae erythrocytes correlated completely with binding of a PfRH5 Mr 28 K fragment from parasites of the cross.
QTL analysis did not identify loci other than PfRH5 that could be linked to A. nancymaae infectivity in 7G8×GB4 progeny recovered from the chimpanzee host. However, several observations suggest that additional genetic determinants may modulate the PfRH5 pathway for A. nancymaae erythrocyte invasion and infection of A. nancymaae monkeys, as: (1) different 7G8×GB4 progeny required less than 10 days to more than 50 days to produce patent A. nancymaae infections; (2) progeny KB8, JC3 and XF12 produced no infections in A. nancymaae even though these clones inherited the GB4 allele of PfRH5 and invaded monkey erythrocytes well in vitro; (3) replacement of the 7G8 PfRH5 allele by the GB4 allele in the JB12 progeny clone resulted in an A. nancymaae erythrocyte invasion rate about two-thirds of that observed for the same replacement in the 7G8 parent; and (4) non-invasive parasites of the unrelated P. falciparum 3D7 clone remained unable to invade A. nancymaae erythrocytes after they were modified to express PfRH5 polymorphisms of the GB4 allele (3D7KV line). Detection of additional determinants involved in these variations may have been obscured by their multiplicity, interactions with partners, or variable effects among the different inherited genetic backgrounds of the 32 recombinant progeny in this study.
The region of highly skewed inheritance on chromosome 7 contains PfEBA175, the gene encoding a 175 kDa parasite protein that binds sialic acid residues of erythrocyte surface glycophorin A (Camus and Hadley, 1985; Sim et al., 1994). All 32 independent recombinants from the chimpanzee host inherited the PfEBA175 allele and nearby genes on chromosome 7 from the 7G8 rather than the GB4 parent. In PfEBA175, the erythrocyte binding domain is defined by a 616 amino acid region (Sim et al., 1994), which differs in the 7G8 and GB4 parasites by nine amino acid residues at polymorphic sites reported previously (data not shown; Liang and Sim, 1997). As the PfEBA175 gene is proposed to be a determinant of primate specificity (Martin et al., 2005), it is possible that inheritance of the 7G8 allele vs. the GB4 allele reflects upon the function of the PfEBA175 protein in primate sialic acid binding.
PfRH5 is the smallest member of the RH family. Its 63 kDa amino acid sequence aligns most closely with that of PrRH5, followed by the N-terminal regions of the 370 kDa PfRH2a and 383 kDa PfRH2b proteins (Rayner et al., 2000). PfRH5 therefore lacks regions corresponding to more than 300 kDa of peptide sequence in the body and C-terminal regions of PfRH2a and PfRH2b, including a predicted transmembrane domain characteristic of other family members. These alignments may help to define binding domains of PfRH2a, PfRH2b and other RH proteins. Our attempts and those of others to disrupt the PfRH5 gene have been unsuccessful (data not shown; Cowman and Crabb, 2006), in marked contrast to the experimental knock-outs achieved for other members of the RH family as well as various P. falciparum EBA genes (Cowman and Crabb, 2006). PfRH5 may therefore have a vital role in the ability of merozoites to recognize and enter various human and non-human erythrocytes. The location of PfRH5 at the apical end of the merozoite presumably makes the protein readily available for processing, discharge and binding with host cell proteins that function in the invasion of erythrocytes.
The ability of parasites in the 7G8×GB4 cross to infect A. nancymaae correlates fully with the binding of a Mr 28K fragment from PfRH5 molecules containing a positively charged lysine residue at position 204. Binding of this fragment may reflect a ligand-receptor function of PfRH5 or its processed forms for parasite entry into the A. nancymaae cells. Neuraminidase removal of negatively charged sialic acid residues from the surface of the monkey erythrocytes completely abrogated binding of the Mr 28K fragment and blocked invasion of the parasites in this work. These results suggest that an ionic bond between the positively charged lysine and a negatively charged sialic acid may promote an important ligand-receptor interaction.
