Abstract
We present a method for examining the relative influence of familial, genetic, and environmental covariate information in flexible nonparametric risk models. Our goal is investigating the relative importance of these three sources of information as they are associated with a particular outcome. To that end, we developed a method for incorporating arbitrary pedigree information in a smoothing spline ANOVA (SS-ANOVA) model. By expressing pedigree data as a positive semidefinite kernel matrix, the SS-ANOVA model is able to estimate a log-odds ratio as a multicomponent function of several variables: one or more functional components representing information from environmental covariates and/or genetic marker data and another representing pedigree relationships. We report a case study on models for retinal pigmentary abnormalities in the Beaver Dam Eye Study. Our model verifies known facts about the epidemiology of this eye lesion—found in eyes with early age-related macular degeneration—and shows significantly increased predictive ability in models that include all three of the genetic, environmental, and familial data sources. The case study also shows that models that contain only two of these data sources, that is, pedigree-environmental covariates, or pedigree-genetic markers, or environmental covariates-genetic markers, have comparable predictive ability, but less than the model with all three. This result is consistent with the notions that genetic marker data encode—at least in part—pedigree data, and that familial correlations encode shared environment data as well.
Keywords: SS-ANOVA, retinal pigmentary abnormalities, RKHS, pedigrees
Smoothing spline ANOVA (SS-ANOVA) models (1–4) have a successful history modeling ocular traits. In particular, an SS-ANOVA model of retinal pigmentary abnormalities,* defined by the presence of retinal depigmentation and increased retinal pigmentation (5, 6), was able to show a nonlinear protective effect of high total serum cholesterol for a cohort of subjects in the Beaver Dam Eye Study (BDES) (2). However, multiple studies have reported that risk variants at two loci, near the CFH and ARMS2 genes, show strong association with the development of age-related macular degeneration (AMD) (7–18), a leading cause of blindness and visual disability (19). Because retinal pigmentary abnormalities are an early sign of age-related macular degeneration, a leading cause of blindness and visual disability in its late stages (19), we want to make use of genotype data for these two genes to extend the SS-ANOVA model for pigmentary abnormalities risk. For example, by extending the SS-ANOVA model of Lin et al. (2) with SNP rs10490924 in the ARMS2 gene region, we were able to see that the protective effect of total serum cholesterol disappears in older subjects that have the risk variant of this SNP. The supporting information (SI) Appendix replicates the model of Lin et al. (2) and shows the extended model, including the SNP data. Smoothing spline logistic regression models are able to tease out these types of complex nonlinear relationships that would not be detected by more traditional parametric models—linear, or of prespecified form.
Beyond genetic and environmental effects, we want to extend the SS-ANOVA model for pigmentary abnormalities with familial data. For instance, pedigrees (see Representing Pedigree Data as Kernels) have been ascertained for a large number of subjects of the BDES. In this article we present a general method that is able to incorporate arbitrary relationships encoded as a graph, e.g., pedigree data, into SS-ANOVA models. This method allows one to examine the importance of relationships between subjects relative to other model terms in a predictive model.
We estimate SS-ANOVA models of the log-odds of pigmentary abnormality risk of the form
where g1 is a term that includes only genetic marker data (e.g., SNPs), g2 is a term containing only environmental covariate data, and h is a smooth function over a space that encodes relationships between subjects. In this relationship space, each subject ti may be thought of as being represented by a “pseudo-attribute” z(ti). In the remainder of the article we will refer to model terms g1 and g2 as S (for SNP) and C (for covariates), respectively, and term h as P (for pedigrees); so, a model containing all three components will be referred to as S+C+P.
Formally, this SS-ANOVA model is defined over the tensor sum of multiple reproducing kernel Hilbert spaces: one or more components representing information from environmental and/or genetic covariates for each subject (corresponding to terms g1 and g2 above) and another representing pedigree relationships. The model is estimated as the solution of a penalized likelihood problem with an additive penalty including a term for each reproducing kernel Hilbert space (RKHS) in the ANOVA decomposition, each weighted by a coefficient. From this decomposition we can measure the relative importance of each model component (S, C, or P). Our main tool in extending SS-ANOVA models with pedigree data is the Regularized Kernel Estimation framework (20). More complex models involving interactions between these three sources of information are possible but beyond the scope of this article.
