Abstract
Amyloid fibrils and partially unfolded intermediates can be distinguished serologically from native amyloidogenic precursor proteins or peptides. In this regard, we previously had reported that mAb 11-1F4, generated by immunizing mice with a thermally denatured variable domain (VL) fragment of the human κ4 Bence Jones protein Len, bound to a non-native conformational epitope located within the N-terminal 18 residues of fibrillar, as well as partially denatured, Ig light chains (O’Nuallain B. et al. (2006) Biochemistry 46, 1240–247). To define further the antibody binding site, we used random peptide phage display and epitope mapping of VL Len using wild-type and alanine-mutated Len peptides where it was shown that the antibody epitope was reliant on up to 10 of the first 15 residues of protein Len. Comparison of Vκ and Vλ N-terminal germline consensus sequences with protein Len and 11-1F4-binding phages indicated that this antibody’s cross-reactivity with light chains was related to an invariant proline at position(s) 7 and/or 8, bulky hydrophobic residues at positions 11 and 13, and additionally, to the ability to accommodate amino acid diversity at positions 1–4. Sequence alignments of the phage peptides revealed a central proline, often flanked by aromatic residues. Taken together, these results have provided evidence for the structural basis of the specificity of 11-1F4 for both κ and λ light chain fibrils. We posit that the associated binding site involves a rare type VI β-turn or touch-turn that is anchored by a cis-proline residue. The identification of an 11-1F4-related mimotope should facilitate development of pan-light chain fibril-reactive antibodies that could be used in the diagnosis and treatment of patients with AL amyloidosis.
The amyloidoses are a group of over 20 protein misfolding disorders associated with often fatal sporadic and hereditary disorders, including Alzheimer’s disease, type II diabetes, and primary (AL) amyloidosis (1–4). All types of amyloid, regardless of amino acid sequence, share tertiary structural and tinctorial features, i.e., they are formed from fibrils composed of extended β-sheets oriented perpendicularly to the long fibril axis (5, 6) and bind the dyes thioflavin T and Congo red (7, 8). Additionally, fibrils, as well as their assembly intermediates, contain generic conformational epitopes not present on the native proteins (9–16). These common characteristics reflect a similar fibrillogenic pathway(s) involving the assembly of soluble higher-order intermediates into fibrils that are structurally dissimilar to their precursors (2, 13, 17).
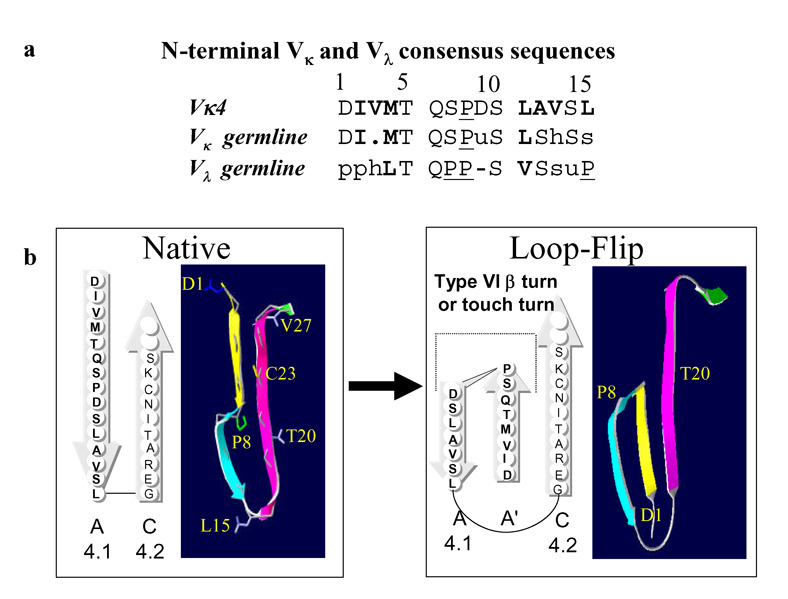
Recently, we determined that the murine mAb 11-1F4, generated by immunizing mice with a thermally denatured variable region (VL) fragment of the human κ4 Bence Jones protein Len (18), bound a cryptic, non-native epitope located within the first 18 residues of Ig light chains (LC) (15, 19) (Figure 1). This antibody has shown therapeutic efficacy in a mouse model, as demonstrated by its ability to accelerate the removal of induced human AL amyloidomas, an effect that was independent of the κ or λ LC isotype or VL subgroup of the injected fibrils (12). Additionally, mAb 11-1F4 inhibited de novo VL fibrillogenesis (19).
FIGURE 1.
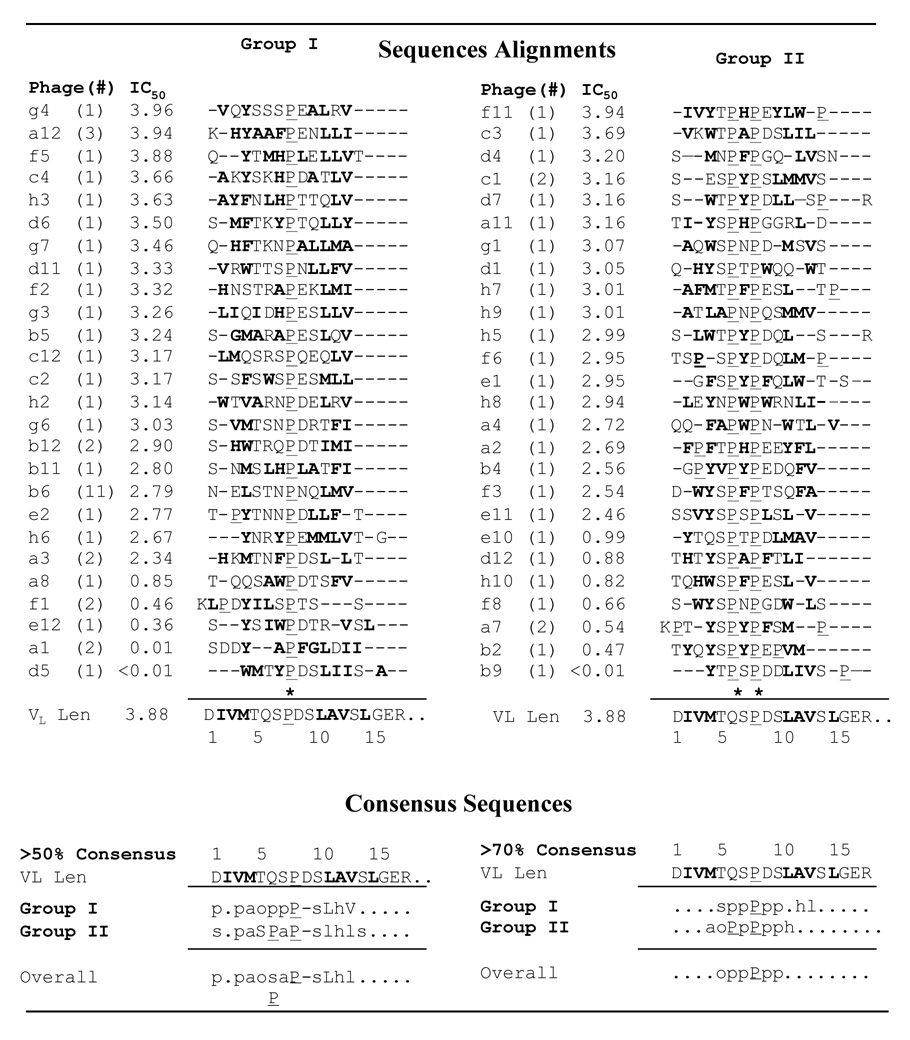
Multiple sequence alignments and consensus sequences for 11-1F4-binding phage peptides. Phage peptides were aligned (28) and divided into Groups I and II, based on the presence of one of the two proline (-X-Pro-X- and Pro-X-Pro) or cysteine (-le/Leu-Cys and Ile/Leu-X-Cys-) motifs, and each group adjusted to give the best alignment with the N-terminal sequence of VL Len. The hydrophobic and/or aromatic residues are bolded and prolines underlined. Phage peptide consensus sequences were determined using the program CONSENSUS (http://www.bork.embl-heidelberg.de/Alignment/). Abbreviations include: a, aromatic (F, H, Y, W); h, hydrophobic, (A, C, F, I, L, M, V); l, aliphatic (I, L, V); o, S or T; p, polar (D, E, G, H, K, N, P, Q, R, S, T, W, Y); s, small side chain (A, C, D, G, N, P, S, T, V) (31), and each uppercase letter represents the particular amino acid.
Using random peptide phage display (20, 21) and epitope mapping with wild-type and alanine mutated (22) Len peptides, we now have defined the nature of the epitope recognized on LC fibrils by mAb 11-1F4. Through phage peptide mimetics, as well as the discovery of a mimotope of 11-1F4’s fibril-related epitope, we have shown that this cross-reactivity involves 10 of the first 15 LC residues. Additionally, our findings indicate that the 11-1F4 conformational epitope involves a rare type VI β-turn or touch-turn that is anchored by a cis-proline residue.
MATERIALS AND METHODS
Proteins, Peptides, and Antibodies
Recombinant (r) Vκ4 Len and Vλ6 Wil were produced in E. coli, as previously described (23). The lyophilized proteins were dissolved in distilled water (~1 mg/ml, ~80 µM), 10× PBS containing 0.5% sodium azide (PBSA) was added to yield a concentration of 1x, and the samples passed through a 0.22 µm PVDF 25 mm Millex®-GV syringe-driven filter unit (Millipore, Beillerica, MA). For competition studies, the proteins were centrifuged at 100,000 ×g for 2 h at room temperature. Protein concentrations were determined spectrophotometrically using 24,535 and 13,075 M−1 cm−1 as the E280s for the Len and Wil proteins, respectively (ProtParam; http://us.expasy. org/tools/protparam.html); the final solutions were aliquoted and stored at −20 °C.