Our results also show that human erythrocytes treated with neuraminidase to remove sialic acid residues can bind the Mr 28K fragment of PfRH5 when it contains I204 but not when it contains K204. Non-treated human erythrocytes, however, can bind both forms of the fragment. We note that different polymorphisms are also present in the PfRH5 sequences of other P. falciparum lines that are able to infect A. nancymaae erythrocytes (Table 1), and many of these polymorphisms involve amino acid class changes, including an I204R substitution in the Palo Alto PfRH5 that may have a functional effect similar to the I204K substitution in GB4. Considering the importance of sialic acid residues to the binding of PfRH5 domains containing K204 but not I204, we suggest that PfRH5 polymorphisms may enable P. falciparum parasites to switch their ligand-receptor interactions and adapt to different erythrocyte surface molecules. Point mutations in the sequences of two other P. falciparum ligands, JESEBL/EBA181 and BAEBL/EBA140, also alter their molecular specificity for different receptors on erythrocytes (Mayer et al., 2002; Mayer et al., 2004). These and other polymorphisms provide an important adaptive ability in the various pathways P. falciparum uses to infect different human erythrocytes. The utility of these pathways is severely restricted in the non-natural host cells of A. nancymaae, accounting for the dependence of LC12 and other 7G8×GB4 parasites on PfRH5 as a single ligand for invasion.
Parasites that infect across species barriers are reported from a number of primate malarias. For example, P. knowlesi parasites that naturally infect monkeys have produced outbreaks of malaria in human populations (Cox-Singh et al., 2008). The close relationship between macaque P. cynomolgi and human P. vivax parasites (Cornejo and Escalante, 2006) points to the existence of shared reservoirs of infection before a recent evolutionary divergence, and the near identity of P. simium to P. vivax may reflect an even more recent host switch back to monkeys from humans (Cornejo and Escalante, 2006). In the present case, there is no evidence that Aotus monkeys carry a reservoir of P. falciparum parasites in South America (Collins, 1994). Some of the P. falciparum strains able to infect A. nancymaae in our study were isolated in Africa (GB4, Ghana; Palo Alto, Uganda) where these parasites had not been under selective pressure to adapt to Aotus monkeys. It seems likely that the ability of these parasites to invade A. nancymaae erythrocytes derives from the diversity of ligand-receptor interactions exploited by P. falciparum in its various pathways to invade human erythrocytes.
Experimental Procedures
Animal Studies
All animal studies were carried out in compliance with National Institutes of Health guidelines under an Animal Care and Use Committee-approved protocol. P. falciparum in A. nancymaae is virulent and can be fatal. Monkeys developing a parasitemia of 5% were promptly cured of their infection with 25 mg/kg mefloquine, as were monkeys showing any signs of dehydration, lethargy, decreased appetite or diarrhea, or anemia with a hematocrit below 25%. A. nancymaae monkeys were either naïve to P. falciparum or had a history of infection more than 6 months before entry in the study. All monkeys were splenectomized. Infections were initiated by the intravenous inoculation of 1 × 107 parasitized erythrocytes from human cell cultures.
The 7G8×GB4 Cross
7G8 and GB4 gametocytes were induced in-vitro as described (Ifediba and Vanderberg, 1981), grown to maturity, and fed through stretched Parafilm membranes to An. freeborni and An. gambiae mosquitoes using a membrane feeder. A splenectomized chimpanzee was infected with sporozoites by the natural biting of ~300 mosquitoes on day 15 (Walliker et al., 1987). Twenty-one days after sporozoite inoculation, the parasitemia in the chimpanzee reached 0.2%, at which time a final blood sample was collected and the asymptomatic infection cured with 15 mg/kg mefloquine. Independent recombinant progeny clones were cloned by limiting dilution, either directly from chimpanzee blood or from parasite lines adapted to culture in human erythrocytes.