In Smoothing-Spline ANOVA Models we discuss the semiparametric risk models we use in this article; in Representing Pedigree Data as Kernels we define pedigrees and introduce our method to include these data in SS-ANOVA risk models; we follow with a case study on the Beaver Dam Eye Study and conclude with a discussion.
Smoothing Spline ANOVA Models
Suppose we are given a dataset of environmental covariates and/or genetic markers for each of n subjects, with measurements for each subject represented as numeric vectors xi, along with measured responses, e.g., presence of pigmentary abnormalities, yi ∈ [0,1], i = 1,…,n. The SS-ANOVA model estimates the log-odds , where p(xi) = Pr(yi = 1|xi), by assuming that f is a function in an RKHS of the form H = H0 ⊕ H1. H0 is a finite dimensional space spanned by a set of functions [ϕ1,…,ϕm], and H1 is an RKHS induced by a given kernel function k(·,·) with the property that 〈k(x,·),g〉H1 = g(x) for g ∈ H1, and thus, 〈k(xi,·),k(xj,·)〉H1 = k(xi,xj). Therefore, f has a semiparametric form given by
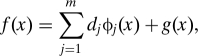 |
for some coefficients dj, where the functions ϕj have a parametric, e.g., linear, form and g ∈ H1. H1 is further decomposed by assuming it is the direct sum of multiple RKHSs, so g ∈H1 is defined as
where [gα] and [gαβ] satisfy side conditions that generalize the standard ANOVA side conditions. Functions gα encode “main effects,” gαβ encode “second-order interactions,” and so on. An RKHS Hα is associated with each component in this sum, along with its corresponding kernel function kα. In this case, we can define a reproducing kernel function for H1 as k(·,·) = ∑αθαkα(·,·) + ∑αβ θαβkαβ(·,·) + ···, where the coefficients θ are tunable hyperparameters that weigh the relative importance of each term in the decomposition.
The SS-ANOVA estimate of f given data (xi,yi), i = 1,…,n, is given by the solution of a penalized likelihood problem of the form:
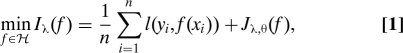 |
where l(yi,f(xi)) = −yif(xi) + log(1 + ef(xi)) is the negative log likelihood of yi given f(xi) and
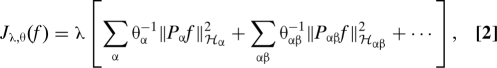 |
with Pαf the projection of f into RKHS Hα and λ a non-negative regularization parameter. The penalty Jλ,α(f) is a seminorm in RKHS H and penalizes the complexity of f using the norm of the RKHS H1 to avoid overfitting f to the training data.
By the representer theorem of Kimeldorf and Wahba (21), the minimizer of the problem in Eq. 1 has a finite representation of the form
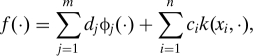 |
in which case ||P1f||H12 = cTKc for matrix K with Kij = k(xi,xj). Thus, for a given value of the regularization parameter λ the minimizer fλ can be estimated by solving the following convex nonlinear optimization problem:
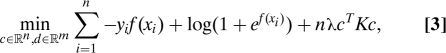 |
where f = [f(x1)…f(xn)]T = Td + Kc with Tij = ϕj(xi). This model requires that hyperparameters, λ, the coefficients θ of the ANOVA decomposition, and any other hyperparameter in kernel functions kα be chosen. In this article, we will use the generalized approximate cross-validation (GACV) method, an approximation to the leave-one-out approximation of the comparative Kullback–Leibler distance between the estimate fλ and the unknown “true” log-odds f (3).
In models that have genetic, environmental, and familial components, the ANOVA decomposition can be used to measure the relative importance of each function component with suitably chosen kernel functions kα. For genetic and environmental components, standard kernel functions can be used to define the corresponding RKHS. However, pedigree data are not represented as feature vectors for which standard kernel functions can be used. However, the optimization problem in Eq. 3 is specified completely by the model matrix T and kernel matrix K. In the next section, we show how to build kernel matrices that encode familial relationships which can then be included in the estimation problem.