Synthetic phage and peptides encompassing all (or part) of the first 22 residues of Len were obtained from the Keck Biotechnology Center at Yale University (http://info.med.yale.edu/wmkeck/; New Haven, CT). Additionally, a peptide consisting of the first 15 Len residues, designated Len(1–15), was purchased from Sigma-Aldrich (St. Louis, Missouri). A custom Len(1–18) peptide library containing single alanine point mutations was generated by solid-phase synthesis at Mimotopes (Raleigh, NC) and the Aβ40 peptide was obtained from Quality controlled Biochemicals (Hopkinton, MA). Mass spectrometric analyses revealed the purity of the phage and Len preparations to be ~50% and that of the Aβ40 peptide, >90%.
Before use, each lyophilized peptide was disaggregated by sequential exposure to TFA and HFIP, as described previously (15, 24). After removal of the organic solvents by evaporation under argon, they were dissolved (0.25–1 mg/mL) in distilled water or 2 mM NaOH (depending on the pI) and 10× PBSA added to a final concentration of 1×. Prior to use, peptides from the Len(1–18) library were diluted from dimethylfluoride into PBSA. Len and phage samples were centrifuged at 20,800 ×g for 20 min at room temperature and Aβ at 50,000 ×g for 18 h at 4 °C. The concentration of peptide in each supernatant was determined, based on a BSA standard curve, in a bicinchoninic acid colorimetric assay (micro-BCA®, Pierce, Rockford, IL). Aβ samples were used immediately; Len and phage peptide preparations were stored at −4 °C for up to 48 h.
Bioreactor generated murine mAb 11-1F4 was furnished by the National Cancer Institute’s Biopharmaceutical Development Program (SAIC-Fredrick, Inc., Fredrick, MD) and its concentration determined (E280, 1.4 M−1 cm−1).
Preparation of Amyloid Fibrils
Fibrils composed of rVκ4 Len and rVλ6 Wil were generated from filtered, soluble precursor proteins by agitation in an orbital shaker, as previously described (25). Briefly, 80 µM of protein in PBSA was placed in a closed 15-ml volume polypropylene tube which was shaken at 225 rpm at 37 °C. Aβ fibrils were generated by incubating the soluble precursor peptide at a concentration of ~30 µM in PBSA at 37 °C, as previously described (14). Maximum fibril formation occurred within 2 wks, as evidenced by ThT fluorescence (26, 27), after which the components were harvested by centrifugation (20,800 ×g for 25 min), sonicated, and stored at −20 °C.
Antibody Production and Characterization
BALB/c mice were immunized x5 with 50-µg injections of an 11-1F4-binding synthetic phage peptide, designated a12 (NH2-KHYAAFPENLLI-CONH2) and conjugated via 1-ethyl-3-[3-dimethylaminopropyl] carbodiimide HCL to keyhole limpet hemocyanin (Pierce). The reactivity of sera obtained 1 wk after the final injection against microtiter plate-immobilized a12 peptide, VL fibrils, and collagen (Sigma) was determined in a europium (Eu3+) based immunosorbant assay (EuLISA) (16, 18), using both biotinyl-goat anti-mouse IgG and anti-IgM for detection.
Selection of 11-1F4 Binding Phage
To remove phage-containing 12-mer peptides that potentially bound nonspecifically to mAb 11-1F4, the Ph.D.-12™ phage-display library (New England Biolabs) was preabsorbed with the murine monoclonal IgGκ protein MOPC-31C (Sigma). Briefly, 200 µL of 2 × 1011 unamplified phage was mixed for 4 h at room temperature with 50 µL of ~20 µg MOPC-31C attached to blocked (PBS containing 0.5% BSA) protein G magnetic beads. The resultant phage complexes were pelleted using a 6-tube magnetic separation rack (New England Biolabs) and the amount of unbound phage in the supernatant (60% recovery) determined by standard plaque titering procedures (28).
Isolation of 11-1F4-bound phage from the absorbed library was achieved by 4 rounds of biopanning. The selection process involved alternating the antibody binding protein (Protein G or A) and the blocking agent (0.5% BSA vs. 1% gelatin in PBS) on the magnetic beads. Antibody-bound phage was eluted using the 11-1F4-binding Len(1–30) peptide (15, 19). The first round consisted of gentle mixing and incubation of 25 nM antibody with 1.2 × 1011 absorbed phage in 0.3 mL of PBS containing 0.05% Tween 20 (PBST) for 1 h at room temperature. The sample then was added to 50 µL protein G beads blocked with 0.5% BSA and, after incubation, the beads were washed x10 with 1 mL PBST; 11-1F4-bound phage was eluted with 3 µM Len(1–30) in 0.3 mL of PBST and then amplified from 3.9 × 106 to ~3 × 1013, using standard amplification and plaque titrating procedures (28). An aliquot (1.2 × 1011) of the amplified phage was used to pan against 11-1F4 bound to gelatin-blocked protein A beads. After 2 additional steps, the antibody-reactive phage were stored in 20% glycerol at −20 °C for up to 6 mo, with no observable loss in antibody reactivity.
Isolation and Sequencing of Clonal Phages
For phage plaque hybridization, a Protran® BA85 0.45 µm nitrocellulose membrane (Schleicher and Schuell), soaked in transfer buffer (PBST), was placed on top of an LB/IPTG/Xgal agar plate containing ~100 plaques and incubated at 37 °C for 1 h. The membrane was lifted and blocked with assay buffer (0.5% gelatin in PBS containing 1% Tween 20), unbound blocking agent removed, and 30 nM of mAb 11-1F4 added. After incubation and washing x3 with PBST, HRP-labeled goat anti-mouse IgG antibody (Jackson Immunoresearch Labs, West Grove, PA) was added and, after washing, phage-bound 11-1F4 was detected by chemiluminescence using ECL™ (Amersham Biosciences Corp., Piscataway, NJ) as the substrate. An 11-1F4 binding phage plaque was defined by the presence of a strong positive signal detected on duplicate hybridizations. In total, 96 positive plaques were picked, amplified, and phage DNA-purified using a QIAprep Spin M13 Kit (QIAGEN Inc., Valencia, CA). Sequencing was performed with an ABI PRISM® 3100 Genetic Analyzer (Applied Biosystems, Foster city, CA) with a Big Dye™ v3.0 Cycle Sequencing kit (Applied Biosystems) at the University of Tennessee's Molecular Biology Resources Facility. Phage peptide sequences were aligned using CLUSTALW (29).
11-1F4 Binding and Competition Assays
The relative strength of mAb 11-1F4 binding with clonal phage, peptides, and rVL Len was determined by EuLISA (15, 30) in the presence or absence of NaSCN (31). The NaSCN assay involved coating 5 × 1010 phage (as determined from A269nm (28)) in PBS onto each well of a high-binding microtiter plate (EIA/RIA Easy Wash, ™COSTAR, Corning, NY) which was covered and incubated overnight at 37 °C. The plate was washed x3 with PBST, blocked with 1% BSA in PBS, and a solution of 1 nM 11-1F4 in assay buffer (blocking buffer containing 0.05 % Tween 20) added. After a 1-h incubation and a washing step, 0.7 M NaSCN in 50 mM phosphate, pH 7.5, was added and the plate incubated for 15 min at 37 °C, followed (after washing) by sequential addition of biotin-labeled goat antimouse IgG (Sigma), Eu3+-streptavidin conjugate (Perkin Elmer), and low-pH Eu3+ releasing enhancement solution (Perkin Elmer) (15). For Len and phage peptide competition EuLISAs, up to 1 mM polypeptide competitor in solution with 2 nM 11-1F4 mAb was included. In the case of Jto and Aβ40, these analyses involved preincubation of 500 nM 11-1F4 with ~0.2 mg/ml competitor for 40 min at 37 °C.
All microtiter plate experiments were done in triplicate in assay buffer (1% BSA in PBS containing 0.05% sodium azide and 0.05% Tween 20). Immediately prior to immobilization, amyloid fibrils and collagen stocks were sonicated (2 × 30 sec bursts) with a probe sonic disruptor (Teledyne/Tekmar, Mason, OH). The error bars in figures represent SD. Values for EC50 and IC50 were determined from sigmoidally fit antibody binding curves (SigmaPlot 2000 ver. 6; Systat Software, Inc.).
Results
Phage Peptides have Position-Specific Sequence Identity with VL Len
Alignment of the amino acid sequence of VL Len with 11-1F4-binding phage peptides indicated that the epitope recognized by this mAb was located within the first 14 residues (Figure 1). With rare exception, the peptides contained at least 1 proline corresponding to Pro8 in VL Len and one half had a second proline. Based on these results, 11-1F4-binding peptides were grouped into two families: -X-Pro-X- or -Pro-X-Pro- (Figure 1, Groups I and II).