Invasion Assays
Erythrocyte invasion assays were performed as described (Dolan et al., 1990). Briefly, erythrocytes infected with mature stage parasites were obtained by gelatin floatation or percoll/sorbitol purification and adjusted to a final concentration of 1 × 107 parasitized erythrocytes/ml. One hundred µl of the parasite suspension were added to 100 µl of target erythrocytes at 2 × 108 cells/ml in complete medium and incubated at 37°C for 12–18 hr. Human and rhesus erythrocytes were included as positive and negative controls, respectively. Invasion was determined by counting the number of ring-infected erythrocytes and expressed as a percent of the invasion counts observed for human erythrocytes. Each assay was performed in triplicate, on at least three independent occasions.
Generation of the 7G8×GB4 Linkage Map and QTL Analysis
Progeny clones were genotyped using polymorphic microsatellites (Su et al., 1999). Additional primers were designed for areas with low coverage or to narrow informative meiosis events from chromosome sequence data for the 3D7 P. falciparum parasite (www.plasmodb.org) (Bahl et al., 2003). The segregation data for all heterozygous loci are provided in Table S1. These data were used to construct a genetic map using the MAPMAKER program and QTL analysis was carried out using the Pseudomarker program as described (Sen and Churchill, 2001; Ferdig et al., 2004).
Allelic Exchange at the PfRH5 Locus
Allelic exchange plasmids were designed to contain a 1396 bp fragment from the PfRH5 sequence of GB4 or 7G8. DNA fragments were amplified with primers containing Not I (5’-TGCGGCCGCATGAAGACTATAAAAATGTGG-3’) and Pst I (5’- ACTGCAGATGCTTTGTCTAATTAGAG-3) restriction sites. These sites facilitated cloning into the pHD22Y vector containing the human dihydrofolate reductase (hDHFR) selectable marker (Fidock and Wellems, 1997). Uninfected human erythrocytes were loaded with plasmid DNA by electroporation and supplied to parasite cultures as described (Deitsch et al., 2001). Parasite populations containing integrated plasmid were identified after approximately 60 days by diagnostic PCR.
Production and Verification of Antisera
A multiple antigen peptide (MAP4) directed against a PfRH5 peptide sequence (LIKCIKNHENDFNKIC) was synthesized by Bio-Synthesis Inc., Lewisville, TX. Anti-peptide sera were produced by immunizing a rabbit with 250 µg peptide in CFA followed by 250 µg peptide in IFA on days 21, 42 and 63 (Spring Valley Laboratories, Inc.).
Two fragments of PfRH5 were amplified from a synthetic bacteria codon-adjusted sequence (a gift from David Garboczi), encoding either a 16 or 28 kDa fragment of PfRH5 (amino acids 304–430 and 304–526). These fragments were cloned into the pNAN T7 expression vector (Su et al., 2006) and expressed in BL21 cells (Stratagene, La Jolla, CA) induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 2 hr.
Immunoblot Analysis and Immunofluorescence (IFA) Microscopy
Sorbitol synchronized ring, trophozoite and schizont stages were lysed with 0.15% saponin in PBS, pelleted, and immediately frozen. Thawed pellets containing 5 × 106 parasitized erythrocytes were resuspended in sample buffer containing 250 mM sucrose, 20 mM HEPES, protease inhibitor cocktail (Sigma-Aldrich, St Louis, MO), and 1x NuPAGE® LDS Sample Buffer (Invitrogen, Carlsbad, CA). After separation by electrophoresis (NuPAGE® Novex system, 4–12% Bis-Tris Gels, Invitrogen), the proteins from the pellets were transferred to a nitrocellulose membrane (Invitrogen). Membranes were blocked overnight in 5% skimmed milk and incubated for 1 hr with rabbit anti-PfRH5 sera (1:250 dilution in PBS), or with rabbit anti-human spectrin to confirm sample loading (Sigma-Aldrich) and processed as described (Gaur et al., 2006).