Representing Pedigree Data as Kernels
A pedigree is an acyclic graph representing a set of genealogical relationships, where each node corresponds to a member of the family, and arcs indicate parental relationships. Thus, each node has two incoming arcs, one for its father and one for its mother (except founder nodes that have no incoming arcs) and an outgoing arc for each offspring. We show an example pedigree in the SI Appendix. We can define a pedigree dissimilarity measure between subjects by using the Malécot kinship coefficient ϕ (22). For individuals i and j in the pedigree this is defined as the probability that a randomly selected pair of alleles, one from each individual, are identical by descent (IBD), that is, they are derived from a common ancestor. For example, the probability of a parent–offspring pair sharing an IBD allele is 1/4: there is a 50% chance that randomly choosing one of the two offspring alleles yields that inherited from the specific parent, and there is a 50% chance that choosing one of the two parental alleles at random yields the allele inherited by the offspring.
Definition 1 (Pedigree Dissimilarity): The pedigree dissimilarity between individuals i and j is defined as dij = −log2(2ϕij), where ϕ is Malécot's kinship coefficient.
In studies such as the BDES, not all family members are subjects of the study; therefore, the graphs we will use to represent pedigrees in our models only include nodes for study subjects rather than the entire pedigree. Furthermore, in our study we want to include exposure to hormone replacement therapy in our pigmentary abnormality risk model, so our relationship graphs will only include female subjects. The SI Appendix shows an example of a relationship graph. The main thrust of our methodology is how to incorporate these relationship graphs—derived from pedigrees and weighted by a pedigree dissimilarity that captures genetic relationship—into predictive risk models. In particular, we want to use nonparametric predictive models that incorporate other data, both genetic and environmental, under the restriction that only a subset of pedigree members are fully observed in both covariates and outcomes. We saw in the previous section that we can do this by defining a kernel matrix K that encodes pedigree relationships between the subjects of interest.
The requirement for a valid kernel matrix to be used in the penalized likelihood estimation problem of Eq. 3 is that the matrix be positive semidefinite: for any vector α ∈ ℝn, αTKα ≥ 0, denoted as K ≽ 0. A property of positive semidefinite matrice is that they can be interpreted as the matrix of inner products between certain functions in an RKHS: the kernel matrix K in Eq. 3 is the matrix of inner products of the evaluation representers k(x,·) of the given data points in H1. Finally, since K ≽ 0 contains the inner products of these representers, we can define a distance metric over these objects as dij2 = Kii + Kjj − 2Kij. We make use of this connection between distances and inner products in the Regularized Kernel Estimation framework to define a kernel based on the pedigree dissimilarity of Definition 1.
The Regularized Kernel Estimation (RKE) framework was introduced by Lu et al. (20) as a robust method for estimating dissimilarity measures between objects from noisy, incomplete, inconsistent, and repetitious dissimilarity data. The RKE framework is useful in settings where object classification or clustering is desired but objects do not easily admit description by fixed-length feature vectors, but instead, there is access to a source of noisy and incomplete dissimilarity information between objects. It estimates a symmetric positive semidefinite kernel matrix K which induces a real squared distance admitting of an inner product as described above.
Assume dissimilarity information is given for a subset Ω of the possible pairs occurring in a training set of n objects, with the dissimilarity between objects i and j denoted as dij ∈ Ω. RKE estimates an n -by- n symmetric positive semidefinite kernel matrix K of size n such that the fitted squared distance between objects induced by K, ij2 = Kii + Kjj − 2Kij, is as close as possible to the square of the observed dissimilarities dij ∈ Ω. Formally, RKE solves the following optimization problem with semidefinite constraints:
The parameter λrke ≥ 0 is a regularization parameter that trades off fit of the dissimilarity data, as given by absolute deviation, and a penalty, trace(K), on the complexity of K. The trace may be seen as a proxy for the rank of K; therefore, RKE is regularized by penalizing high dimensionality of the space spanned by K. RKE requires that Ω satisfies a connectivity constraint: the undirected graph consisting of objects as nodes and edges between them, such that an edge between nodes i and j is included if dij ∈ Ω, is connected. Additionally, optional weights wij may be associated with each dij ∈ Ω. A method for choosing the regularization parameter λrke is required, but, by treating λrke as a hyperparameter to the kernel matrix of the SS-ANOVA problem we can tune by using the GACV criterion.
The fact that RKE operates on inconsistent dissimilarity data, rather than distances, is significant in this context. The pedigree dissimilarity of Definition 1 is not a distance since it does not satisfy the triangle inequality for general pedigrees. We show an example where this is the case in SI Appendix.