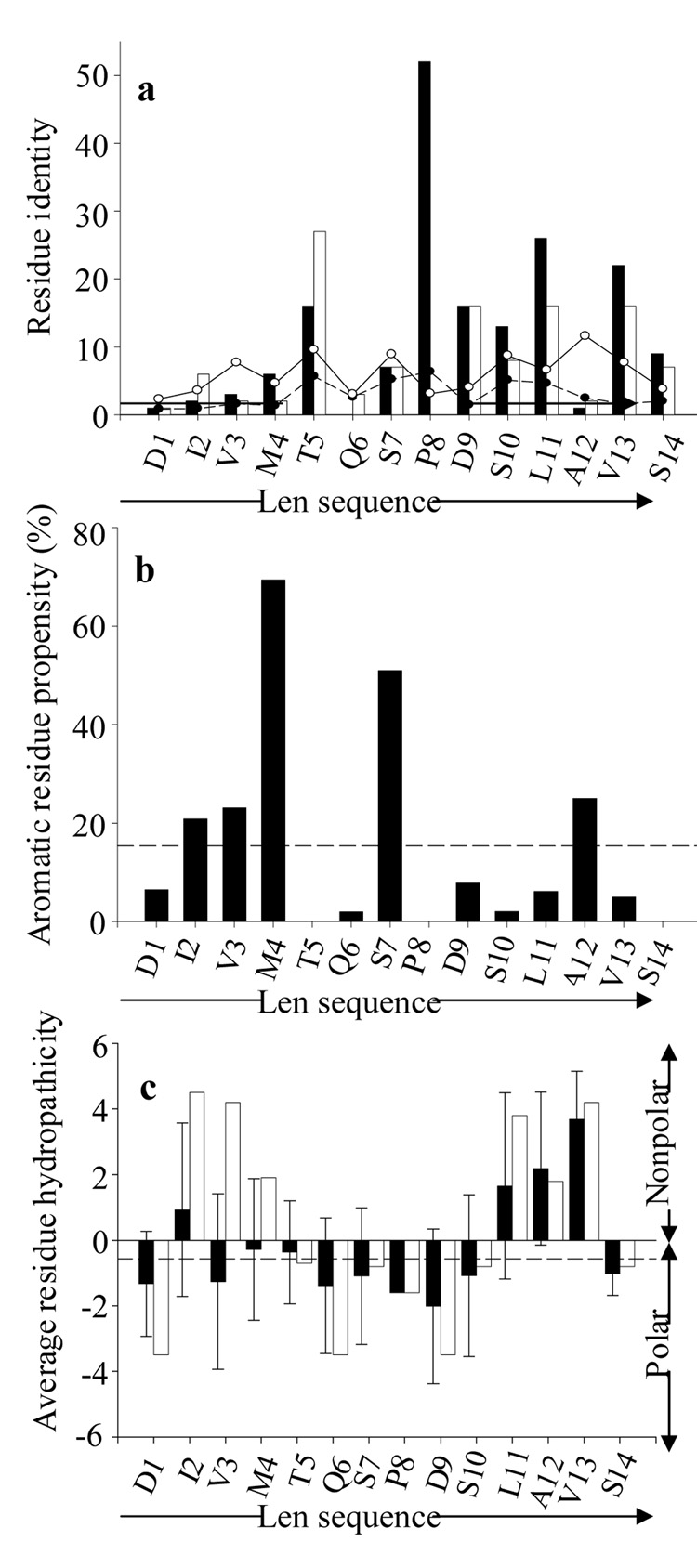
Analyses of consensus sequences for phage peptides assigned to Groups I or II (50% and 70% threshold values, respectively) showed that there was a high proportion of identical or physiochemically similar amino acids (32) at 10 of the first 14 positions of protein Len (Figure 1 and Figure 2a), with maximum conserved identity occurring at positions 5, 8, 9, 11, and 13 (Thr, Pro, Asp, Leu, and Val, respectively). This correlation was greater than would be expected, based on the observed frequency of amino acids in the random peptide phage display library (http://www.neb.com/nebecomm/ManualFiles/ manualE8110.pdf). The overall consensus sequence, incorporating peptides from both Groups I and II, revealed that the region of greatest homology spanned a 6-polar residue stretch between positions 5 and 10 (Figure 1). Similarity also was observed within the first 4 positions of the phage peptide sequences that contained a high percentage of aromatic resides at position 4; however, these amino acids were dissimilar to those found in the Len molecule (Figure 1 and Figure 2a, b). Three additional positions -- 6, 7, and 12 (Gln, Ser, and Ala) -- also exhibited considerable sequence heterogeneity, with a predominance of aromatic residues at position 7.
FIGURE 2.
Sequence-position comparison of 11-1F4-binding phage peptides with the first 14 residues of VL Len. (a) Residue identity between phage peptides and VL Len; the number of identical (closed bars) and chemically similar (open bars) residues for each position are shown. The solid and open circles indicate the number of identical and chemically similar phage peptide residues expected to occur by chance, based on the observed frequency of amino acids in the random peptide phage display library (http://www.neb.com/nebecomm/ManualFiles/manualE8110.pdf). (b) Percentage of aromatic residues in the phage peptides at each position. The line dashed represents the percentage abundance of aromatic amino acids relative to all other residues in the random peptide phage display library. (c) Average residue hydropathicity values (33) for phage peptides (closed bars) relative to those of the Len(1–14) peptide (open bars). Each sequence position average hydropathicity value was determined from the sum of residue hydropathicity values divided by the number of total residues. The dashed line shows the average hydropathicity value for all 20 amino acids, corrected for the abundance of each in the random peptide phage display library. All sequence-position comparisons were determined using the multiple sequence alignments shown in Figure 1.
In contrast to VL Len, the presence of more polar residues within the first 4 positions of the phage peptides resulted in a hydrophilic N-terminal (Figure 2c). Further characterization of their physiochemical properties, such as pI, electrostatic charge, stability, aliphaticity, and hydropathicity (http://us.expasy.org/cgi-bin/protparam) (33), indicated that these molecules were more basic [pI values = 5.60 ± 1.5 vs 3.56 for Len(1–15)].
The 11-1F4 Epitope is Within the First 15 Residues of VL Len
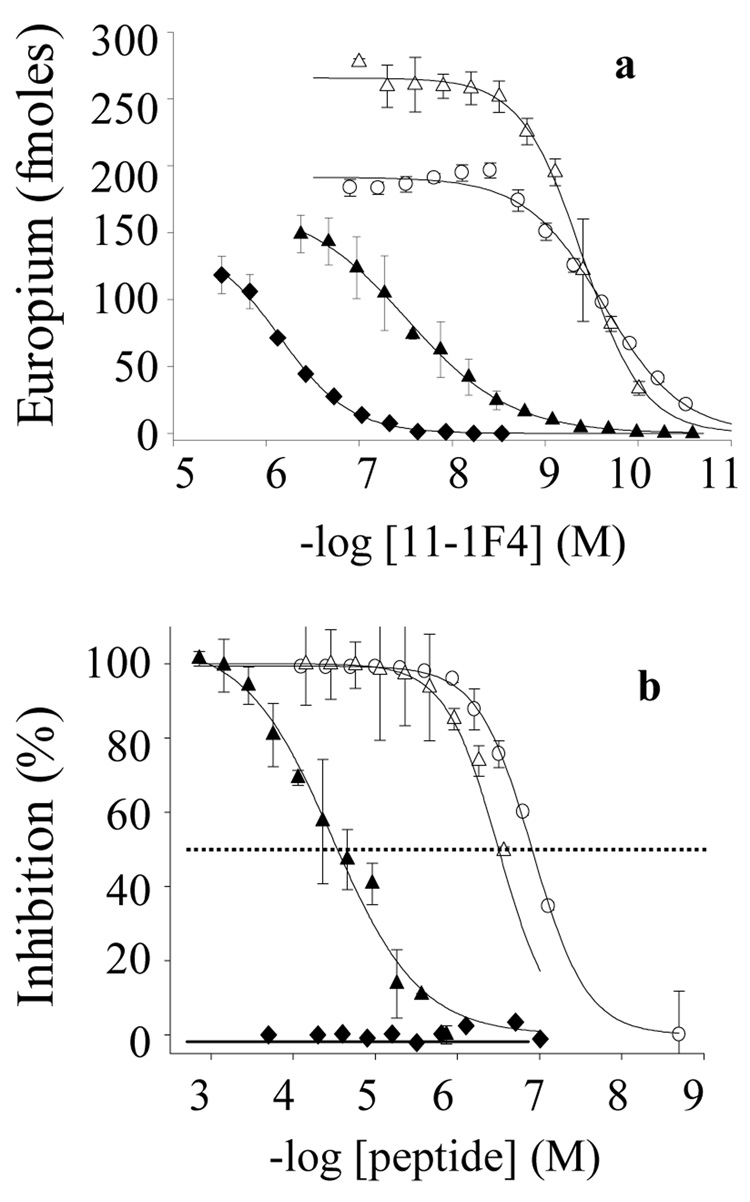
mAb 11-1F4 bound equally to plate-immobilized VL Len and Len peptides encompassing the first 15 residues, with EC50 values in the subnanomolar range (Figure 3a, Table 1). In contrast, the reactivity of 11-1F4 with Len(1–13) was ~50× weaker, thus indicating the importance of Ser14 and/or Leu15 for this interaction. Consistent with these observations, both the Len(1–22) and Len(1–15) peptides were similarly effective at inhibiting 11-1F4 binding to plate-immobilized Len(1–22), whereas Len(1–13) was ~30× less effective (Figure 3b, Table 1).