For IFA, schizont preparations were fixed either with chilled acetone or 1% formaldehyde for 5 min at room temperature, incubated in blocking buffer (PBS with 0.1% Triton X-100, 2.5 mg/ml goat serum) for 5 min and then probed for 1 hr at 37°C with rabbit anti-PfRH5 sera plus either rat anti-PfEBA175 sera or mouse anti-PfRAP1 sera (Stowers et al., 1996) (1:50 dilution in blocking buffer). Samples were processed and microscopic images were collected and analysed as described (Gaur et al., 2007).
Erythrocyte Binding Assay
Erythrocyte binding assays were performed as described (Gaur et al., 2007).
Supplementary Material
Acknowledgements
We thank Andre Laughinghouse, Kevin Lee, Robert Gwadz, Brian Keegan, Josh Reece, Cheryl Kothe, Mark Szarowicz, Angela Lunger, Kelly Magee and Carole Long for help with mosquito and A. nancymaae infections, Marisa St. Claire, Max Shapiro, Charlene Shaver and Rick Fairhurst for assistance producing the cross, Allan Saul for RAP-1 monoclonal antibodies, the Malaria Research and Reference Reagent Resource Center for anti-EBA-175 (deposited by John Adams) and various parasites, Juliana Sá for 7G8 cDNA, David Garboczi and Hua-Poo Su for codon-adjusted RH5, Julian Rayner for the full-length P. reichenowi RH5 sequence, Mrinal Bhattacharyya, Lisa Ranford-Cartwright, Michael Ferdig, Jigar Patel, Janni Papakrivos, Owen Schwartz and Juraj Kabat for helpful advice, and Bill Collins, Akhil Vaidya and Xin-zhuan Su for discussions. This work was supported by the intramural research program of the NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams JH, Hudson DE, Torii M, Ward GE, Wellems TE, Aikawa M, Miller LH. The Duffy receptor family of Plasmodium knowlesi is located within the micronemes of invasive malaria merozoites. Cell. 1990;63:141–153. doi: 10.1016/0092-8674(90)90295-p. [DOI] [PubMed] [Google Scholar]
- Aikawa M, Miller LH, Johnson J, Rabbege J. Erythrocyte entry by malarial parasites. A moving junction between erythrocyte and parasite. J. Cell Biol. 1978;77:72–82. doi: 10.1083/jcb.77.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl A, Brunk B, Crabtree J, Fraunholz MJ, Gajria B, Grant GR, Ginsburg H, Gupta D, Kissinger JC, Labo P, et al. PlasmoDB: the Plasmodium genome resource. A database integrating experimental and computational data. Nucleic Acids Res. 2003;31:212–215. doi: 10.1093/nar/gkg081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von HG, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Breuer WV, Ginsburg H, Cabantchik ZI. An assay of malaria parasite invasion into human erythrocytes. The effects of chemical and enzymatic modification of erythrocyte membrane components. Biochim. Biophys. Acta. 1983;755:263–271. doi: 10.1016/0304-4165(83)90213-1. [DOI] [PubMed] [Google Scholar]
- Burkot TR, Williams JL, Schneider I. Infectivity to mosquitoes of Plasmodium falciparum clones grown in vitro from the same isolate. Trans. R. Soc. Trop. Med. Hyg. 1984;78:339–341. doi: 10.1016/0035-9203(84)90114-7. [DOI] [PubMed] [Google Scholar]
- Camus D, Hadley TJ. A Plasmodium falciparum antigen that binds to host erythrocytes and merozoites. Science. 1985;230:553–556. doi: 10.1126/science.3901257. [DOI] [PubMed] [Google Scholar]
- Collins WE. The Owl Monkey as a Model for Malaria. In: Bear JF, Weller RE, Kakoma I, editors. Aotus: The Owl Monkey. San Diego: Academic Press, Inc.; 1994. pp. 217–244. [Google Scholar]