The solution to the RKE problem is a symmetric positive semidefinite matrix K from which an embedding Z ∈ ℝn×r in r-dimensional Euclidean space is obtained by decomposing K as K = ZZT with Z = ΓrΛr1/2, where the n × r matrix Γr and the r × r diagonal matrix Λr contain the r leading eigenvalues and eigenvectors of K, respectively. We refer to the ith row of Z as the vector of “pseudo”-attributes z(i) for subject i. We show an example embedding from RKE in SI Appendix. A method for choosing r is required and we discuss one in Materials and Methods. We may consider the embedding resulting from RKE as providing a set of “pseudo”-attributes z(i) for each subject in this pedigree space and a smooth predictive function may be estimated in this space. In principle, we should impose a rotational invariance when defining this smooth function since only distance information was used to create the embedding, e.g., by using a Matérn family kernel (see SI Appendix).
Case Study: Beaver Dam Eye Study
The Beaver Dam Eye Study (BDES) is an ongoing population-based study of age-related ocular disorders. Subjects at baseline, examined between 1988 and 1990, were a group of 4,926 people aged 43-86 years who lived in Beaver Dam, WI. A description of the population and details of the study at baseline may be found in Klein et al. (23). Although we will only use data from this baseline study for our experiments, 5-, 10-, and 15-year follow-up data were also obtained (24–26). Familial relationships of participants were ascertained and pedigrees constructed (27) for the subset of subjects who had at least one relative in the cohort. Genotype data for specific SNPs was subsequently generated for those participants included in the pedigree data.
Our goal in this case study is to use genetic and pedigree data to extend the work of Lin et al. (2) that studies the association between retinal pigmentary abnormalities and a number of environmental covariates. We estimated SS-ANOVA models of the form
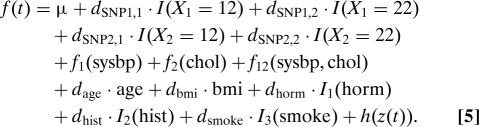 |
The terms in the first two lines encode the effect of the two genetic markers (SNPs), the next few terms encode the effect of the environmental covariates listed in Table 1, and the term h(z(t)) encodes familial effects and is estimated by the methods presented above. We denote these model components, respectively, as P (for pedigree), S (for SNP), and C (for covariates). Our goal was to compare different models containing different combinations of these components. For example, P-only refers to a model containing only a pedigree component; S+C, to a model containing components for genetic markers and environmental covariates; C-only was the original SS-ANOVA model for pigmentary abnormalities (2); and P+S+C refers to a model containing components for all three data sources.
Table 1.
Environmental covariates for BDES pigmentary abnormality risk SS-ANOVA model
| Code | Units | Description |
|---|---|---|
| horm | yes/no | Current usage of hormone replacement therapy |
| hist | yes/no | History of heavy drinking |
| bmi | kg/m2 | Body mass index |
| age | years | Age at baseline |
| sysbp | mmHg | Systolic blood pressure |
| chol | mg/dL | Total serum cholesterol |
| smoke | yes/no | History of smoking |
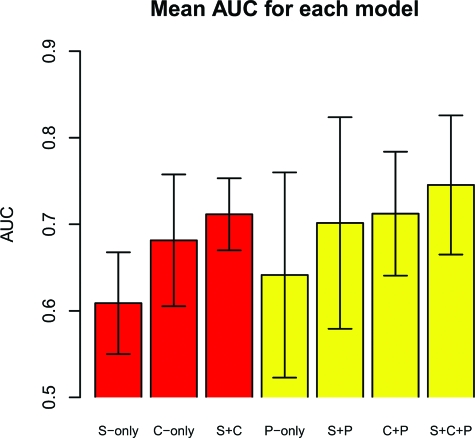
We used area under the receiver operating characteristic (ROC) curve (28, referred to as AUC) estimated by using 10-fold cross-validation to compare predictive performance of models with various component combinations. Fig. 1 summarizes our results by plotting the mean AUC of each model. For pedigree models, Fig. 1 shows the best AUC of either using RKE to define the pedigree kernel or an alternative method described in Materials and Methods. We include full results in SI Appendix. We can observe large variation in the AUC reported for most model/method combinations over the cross-validation folds, but some features are apparent: for example, the model with highest overall mean AUC is the S+C+P model. We carried out pairwise t tests on a few model comparisons and report P values from estimates where variance is calculated from the differences in AUC between the pair of models being compared over the 10 cross-validation folds. Although there was high variability in AUC over the 10 cross-validation folds for most individual models, in general, there was much less variation in the difference in AUC between the pairs of models we compare below across the 10 cross-validation folds. The next few paragraphs summarize and discuss the results from these tests.