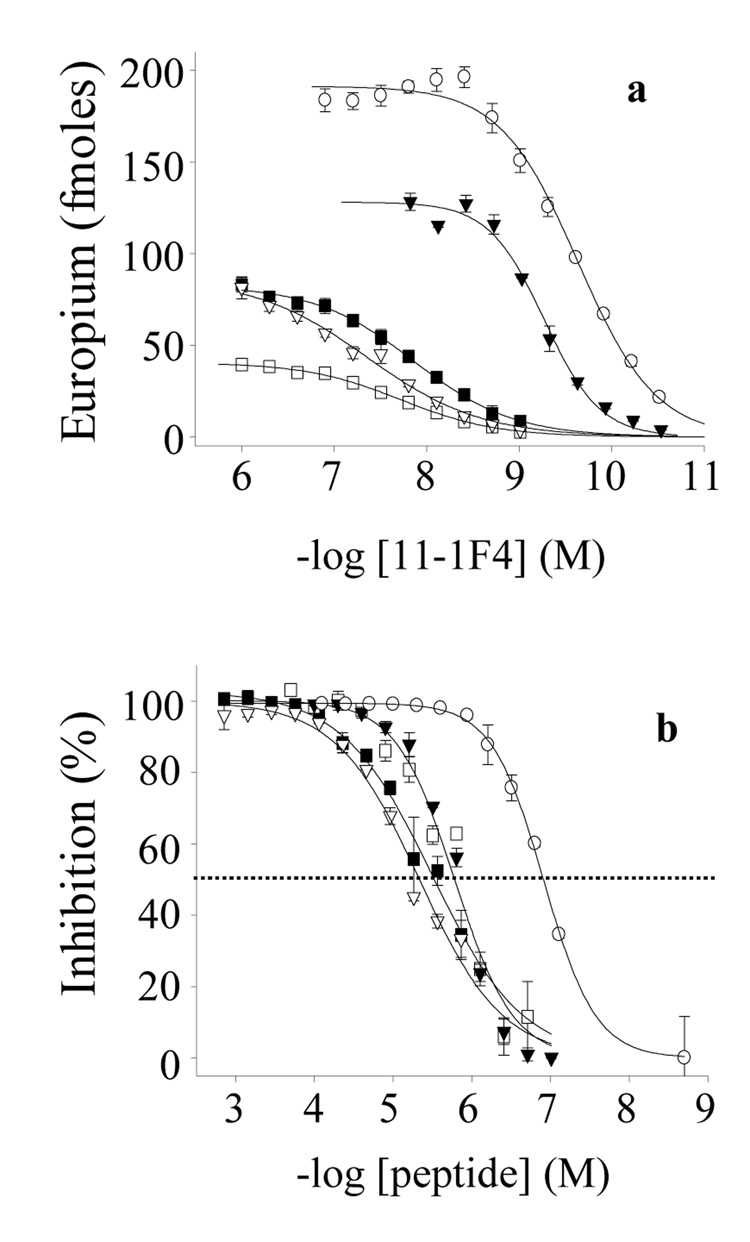
FIGURE 3.
Binding of mAb 11-1F4 to plate-immobilized Len peptides. (a) Binding of 11-1F4 to Len peptides. (b) Competitive inhibition of 2 nM 11-1F4 binding to Len(1–22) in the presence or absence of Len peptides. Len(1–15), (△); Len(1–13), (▲); LenP8S(1–22), (♦); and Len(1–22), (○). The dashed line indicates 50% inhibition.
Table 1.
11-1F4 binding to plate immobilized phage and Len peptides and the inhibitory effects of these molecules on antibody binding to immobilized Len(1–22).
| Immobilized Polypeptide | Solution-Phase Competition | ||||
|---|---|---|---|---|---|
| Polypeptide | Phage Peptide Groupa | Sequenceb | EC50c(nM) | NaSCN assay IC50d(µM) | IC50e(µM) |
| Len VL | n.a.f | DIVMTQSPDSLAVSLGERATIN… | 0.18 ± 0.03 | 3.9 | ≫30 |
| a12 | I | K-HYAAFPENLLI | 0.54 ± 0.3 | 3.7 (3.9) | 1.5 ± 0.1 |
| g4 | I | -VQYSSSPEALRV | 14 ± 6.0 | 3.7 (4.0) | 3.2 ± 0.0 |
| f11 | II | -IVYTPHPEYLW-P | 32 ± 0.1 | 3.3 (3.9) | 6.7 ± 0.1 |
| b2 | II | TYQYSPYPEPVM | 44 ± 0.1 | 0.5 (0.5) | 4.6 ± 0.1 |
| a7 | II | KPT-YSPYPFSM--P | 18 ± 0.1 | 2.3 (0.5) | 1.7 ± 0.1 |
| Len(1–22) | n.a. | DIVMTQSPDSLAVSLGERATIN | 0.24 ± 0.1 | 1.37 | 0.07 ± 0.01 |
| Len(1–15) | n.a. | DIVMTQSPDSLAVSL | 0.39 ± 0.05 | n.d.g | 0.27 ± 0.01 |
| Len(1–13) | n.a. | DIVMTQSPDSLAV | 16 ± 0.1 | n.d. | 33 ± 0.07 |
Phage peptides were grouped based on the presence of highly conserved proline residues, as shown in Fig. 1.
Peptide sequences were aligned by CLUSTALW (29); hydrophobic and/or aromatic residues are bolded and proline is underlined.
Each EC50 value was determined from the midpoint of 11-1F4 titration curves, as shown in Figure 4a and Figure 7a.
Each IC50 value was determined from the midpoint of NaSCN titration curves (see Figure 3a and b). IC50 values with or without brackets were determined for 11-1F4 binding to phage peptides alone or expressed on phage particles, respectively.
Each IC50 value was determined from the midpoint of peptide competition curves shown in Figure 4b and Figure 7b.
n.a., not applicable.
n.d., not determined.
11-1F4 Binding to Non-Native VL Len, Len LC, Len Peptides, and Phages
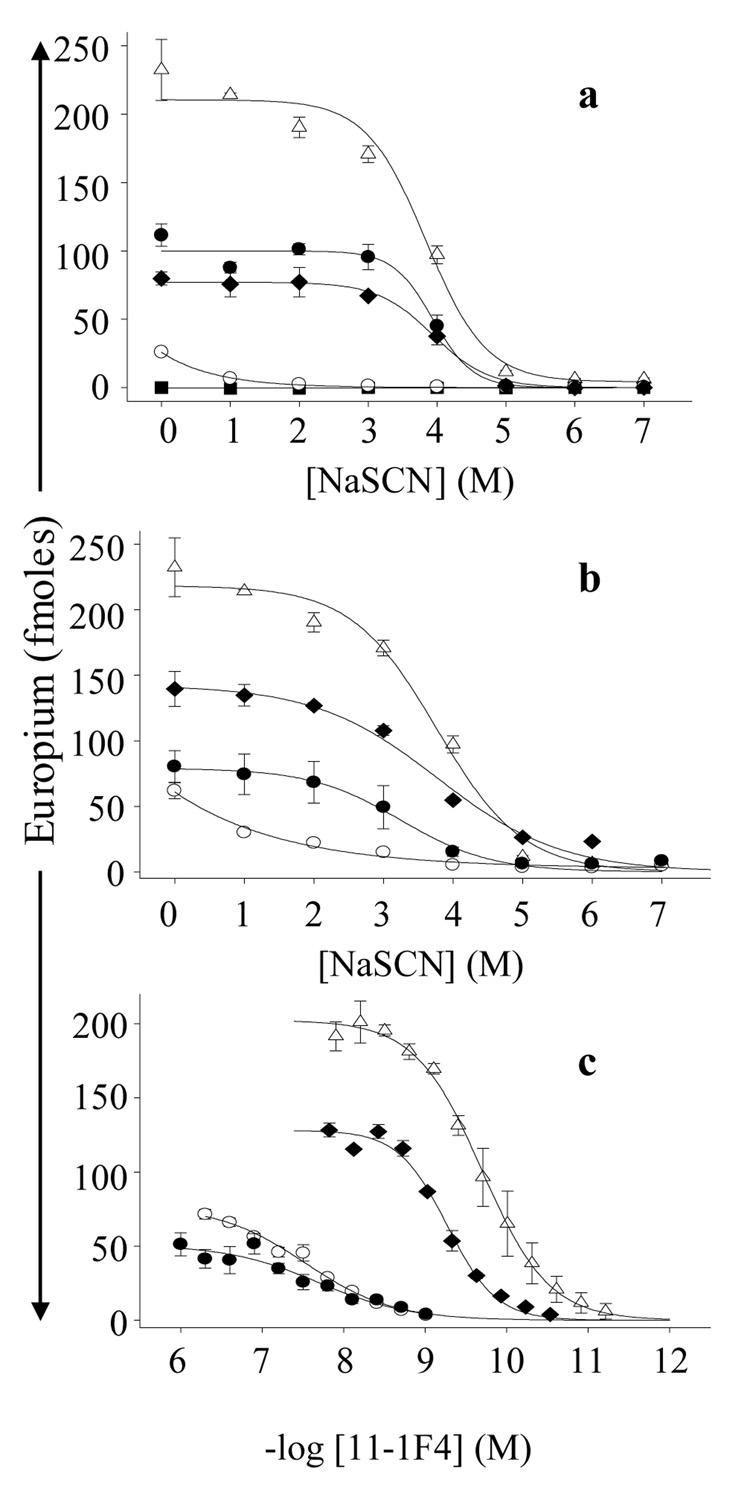
A broad range of IC50 values (0.5 - <0.01 M) were obtained when ~50 11-1F4-binding phages were evaluated (Figure 1). Notably, the strongest antibody complexes (IC50, ~4 M) occurred with phages a12, f11, c3, c4, g4, f5, and h3 (Figure 1 and Figure 4a). In contrast, the b2 phage and several others that were isolated based on positive interactions with the antibody bound weakly in the presence of NaSCN. As expected, no or very weak binding was observed with polyclonal phages from the unselected library. Presumably, the lower assay signal that was found for 11-1F4 binding to the immobilized phage (as compared to that for protein Len) reflected differences in epitope density that resulted from antigen immobilization.
FIGURE 4.
11-1F4-binding to plate-immobilized phage, VL Len, and synthetic phage peptides. NaSCN titration curves for antibody binding to (a) phage, protein Len, or (b) synthetic phage peptides. (c) 11-1F4 titration curves for binding to synthetic phage peptides. IC50 and EC50 values were determined from the midpoint of each titration curve. Len, (△); f11, (●); b2, (○); and a12, (♦) phage particles or peptide; and unamplified phage display library control (■).
To confirm that mAb 11-1F4 reactivity with the phage was via the 12-mer peptide mimetics expressed on its surface, synthetic peptides representing high-affinity (a12, g4, and f11) or weak binding (b2 and a7) phage were tested in the NaSCN assay. With the exception of the a7 molecule, the antibody reacted with the isolated peptides in a fashion similar to that of the entire phage. Three -- a12, g4 and f11 -- bound the mAb 11-1F4 in the presence of ~3.5 M NaSCN, comparable to that seen when VL Len was used as substrate (Figure 4a, b, Table 1). In contrast, the a7 synthetic peptide interacted with 11-1F4 at much greater concentrations of chaotrope, as compared to the phage-expressed a7 derivative (IC50, 2.3 M vs 0.54 M), indicating an increase in stabilizing hydrophobic and/or electrostatic interactions between the antibody and the synthetic peptide (Table 1).