- Collins WE, Grady KK, Millet P, Sullivan JS, Morris CL, Galland GG, Richardson BB, Yang C. Adaptation of a strain of Plasmodium falciparum from a Montagnard refugee to Aotus monkeys. J. Parasitol. 1997;83:1174–1177. [PubMed] [Google Scholar]
- Cornejo OE, Escalante AA. The origin and age of Plasmodium vivax. Trends Parasitol. 2006;22:558–563. doi: 10.1016/j.pt.2006.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowman AF, Crabb BS. Invasion of red blood cells by malaria parasites. Cell. 2006;124:755–766. doi: 10.1016/j.cell.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Cox-Singh J, Davis TM, Lee KS, Shamsul SS, Matusop A, Ratnam S, Rahman HA, Conway DJ, Singh B. Plasmodium knowlesi malaria in humans is widely distributed and potentially life threatening. Clin. Infect. Dis. 2008;46:165–171. doi: 10.1086/524888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch K, Driskill C, Wellems T. Transformation of malaria parasites by the spontaneous uptake and expression of DNA from human erythrocytes. Nucleic Acids Res. 2001;29:850–853. doi: 10.1093/nar/29.3.850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan SA, Miller LH, Wellems TE. Evidence for a switching mechanism in the invasion of erythrocytes by Plasmodium falciparum. J. Clin. Invest. 1990;86:618–624. doi: 10.1172/JCI114753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Triglia T, Ralph SA, Rayner JC, Barnwell JW, McFadden GI, Cowman AF. Phenotypic variation of Plasmodium falciparum merozoite proteins directs receptor targeting for invasion of human erythrocytes. EMBO J. 2003;22:1047–1057. doi: 10.1093/emboj/cdg096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dvorak JA, Miller LH, Whitehouse WC, Shiroishi T. Invasion of erythrocytes by malaria merozoites. Science. 1975;187:748–750. doi: 10.1126/science.803712. [DOI] [PubMed] [Google Scholar]
- Fajfar-Whetstone CJ, Collins WE, Ristic M. In vitro and in vivo adaptation of the Geneve/SGE-1 strain of Plasmodium falciparum to growth in a squirrel monkey (Saimiri sciureus) model. Am. J. Trop. Med. Hyg. 1987;36:221–227. doi: 10.4269/ajtmh.1987.36.221. [DOI] [PubMed] [Google Scholar]
- Ferdig MT, Cooper RA, Mu J, Deng B, Joy DA, Su XZ, Wellems TE. Dissecting the loci of low-level quinine resistance in malaria parasites. Mol. Microbiol. 2004;52:985–997. doi: 10.1111/j.1365-2958.2004.04035.x. [DOI] [PubMed] [Google Scholar]
- Fidock DA, Wellems TE. Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. U. S. A. 1997;94:10931–10936. doi: 10.1073/pnas.94.20.10931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaur D, Furuya T, Mu J, Jiang LB, Su XZ, Miller LH. Upregulation of expression of the reticulocyte homology gene 4 in the Plasmodium falciparum clone Dd2 is associated with a switch in the erythrocyte invasion pathway. Mol. Biochem. Parasitol. 2006;145:205–215. doi: 10.1016/j.molbiopara.2005.10.004. [DOI] [PubMed] [Google Scholar]
- Gaur D, Singh S, Jiang L, Diouf A, Miller LH. Recombinant Plasmodium falciparum reticulocyte homology protein 4 binds to erythrocytes and blocks invasion. Proc. Natl. Acad. Sci. U. S. A. 2007;104:17789–17794. doi: 10.1073/pnas.0708772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiman QM, Meagher MJ. Susceptibility of a New World monkey to Plasmodium falciparum from man. Nature. 1967;215:437–439. doi: 10.1038/215437a0. [DOI] [PubMed] [Google Scholar]