Consistent with results obtained using the NaSCN-titration assay, antibody binding curves showed that mAb 11-F4 bound with similar affinity to both the a12 synthetic peptide and VL Len with EC50s of 0.5 and 0.2 nM, respectively (Figure 4c). However, antibody binding to the a7, g4, and f11 synthetic peptides was much weaker than with the respective phage and was comparable to the interaction with the b2 peptide that formed a relatively weak complex with 11-1F4 in the presence of NaSCN (Figure 4c, Table 1). Presumably, these assay-related discrepancies reflect differences in the methods used to measure antibody binding.
To ascertain if the epitope recognized by mAb 11-1F4 was retained when the synthetic phage peptides were in solution, their ability to inhibit the reactivity of 11-1F4 with plateimmobilized Len(1–22) was determined. These studies showed that each had a similar capacity with IC50s in the 1.5 – 6.7 µM range (Figure 5, Table 1), values ~20-fold higher than that for Len(1–22); in contrast, the native protein Len failed to suppress the reaction (IC50, > 30.0 µM).
FIGURE 5.
Binding of mAb 11-1F4 to plate-immobilized Len(1–22) and phage peptides in the presence and absence of peptide inhibitors. (a) Binding of 11-1F4 to phage peptides or Len(1–22). (b) Competitive inhibition of 2 nM 11-1F4 binding to Len(1–22) in the presence of phage peptides or Len(1–22). The dashed line indicates 50% inhibition. a12, (▼); g4, (■); b2, (▽); a7, (□); and Len(1–22), (○).
Alanine Scanning Mutagenesis
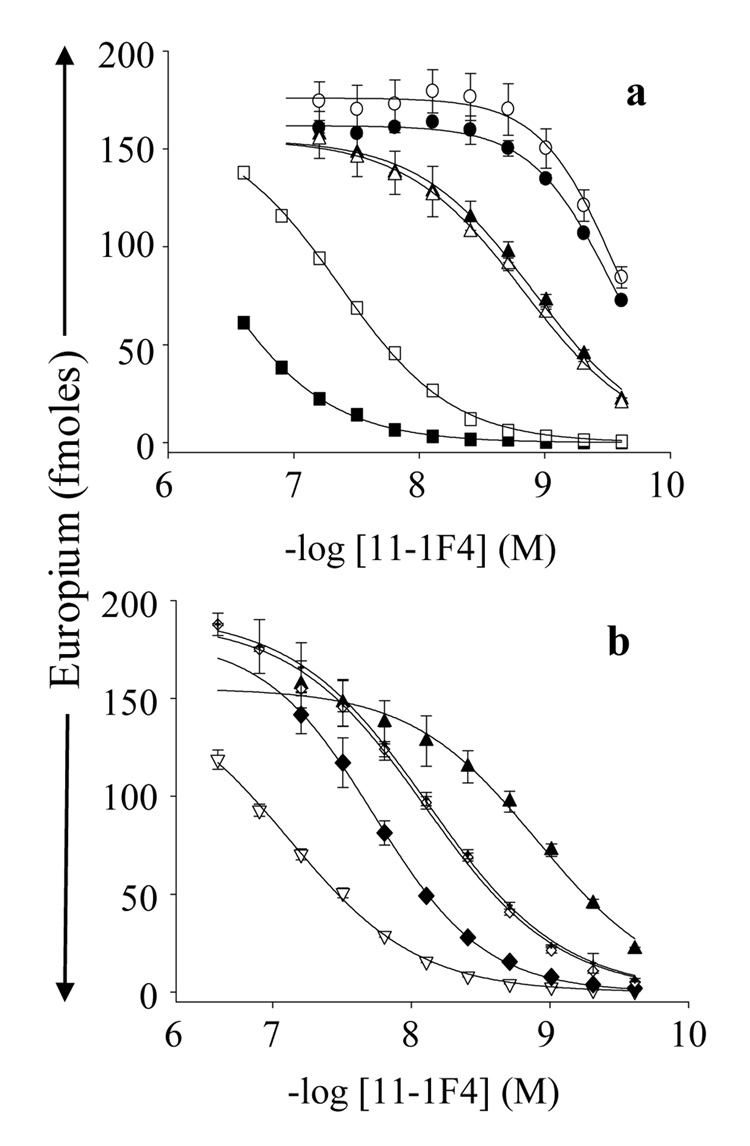
The substitution of alanine for proline, asparagine, leucine, or valine at positions, 8, 9, 11, and 13, respectively, markedly hindered the interaction of the Len(1–18) with mAb 11-1F4, yielding ~20-400-fold higher EC50 values (Figure 6, Table 2). The greatest effect was observed with Pro8Ala. These results were consistent with the significant sequence identity that occurred at these 4 positions in the phage peptides and VL Len, as well as our previous finding that 11-1F4-binding to Len peptides was drastically reduced by the substitution of serine for proline at position 8 (15). In addition, the relatively large decrease in antibody binding with Asp9Ala, Leu11Ala, and Val13Ala substitutions correlated with the lack of an alanine residue at these corresponding positions in the phage peptides (Table 2).
FIGURE 6.
Interaction of mAb 11-1F4 with plate-immobilized, Ala-substituted Len(1–18) peptides. (a) Binding of 11-1F4 to Len(1–18), (▲); IA2, (○); V3A, (●); T5A, (Δ); D9A, (□); and P8A, (■). (b) Antibody binding to Len(1–18), (▲); I11A, (▽); V13A, (♦); S14A, (◊); and I15A, (×).
Table 2.
Peptide phage display and alanine scanning analysis of N-terminal VL Len residues implicated in 11-1F4 binding.
| VL Len residue position | Phage peptide | Len(1–18) Alanine scanning | Phage and Len(1–18) Residue Preferencesd | ||
|---|---|---|---|---|---|
| Identitya(%) | Ala (%) | EC50b(nM) | EC50 WT EC50c | ||
| D1 | 3.8 | 0 | 3.91 ± 0.001 | 2.7 | S or T |
| I2 | 15.4 | 19.2 | 0.27 ± 0.001 | 0.22 | Ala or any amino acid |
| V3 | 9.6 | 0 | 0.29 ± 0.004 | 0.24 | Polar |
| M4 | 15.4 | 0 | 2.51 ± 0.01 | 2.1 | Aromatic (preferably Tyr)e |
| T5 | 82.7 | 10.0 | 1.48 ± 0.01 | 1.2 | Small (preferably S or T) |
| Q6 | 5.8 | 4.0 | 2.33 ± 0.01 | 1.9 | Polar |
| S7 | 26.9 | 8.0 | 0.62 ± 0.01 | 0.51 | Polar (preferably aromatic) |
| P8 | 100 | 0 | >500 ± 10 | >400 | Invariant proline |
| D9 | 61.5 | 2.0 | 43.2 ± 0.2 | 36 | Negatively charged, D/E |
| S10 | 40.4 | 6.3 | 0.85 ± 0.003 | 0.71 | Small polar (preferably S or T) |
| L11 | 80.8 | 0 | 77.1 ± 0.7 | 64 | L or bulky hydrophobic |
| A12 | 5.8 | 0 | 2.86 ± 0.02d | 2.4 | Bulky hydrophobic |
| V13 | 73.0 | 2.6 | 18.6 ± 0.1 | 16 | V or aliphatic |
| S14 | 30.7 | 0 | 7.96 ± 0.04 | 6.6 | S or T |
| L15 | n.a.f | n.a. | 7.64 ± 0.02 | 6.4 | (L) |
| G16 | n.a. | n.a. | 2.12 ± 0.01 | 1.8 | n.a. |
| E17 | n.a. | n.a. | 2.48 ± 0.01 | 2.1 | n.a. |
| R18 | n.a. | n.a. | 0.55 ± 0.002 | 0.5 | n.a. |
Sequence identity for each phage peptide compared to VL Len was determined as the sum of the percentages of identical and chemically similar (32) residues at each position from the multiple sequence alignments of phage peptide groups I and II shown in Figure 1.
EC50 values were determined from the midpoint of 11-1F4 titration curves shown in Figure 6.
WT EC50 is the EC50 value for 11-1F4 binding to plate-immobilized wild-type Len(1–18).
Residue preference at each position in Len(1–18) was determined from the multiple sequence alignments of phage peptide groups I and II with VL Len, and the binding of 11-1F4 to Alasubstituted Len(1–18) peptides (Figure 1, Figure 2, and Figure 6). Residue preferences with or without brackets are those that were preferred for Len and phage peptides, respectively.
Residue preference in brackets are those specific to the Len(1–18) peptide.
n.a., not applicable.
The substitution of alanine for any of the first 4 residues of Len(1–18) had little effect on 11-1F4 binding (Table 2); however, stronger interactions occurred with the Ile2Ala and Val3Ala variants that generated EC50 values ~4× greater than that for the wild-type peptide (Figure 6, Table 2). Increased antibody binding to the Ile2Ala variant was in agreement with the frequent occurrence (19.2%) of alanine at this position in the phage peptides (Table 2). In contrast, alanine was never observed in the location equivalent to Val3. Instead, polar residues were abundant, including a high percentage of aromatic amino acids (Table 2), thus suggesting that these differences reflected variations in side-chain usage for maintaining the conformational epitope recognized by 11-1F4 in the Len and the phage-derived peptides.
Several other variations were observed between the Ala scanning and phage peptide alignment analyses (Table 2). The Leu15Ala substitution in Len(1–18) resulted in ~6× weaker peptide interaction with 11-1F4, implying an important role of leucine for this reactivity; however, this residue was absent at this position in the 11-1F4 binding phage peptides (Figure 1). In addition, the Thr5Ala and Ser10Ala substitutions in Len(1–18) had little effect on antibody binding, although there was a preference for hydroxyl side chains at these positions in the phage peptides (Figure 2 and Figure 7, Table 2).
FIGURE 7.
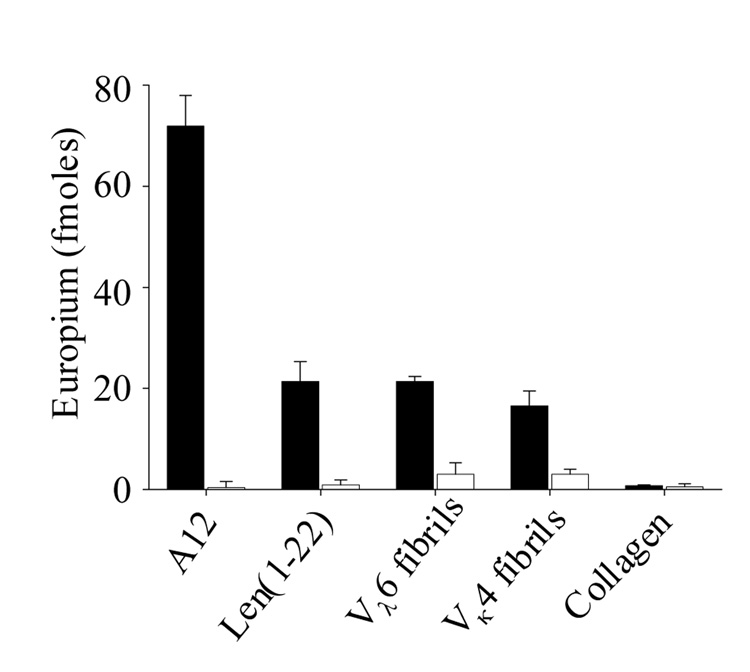
Characterization of the specificity of anti-phage peptide immune serum. Reactivity of serum from an A12-immunized mouse (solid bars) and non-immunized mouse (open bars) against the immunogen, Len(1–22), as well as VL Len, and collagen fibrils.
Immunizing Mice with the a12 Peptide Generated Anti-Light Chain Fibril Antibodies
To validate that the phage peptides mimicked the structure of the fibril-associated epitope recognized by 11-1F4, mice were immunized with KLH-conjugated a12 peptide. The resultant immune sera contained antibodies that had reactivity similar to that for mAb 11-1F4; namely, they had specificity for the a12 and Len(1–22) peptides, as well as synthetic fibrils composed of Vκ6 Wil and Vλ4 Len (Figure 7). Neither 11-1F4 nor the a12-immune serum recognized non-amyloid triple-helix collagen fibrils. In addition, sera from non-immunized mice were virtually unreactive with the test molecules.
11-1F4 Binding to Aβ40 Fibrils
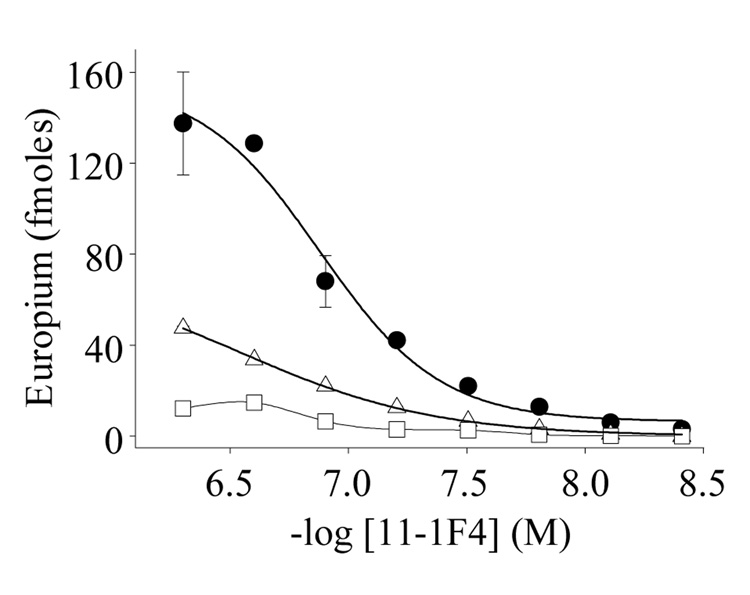
The reactivity of mAb 11-1F4 with Aβ40 fibrils was demonstrated by EuLISA. As shown in Figure 8, the EC50 of this interaction was ~100 nM, a value comparable to that obtained with LC fibrils (15). To determine if the binding could be inhibited by LC fibrils, we carried out competition studies where we found that sonicated Vλ6 (Jto) and Aβ40 fibrils, at 50-fold excesses, were equally potent as inhibitors, indicating that the antibody used the same paratope for binding to heterologous fibrils.
FIGURE 8.
Competitive inhibition of mAb 11-1F4 binding to Aβ40 fibrils. Binding of antibody to plate-immobilized Aβ40 fibrils in the absence of competitor (●) or in the presence of Jto fibrils (□) or Aβ40 fibrils (△). Competition experiments were performed by pre-incubating 11-1F4 with ~0.2 mg/ml fibrils.
DISCUSSION
The aggregation of amyloidogenic proteins into insoluble fibrils is associated with a series of conformational transitions that result in the exposure of neo-antigenic sites that are associated with amyloid fibrils and assembly intermediates (10–14, 16, 34, 35), as well as the loss of native epitopes (36, 37). We previously had demonstrated that mAb 11-1F4 bound to a cryptic epitope present on κ and λ LC amyloid fibrils, as well as partially denatured precursor proteins, but did not when these proteins were in their native state (15, 19). Using overlapping peptide mapping techniques, the epitope was localized within the first 18 amino acids of the Vκ4 Len immunogen. We hypothesized that the LC fibril binding site involved an edge-strand perturbation, similar to the neo-epitope formed by TTR fibrils (38). Additionally, the requirement of Pro8 (conserved within virtually all κ and λ LCs (39, 40)) for this interaction to occur led to our postulation that this residue anchored a β-turn and led to a peptide chain reversal, analogous to that formed from trans-proline in β2-microglobulin fibrils (41, 42). To understand more fully the molecular basis of mAb 11-1F4’s reactivity with both κ and λ LCs, as well as the nature of the fibril-specific epitope recognized by this reagent, we probed the structural characteristics of this determinant using peptide phage display and peptide epitope mapping.
Our studies indicated that 11-1F4 binding to Vκ4 Len and non-κ4 light chains was reliant on an invariant proline and up to 9 other residues (Asp1, Ile2, Val3, Met4, Asp9, Leu11, Val13, Ser14, and Leu15). We attribute the stronger 11-1F4 interactions with κ4 fibrils, as compared with those formed from other LC isotypes (EC50s, 0.2 nM vs. ~ 100 nM, respectively (15)), to the presence of an Asp at position 9, given that the single germline gene that encodes Vκ4 proteins specifies this amino acid at this location (39); additionally, the Asp9 substitution in Len(1–18) resulted in a 60-fold weaker antibody binding with an EC50 value similar to that for non-κ4 LCs. Further, ~60% of the 11-1F4-binding phage peptides contained a negatively charged amino acid (Asp or Glu) at position 9.
Inspection of germline Vκ and Vλ N-terminal consensus sequences revealed identity, compared to Vκ4 LCs, at 9 positions (1, 4, 5, 6, 8, 10, 11, 12, and 14) that we had implicated in mAb 11-1F4 binding (Figure 9a). Notably, this antibody reacts with similar affinity to all non-κ4 AL fibrils, partially denatured LCs, and non-LC fibrils (12, 15). Presumably, this reflects an ability to accommodate significant side-chain diversity within the 11-1F4 binding site that we attribute to an unusually large pocket resulting from an uncommonly short (i.e., 2-residue) heavy chain CDR3 (C. Dealwis and A.S., unpublished data).
FIGURE 9.
Consensus sequence analyses and schematic representation of the conformational fibril-associated epitope recognized by mAb 11-1F4. (a) Comparison of Vκ4 with non-κ4 germ line consensus sequences (bolded residues are hydrophobic.). Abbreviations used in consensus sequences are: h, hydrophobic (A, C, F, I, L, M, V); o, S or T; p, polar (D, E, G, H, K, N, P, Q, R, S, T, W, Y); s, small residues (A, C, D, G, N, P, S, T, V), and u, minute residues (A, C, G, S). Consensus sequences were determined from VL germline sequences using CONSENSUS (http://us.expasy.org/tools/) analysis of CLUSTALW (28) multiple aligned sequences with 70% threshold. (b) Proposed model for the conformational epitope recognized by 11-1F4 involves a rearrangement of the first 9 VL residues centered on a cis-proline type VI β-turn or touch-turn induced by partial denaturation via surface adsorption or by fibrillogenesis. The N-terminal 15 residues are bolded.
Phage display analyses and alanine-scanning mutagenesis of the Len(1–18) indicated that mAb 11-1F4 specificity depended primarily on the presence of an invariant prolyl residue at position(s) 7 and/or 8. The requirement of a ubiquitous proline for 11-1F4 binding is unlikely to be due to direct interactions with this residue, since the antibody does not recognize proline-rich collagen (15) (Figure 7). Antibody binding also was highly dependent on the presence of a leucine and valine at positions 11 and 13, respectively. Notably, there were high numbers of Leu11 and Val13 within the 11-1F4-binding phage peptides and these residues also are specified by Vκ2, Vκ4, and Vλ7 germline genes (39).