- Heppner DG, Cummings JF, Ockenhouse C, Kester KE, Lyon JA, Gordon DM. New World monkey efficacy trials for malaria vaccine development: critical path or detour? Trends Parasitol. 2001;17:419–425. doi: 10.1016/s1471-4922(01)02012-8. [DOI] [PubMed] [Google Scholar]
- Ifediba T, Vanderberg JP. Complete in vitro maturation of Plasmodium falciparum gametocytes. Nature. 1981;294:364–366. doi: 10.1038/294364a0. [DOI] [PubMed] [Google Scholar]
- Iyer J, Gruner AC, Renia L, Snounou G, Preiser PR. Invasion of host cells by malaria parasites: a tale of two protein families. Mol. Microbiol. 2007;65:231–249. doi: 10.1111/j.1365-2958.2007.05791.x. [DOI] [PubMed] [Google Scholar]
- Kaneko O, Soubes SC, Miller LH. Plasmodium falciparum: invasion of Aotus monkey red blood cells and adaptation to Aotus monkeys. Exp. Parasitol. 1999;93:116–119. doi: 10.1006/expr.1999.4441. [DOI] [PubMed] [Google Scholar]
- Kooij TW, Carlton JM, Bidwell SL, Hall N, Ramesar J, Janse CJ, Waters AP. A Plasmodium whole-genome synteny map: indels and synteny breakpoints as foci for species-specific genes. PLoS. Pathog. 2005;1:e44. doi: 10.1371/journal.ppat.0010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H, Sim BK. Conservation of structure and function of the erythrocyte-binding domain of Plasmodium falciparum EBA-175. Mol. Biochem. Parasitol. 1997;84:241–245. doi: 10.1016/s0166-6851(96)02791-0. [DOI] [PubMed] [Google Scholar]
- Martin MJ, Rayner JC, Gagneux P, Barnwell JW, Varki A. Evolution of human-chimpanzee differences in malaria susceptibility: relationship to human genetic loss of N-glycolylneuraminic acid. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12819–12824. doi: 10.1073/pnas.0503819102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer DC, Mu JB, Feng X, Su XZ, Miller LH. Polymorphism in a Plasmodium falciparum erythrocyte-binding ligand changes its receptor specificity. J. Exp. Med. 2002;196:1523–1528. doi: 10.1084/jem.20020750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer DC, Mu JB, Kaneko O, Duan J, Su XZ, Miller LH. Polymorphism in the Plasmodium falciparum erythrocyte-binding ligand JESEBL/EBA- 181 alters its receptor specificity. Proc. Natl. Acad. Sci. U. S. A. 2004;101:2518–2523. doi: 10.1073/pnas.0307318101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell GH, Hadley TJ, McGinniss MH, Klotz FW, Miller LH. Invasion of erythrocytes by Plasmodium falciparum malaria parasites: evidence for receptor heterogeneity and two receptors. Blood. 1986;67:1519–1521. [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J. T-Coffee: A novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- Ogun SA, Holder AA. A high molecular mass Plasmodium yoelii rhoptry protein binds to erythrocytes. Mol. Biochem. Parasitol. 1996;76:321–324. doi: 10.1016/0166-6851(95)02540-5. [DOI] [PubMed] [Google Scholar]
- Perkins ME, Holt EH. Erythrocyte receptor recognition varies in Plasmodium falciparum isolates. Mol. Biochem. Parasitol. 1988;27:23–34. doi: 10.1016/0166-6851(88)90021-7. [DOI] [PubMed] [Google Scholar]