Although all κ LCs exhibit significant sequence identity with the first 4 residues of protein Len (a region important for 11-1F4 binding (15)), λ proteins are dissimilar. Nevertheless, these differences did not affect antibody reactivity since 11-1F4 bound equally to κ (non-κ4) and λ LCs (15). The ability of 11-1F4 to tolerate LC side-chain diversity within this region was demonstrated by the formation of stable antibody-phage complexes that occurred with phage peptides containing an N-terminal glutamine or serine (residues typically found in λ LCs, in contrast to aspartic or glutamic acid common to κ proteins). Additionally, the presence of an aromatic amino acid at the third position in ~20% of the 11-1F4-binding phage peptides (and present in one-third of all λ proteins) indicates the capability of this antibody to accommodate side-chain diversity within this region.
Confirmation of the Structure of the Neo-Epitope Bound by 11-1F4
Based on x-ray crystallographic analysis of the VL dimer Len (43), we posited that 11-1F4 bound a non-native structure present within the first 18 LC residues (15) and that this interaction required a peptide chain reversal that moved a portion of strand A (4.1) adjacent to strand B (4.2), following the formation of a Pro8-anchored type I or cis-Pro8 type VI-β turn (44) or touch-turn (45). In native LCs, these residues are involved in β-sheet formation and, further, a cis-Pro8 anchors an open turn or bend within strand A (Figure 9b); thus, a chain reversal would reposition the first 4 residues, shown to be critical for maintaining antibody-binding, in close proximity to the equally essential amino acids between positions 11 and 15.
The phage peptide analyses provide further support for a β-turn epitope, as evidenced by the characteristic distribution of amino acids next to the ubiquitous central prolyl residue. Such turns generally consist of a stretch of polar residues flanking a central proline(s) which, in turn, is bordered on either side by amino acids frequently found in the β-strands of β-hairpin peptides (46, 47). Additionally, the clustering of aromatic and hydrophobic residues at the N-and C-termini of the phage peptides presumably provides the driving force for β-hairpin folding (48). Our experimental data also indicate that the epitope is likely to be anchored by a type VI β-turn or touch turn, since the unusually high propensity for an aromatic residue to precede the central proline in the phage peptides is consistent with a cis-proline conformation (49–51). Additionally, the large decrease in 11-1F4 binding that occurred with the substitution of proline for alanine at position 8 of Len(1–18) is consistent with a cis-proline β-turn, as such a dramatic effect would not occur in β-turns anchored by a trans-prolyl residue (44, 45). The presence of 2 adjacent prolines, which are found in rare type VI β-turns (52–54), also was seen in ~50% of all phage peptide sequences and occurs in the majority of proteins encoded by Vλgenes. We posit that mAb 11-1F4’s unusually large binding pocket enables interaction with Aβ fibrils, which, although lacking a prolyl residue, putatively contain a type I β-turn (55).
Generation of Pan-LC Fibril-Reactive Antibodies Using the a12 Mimotope
The finding that the a12 peptide is a mimotope (56) (i.e., induced a murine antibody response that mimicked 11-1F4 cross-reactivity) suggests that this molecule may serve as a vaccine to generate therapeutic antibodies in patients with AL amyloidosis. Recently, phage peptide mimotopes also have been identified against conformational epitopes on Aβ42 fibrils (57); presumably, this class of molecules has several advantages over immunization with soluble or fibrillar amyloidogenic proteins, including enhanced immunogenicity compared to self-antigens and reduced toxicity. Additionally, it is more likely that a pan-LC fibril-reactive antibody response that is not biased towards a κ or λ isotype would be obtained using the a12 peptide.
Our future efforts will be devoted towards the generation of pan-LC fibril-reactive mAbs using the a12 immunogen and confirmation of the structure of the neo-epitope bound by mAb 11-1F4 through x-ray crystallographic studies of an 11-1F4-Len(1–15) complex, as well as determination of the solution structure of wild-type and chemically modified Len(1–15) (with stabilizers of the central cis-proline (58, 59)) using CD, NMR, and/or FRET analysis. These studies should provide the necessary information for generation of novel reagents to be used as therapeutic and/or diagnostic tools in patients with AL amyloidosis.
Footnotes
This work was supported in part by the University of Tennessee Medical Center’s Physicians Medical Education and Research Foundation, a NIBIB/NINDS Bioengineering Research Partnership Award EB 00789, USPHS Research Grant CA 100506 from the National Cancer Institute, and the Aslan Foundation. A.S. is an American Cancer Society Clinical Research Professor.
Abbreviations: Aβ, amyloid β peptide consisting of the first 40 amino acids; AL amyloidosis, light chain-associated amyloidosis; LC, light chain; EuLISA, europium (Eu3+)-linked immunosorbant assay; mAb, monoclonal antibody; PBSA, phosphate buffered saline containing 0.05% sodium azide; ThT, thioflavin T; VL, light chain variable region.
REFERENCES
- 1.Comenzo RL. Current and emerging views and treatments of systemic immunoglobulin light-chain (Al) amyloidosis. Contrib. Nephrol. 2007;153:195–210. doi: 10.1159/000096768. [DOI] [PubMed] [Google Scholar]
- 2.Stefani M. Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim. Biophys. Acta. 2004;1739:5–25. doi: 10.1016/j.bbadis.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Westermark P, Benson MD, Buxbaum JN, Cohen AS, Frangione B, Ikeda S, Masters CL, Merlini G, Saraiva MJ, Sipe JD. Amyloid: toward terminology clarification. Report from the Nomenclature Committee of the International Society of Amyloidosis. Amyloid. 2005;12:1–4. doi: 10.1080/13506120500032196. [DOI] [PubMed] [Google Scholar]
- 4.Ross CA, Poirier MA. Protein aggregation and neurodegenerative disease. Nat. Med. 2004 10 Suppl:S10–S17. doi: 10.1038/nm1066. [DOI] [PubMed] [Google Scholar]
- 5.Makin OS, Atkins E, Sikorski P, Johansson J, Serpell LC. Molecular basis for amyloid fibril formation and stability. Proc. Natl. Acad. Sci. U.S.A. 2005;102:315–320. doi: 10.1073/pnas.0406847102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Makin OS, Serpell LC. Structures for amyloid fibrils. FEBS J. 2005;272:5950–5961. doi: 10.1111/j.1742-4658.2005.05025.x. [DOI] [PubMed] [Google Scholar]
- 7.LeVine H., 3rd Thioflavine T interaction with synthetic Alzheimer's disease beta-amyloid peptides: detection of amyloid aggregation in solution. Protein Sci. 1993;2:404–410. doi: 10.1002/pro.5560020312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Westermark GT, Johnson KH, Westermark P. Staining methods for identification of amyloid in tissue. Methods Enzymol. 1999;309:3–25. doi: 10.1016/s0076-6879(99)09003-5. [DOI] [PubMed] [Google Scholar]
- 9.Dumoulin M, Dobson CM. Probing the origins, diagnosis and treatment of amyloid diseases using antibodies. Biochimie. 2004;86:589–600. doi: 10.1016/j.biochi.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 10.Franklin EC, Zucker-Franklin D. Antisera specific for human amyloid reactive with conformational antigens. Proc. Soc. Exp. Biol. Med. 1972;140:565–568. doi: 10.3181/00379727-140-36504. [DOI] [PubMed] [Google Scholar]
- 11.Glabe CG. Conformation-dependent antibodies target diseases of protein misfolding. Trends Biochem. Sci. 2004;29:542–547. doi: 10.1016/j.tibs.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 12.Hrncic R, Wall J, Wolfenbarger DA, Murphy CL, Schell M, Weiss DT, Solomon A. Antibody-mediated resolution of light chain-associated amyloid deposits. Am. J. Pathol. 2000;157:1239–1246. doi: 10.1016/S0002-9440(10)64639-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 14.O'Nuallain B, Wetzel R. Conformational Abs recognizing a generic amyloid fibril epitope. Proc. Natl. Acad. Sci. U.S.A. 2002;99:1485–1490. doi: 10.1073/pnas.022662599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O'Nuallain B, Allen A, Kennel SJ, Weiss DT, Solomon A, Wall JS. Localization of a conformational epitope common to non-native and fibrillar immunoglobulin light chains. Biochemistry. 2007;46:1240–1247. doi: 10.1021/bi0616605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Nuallain B, Hrncic R, Wall JS, Weiss DT, Solomon A. Diagnostic and therapeutic potential of amyloid-reactive IgG antibodies contained in human sera. J. Immunol. 2006;176:7071–7078. doi: 10.4049/jimmunol.176.11.7071. [DOI] [PubMed] [Google Scholar]
- 17.Kourie JI, Henry CL. Ion channel formation and membrane-linked pathologies of misfolded hydrophobic proteins: the role of dangerous unchaperoned molecules. Clin. Exp. Pharmacol. Physiol. 2002;29:741–753. doi: 10.1046/j.1440-1681.2002.03737.x. [DOI] [PubMed] [Google Scholar]