- Rayner JC, Galinski MR, Ingravallo P, Barnwell JW. Two Plasmodium falciparum genes express merozoite proteins that are related to Plasmodium vivax and Plasmodium yoelii adhesive proteins involved in host cell selection and invasion. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9648–9653. doi: 10.1073/pnas.160469097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner JC, Huber CS, Galinski MR, Barnwell JW. Rapid evolution of an erythrocyte invasion gene family: the Plasmodium reichenowi Reticulocyte Binding Like (RBL) genes. Mol. Biochem. Parasitol. 2004;133:287–296. doi: 10.1016/j.molbiopara.2003.10.017. [DOI] [PubMed] [Google Scholar]
- Sen S, Churchill GA. A statistical framework for quantitative trait mapping. Genetics. 2001;159:371–387. doi: 10.1093/genetics/159.1.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim BK, Chitnis CE, Wasniowska K, Hadley TJ, Miller LH. Receptor and ligand domains for invasion of erythrocytes by Plasmodium falciparum. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- Stowers AW, Cooper JA, Ehrhardt T, Saul A. A peptide derived from a B cell epitope of Plasmodium falciparum rhoptry associated protein 2 specifically raises antibodies to rhoptry associated protein 1. Mol. Biochem. Parasitol. 1996;82:167–180. doi: 10.1016/0166-6851(96)02730-2. [DOI] [PubMed] [Google Scholar]
- Stubbs J, Simpson KM, Triglia T, Plouffe D, Tonkin CJ, Duraisingh MT, Maier AG, Winzeler EA, Cowman AF. Molecular mechanism for switching of P. falciparum invasion pathways into human erythrocytes. Science. 2005;309:1384–1387. doi: 10.1126/science.1115257. [DOI] [PubMed] [Google Scholar]
- Su HP, Lin DY, Garboczi DN. The structure of G4, the poxvirus disulfide oxidoreductase essential for virus maturation and infectivity. J. Virol. 2006;80:7706–7713. doi: 10.1128/JVI.00521-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X-Z, Ferdig M, Huang Y, Huynh CQ, Liu A, You J, Wootton JC, Wellems TE. A genetic map and recombination parameters of the human malaria parasite Plasmodium falciparum. Science. 1999;286:1351–1353. doi: 10.1126/science.286.5443.1351. [DOI] [PubMed] [Google Scholar]
- Sullivan JS, Sullivan JJ, Williams A, Grady KK, Bounngaseng A, Huber CS, Nace D, Williams T, Galland GG, Barnwell JW, Collins WE. Adaptation of a strain of Plasmodium falciparum from Ghana to Aotus lemurinus griseimembra, A. nancymai, and A. vociferans monkeys. Am. J. Trop. Med. Hyg. 2003;69:593–600. [PubMed] [Google Scholar]
- Trager W, Jensen JB. Human malaria parasites in continuous culture. Science. 1976;193:673–675. doi: 10.1126/science.781840. [DOI] [PubMed] [Google Scholar]
- Triglia T, Thompson JK, Cowman AF. An EBA175 homologue which is transcribed but not translated in erythrocytic stages of Plasmodium falciparum. Mol. Biochem. Parasitol. 2001;116:55–63. doi: 10.1016/s0166-6851(01)00303-6. [DOI] [PubMed] [Google Scholar]
- Walliker D, Quakyi IA, Wellems TE, McCutchan TF, Szarfman A, London WT, Corcoran LM, Burkot TR, Carter R. Genetic analysis of the human malaria parasite Plasmodium falciparum. Science. 1987;236:1661–1666. doi: 10.1126/science.3299700. [DOI] [PubMed] [Google Scholar]
- Wootton JC, Feng X, Ferdig MT, Cooper RA, Mu J, Baruch DI, Magill AJ, Su XZ. Genetic diversity and chloroquine selective sweeps in Plasmodium falciparum. Nature. 2002;418:320–323. doi: 10.1038/nature00813. [DOI] [PubMed] [Google Scholar]
- Young MD, Baerg DC, Rossan RN. Parasitological review. Experimental monkey hosts for human plasmodia. Exp. Parasitol. 1975;38:136–152. doi: 10.1016/0014-4894(75)90046-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.