- 18.Schneider M, Hilschmann N. The primary structure of a monoclonal immunoglobulin L-chain of kappa-type, subgroup IV (Bence-Jones protein Len). A new subgroup of the kappa-type L-chain. Hoppe Seylers Z. Physiol. Chem. 1974;355:1164–1168. [PubMed] [Google Scholar]
- 19.O'Nuallain B, Murphy CL, Wolfenbarger DA, Kennel S, Solomon A, Wall JS. The amyloid-reactive monoclonal antibody 11-1F4 binds a cryptic epitope on fibrils and partially denatured immunoglobulin light chains and inhibits fibrillogenesis. In: Grateau G, Kyle RA, Skinner M, editors. Xth International symposium on Amyloid and Amyloidosis: from molecular dissection to therapeutics. Boca Raton, Florida, USA, Tours, Loire Valley: CRC Press; 2004. pp. 482–484. [Google Scholar]
- 20.Benhar I. Biotechnological applications of phage and cell display. Biotechnol. Adv. 2001;19:1–33. doi: 10.1016/s0734-9750(00)00054-9. [DOI] [PubMed] [Google Scholar]
- 21.Wang LF, Yu M. Epitope identification and discovery using phage display libraries: applications in vaccine development and diagnostics. Curr. Drug Targets. 2004;5:1–15. doi: 10.2174/1389450043490668. [DOI] [PubMed] [Google Scholar]
- 22.Morrison KL, Weiss GA. Combinatorial alanine-scanning. Curr. Opin. Chem. Biol. 2001;5:302–307. doi: 10.1016/s1367-5931(00)00206-4. [DOI] [PubMed] [Google Scholar]
- 23.Wall J, Schell M, Murphy C, Hrncic R, Stevens FJ, Solomon A. Thermodynamic instability of human lambda 6 light chains: correlation with fibrillogenicity. Biochemistry. 1999;38:14101–14108. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- 24.O'Nuallain B, Williams AD, Westermark P, Wetzel R. Seeding specificity in amyloid growth induced by heterologous fibrils. J. Biol. Chem. 2004;279:17490–17499. doi: 10.1074/jbc.M311300200. [DOI] [PubMed] [Google Scholar]
- 25.Wall J, Murphy CL, Solomon A. In vitro immunoglobulin light chain fibrillogenesis. Methods Enzymol. 1999;309:204–217. doi: 10.1016/s0076-6879(99)09016-3. [DOI] [PubMed] [Google Scholar]
- 26.Naiki H, Gejyo F. Kinetic analysis of amyloid fibril formation. Methods Enzymol. 1999;309:305–318. doi: 10.1016/s0076-6879(99)09022-9. [DOI] [PubMed] [Google Scholar]
- 27.Levine H., 3rd Qunatification of beta-sheet amyloid fibril structures with thioflavin T methods. Methods Enzymol. 1999:274–284. doi: 10.1016/s0076-6879(99)09020-5. [DOI] [PubMed] [Google Scholar]
- 28.Bonnycastle LLC, Menendez A, Scott JK. General phage methods. In: Barbas CF, Burton DR, Scott JK, Silverman GJ, editors. Phage Display: A Laboratory Manual. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 29.Diamandis EP. Immunoassays with time-resolved fluorescence spectroscopy: principles and applications. Clin. Biochem. 1988;21:139–150. doi: 10.1016/0009-9120(88)90001-x. [DOI] [PubMed] [Google Scholar]
- 30.Goldblatt D. Simple solid phase assays of avidity. In: Johnstone AP, Turner MW, editors. Immunochemistry 2: A pratical approach. Oxford: Oxford University press; 1997. pp. 31–51. [Google Scholar]
- 31.Taylor WR. The classification of amino acid conservation. J. Theor. Biol. 1986;119:205–218. doi: 10.1016/s0022-5193(86)80075-3. [DOI] [PubMed] [Google Scholar]
- 32.Goldsteins G, Persson H, Andersson K, Olofsson A, Dacklin I, Edvinsson A, Saraiva MJ, Lundgren E. Exposure of cryptic epitopes on transthyretin only in amyloid and in amyloidogenic mutants. Proc. Natl. Acad. Sci. U.S.A. 1999;96:3108–3113. doi: 10.1073/pnas.96.6.3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Linke RP, Zucker-Franklin D, Franklin ED. Morphologic, chemical, and immunologic studies of amyloid-like fibrils formed from Bence Jones Proteins by proteolysis. J. Immunol. 1973;111:10–23. [PubMed] [Google Scholar]
- 34.Franklin EC, Pras M. Immunologic studies of water-soluble human amyloid fibrils. Comparative studies of eight amyloid preparations. J. Exp. Med. 1969;130:797–808. doi: 10.1084/jem.130.4.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan FF, Biffin S, Brazier MW, Suarez M, Cappai R, Hill AF, Collins SJ, Sullivan JS, Middleton D, Multhaup G, Geczy AF, Masters CL. Detection of prion epitopes on PrP and PrP of transmissible spongiform encephalopathies using specific monoclonal antibodies to PrP. Immunol. Cell Biol. 2005;83:632–637. doi: 10.1111/j.1440-1711.2005.01384.x. [DOI] [PubMed] [Google Scholar]
- 36.Costa PM, Teixeira A, Saraiva MJ, Costa PP. Immunoassay for transthyretin variants associated with amyloid neuropathy. Scand. J. Immunol. 1993;38:177–182. doi: 10.1111/j.1365-3083.1993.tb01710.x. [DOI] [PubMed] [Google Scholar]
- 37.Kabat EA, Wu TT, Perry HM, Gottesman KS, Foeller C. NIH, editor. NIH; Sequence of proteins of immunological interest. (5 ed.) 1992
- 38.Huber R, Steigemann W. Two cis-prolines in the Bence-Jones protein Rei and the cis-pro-bend. FEBS Lett. 1974;48:235–237. doi: 10.1016/0014-5793(74)80475-8. [DOI] [PubMed] [Google Scholar]
- 39.Ivanova MI, Sawaya MR, Gingery M, Attinger A, Eisenberg D. An amyloid-forming segment of beta2-microglobulin suggests a molecular model for the fibril. Proc. Natl. Acad. Sci. U.S.A. 2004;101:10584–10589. doi: 10.1073/pnas.0403756101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat. Struct. Mol. Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 41.Huang DB, Chang CH, Ainsworth C, Johnson G, Solomon A, Stevens FJ, Schiffer M. Variable domain structure of kappa IV human light chain Len: high homology to the murine light chain McPC603. Mol. Immunol. 1997;34:1291–1301. doi: 10.1016/s0161-5890(98)00002-9. [DOI] [PubMed] [Google Scholar]
- 42.Rose GD, Gierasch LM, Smith JA. Turns in peptides and proteins. Adv. Protein Chem. 1985;37:1–109. doi: 10.1016/s0065-3233(08)60063-7. [DOI] [PubMed] [Google Scholar]
- 43.Videau LL, Arendall WB, 3rd, Richardson JS. The cis-Pro touch-turn: a rare motif preferred at functional sites. Proteins. 2004;56:298–309. doi: 10.1002/prot.20101. [DOI] [PubMed] [Google Scholar]
- 44.Gellman SH. Minimal model systems for beta sheet secondary structure in proteins. Curr. Opin. Chem. Biol. 1998;2:717–725. doi: 10.1016/s1367-5931(98)80109-9. [DOI] [PubMed] [Google Scholar]
- 45.Russell SJ, Blandl T, Skelton NJ, Cochran AG. Stability of cyclic beta-hairpins: asymmetric contributions from side chains of a hydrogen-bonded cross-strand residue pair. J. Am. Chem. Soc. 2003;125:388–395. doi: 10.1021/ja028075l. [DOI] [PubMed] [Google Scholar]
- 46.Maynard AJ, Sharman GJ, Searle MS. Origin of beta-hairpin stability in solution: structural and thermodynamic analysis of the folding of a model peptide supports hydrophobic stabilization in water. J. Am. Chem. Soc. 1998;120:1996–2007. [Google Scholar]
- 47.Demchuk E, Bashford D, Case DA. Dynamics of a type VI reverse turn in a linear peptide in aqueous solution. Fold. Des. 1997;2:35–46. doi: 10.1016/S1359-0278(97)00004-7. [DOI] [PubMed] [Google Scholar]
- 48.Wu WJ, Raleigh DP. Local control of peptide conformation: stabilization of cis proline peptide bonds by aromatic proline interactions. Biopolymers. 1998;45:381–394. doi: 10.1002/(SICI)1097-0282(19980415)45:5<381::AID-BIP6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 49.Yao J, Dyson HJ, Wright PE. Three-dimensional structure of a type VI turn in a linear peptide in water solution. Evidence for stacking of aromatic rings as a major stabilizing factor. J. Mol. Biol. 1994;243:754–766. doi: 10.1016/0022-2836(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 50.Meng HY, Thomas KM, Lee AE, Zondlo NJ. Effects of i and i+3 residue identity on cis-trans isomerism of the aromatic(i+1)-prolyl(i+2) amide bond: implications for type VI beta-turn formation. Biopolymers. 2006;84:192–204. doi: 10.1002/bip.20382. [DOI] [PubMed] [Google Scholar]
- 51.Guruprasad K, Rajkumar S. Beta-and gamma-turns in proteins revisited: a new set of amino acid turn-type dependent positional preferences and potentials. J. Biosci. 2000;25:143–156. [PubMed] [Google Scholar]
- 52.Balamurugan B, Samaya Mohan K, Ramesh J, Roshan MN, Sumathi K, Sekar K. SSEP-2.0: Secondary Structural Elements of Proteins. Acta Crystallogr. D. Biol. Crystallogr. 2005;61:634–636. doi: 10.1107/S0907444905005883. [DOI] [PubMed] [Google Scholar]
- 53.Che Y, Marshall GR. Impact of cis-proline analogs on peptide conformation. Biopolymers. 2006;81:392–406. doi: 10.1002/bip.20431. [DOI] [PubMed] [Google Scholar]
- 54.Che Y, Marshall GR. Impact of azaproline on Peptide conformation. J. Org. Chem. 2004;69:9030–9042. doi: 10.1021/jo0487303. [DOI] [PubMed] [Google Scholar]
- 55.Li L, Darden TA, Bartolotti L, Kominos D, Pedersen LG. An atomic model for the pleated β-sheet structure of Aβ amyloid protofilaments. Biophys. J. 1999;76:2871–2878. doi: 10.1016/S0006-3495(99)77442-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 58.Meloen RH, Puijk WC, Slootstra JW. Mimotopes: realization of an unlikely concept. J. Mol. Recognit. 2000;13:352–359. doi: 10.1002/1099-1352(200011/12)13:6<352::AID-JMR509>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 59.Gevorkian G, Petrushina I, Manoutcharian K, Ghochikyan A, Acero G, Vasilevko V, Cribbs DH, Agadjanyan MG. Mimotopes of conformational epitopes in fibrillar β-amyloid. J. Neuroimmunol. 2004;156:10–20. doi: 10.1016/j.jneuroim.2004.06.004. [DOI] [PubMed] [Google Scholar]