Fig. 1.
AUC comparison of models. Model labels are explained in the text. Error bars are one standard deviation from the mean. Yellow bars indicate models containing pedigree data. Full AUC scores are given in SI Appendix, Table S1.
For pedigree-less models, the S+C model containing both markers and covariates had better AUC than either the S-only or C-only models (P values, 0.00250 and 0.065, respectively). This means that combining genetic markers and environmental covariates yields a better model than either data source by itself, a result consistent with the known epidemiology of pigmentary abnormalities, where risk is associated with both the genetic markers and environmental covariates included in this model.
The model with the highest overall mean AUC was the S+C+P model, with statistically significant differences at the 90% level for all except the S+P (P value, 0.108) model. This is the main result of our article: a full model containing genetic marker data along with environmental covariates and pedigree data is the best performing model for predictive purposes over models that only contain a subset of these data sources.
Models with only two data sources, i.e., S+C, S+P, and C+P, performed statistically similarly. That is, there were no statistically significant differences in the predictive performance of these models, although the C+P model performed slightly better. This finding is consistent with the notions that SNP data—at least in part—encode pedigree data and that familial correlations encode shared environment data as well. We can see the former since SNP and pedigree data each add, relatively speaking, about the same amount of information when combined with covariate data; neither is strongly more informative than the other in the present context. In contrast, we can see the latter since combining SNP and pedigree data is as informative as combining SNP and covariate data. In summary, models that contain only two of these data sources, that is, pedigree-environmental covariates, or pedigree-genetic markers, or environmental covariates-genetic markers, have comparable predictive ability, while less than the model with all three.
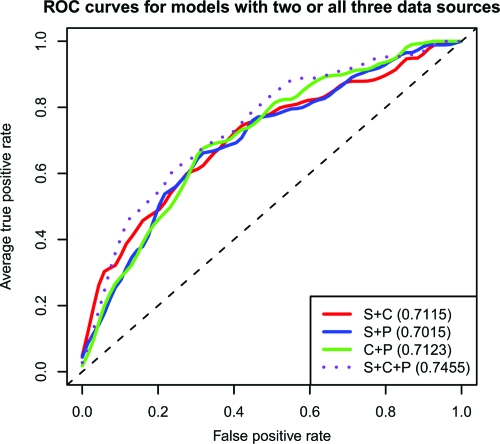
The ROC curves for models using only two data sources (Fig. 2) show an interesting trend. We can see that in the high-sensitivity portion of the curve (false-positive rate between 0 and 0.2), the S+C model, which does not contain any pedigree data, dominated the other two models. However, we see that the pedigree models dominated the S+C model on the other extreme portion of the curve (true positive rate higher than 0.8). The ROC curve for the S+C+P model dominates these three curves throughout ROC space.
Fig. 2.
ROC curves for models with two or all three data sources. The legend includes AUC values of each model in parentheses.
Another observation we can make is that the P-only model had greater AUC than the S-only model but without a statistically significant difference (P value, 0.207). However, combining the two terms improves predictive performance significantly, indicating that the genetic influence on pigmentary abnormality risk is not properly modeled by either data source alone.
We conclude this case study by looking at diagnostics of the resulting models to illustrate the effect of including pedigree data in the pigmentary abnormalities risk model. Cosine diagnostics (4, 29) are an illustrative way of displaying the relative weight of model terms in the SS-ANOVA decomposition. We used the π diagnostic to compare the S+C+P and S+C models (SI Appendix, Table S2). We saw that in the pedigree-less S+C model, the environmental covariates (C) have 0.66 decomposition weight. However, in the full S+C+P model, 0.53 of the decomposition weight is in the pedigree term, while the relative weight of the other two terms are essentially unchanged: for example, the SNP terms (S) have 0.17/(0.17 + 0.26) = 0.39 of the weights of the S and C terms in the S+C+P model and 0.34 of the weight in the S+C model. The fact that the C term in the full-data S+C+P model has more weight than the S term may explain why the C+P model slightly outperforms the S+P model.
Considering that the full S+C+P model had the best prediction performance and that the pedigree term had a large relative weight in the model, we may conclude that incorporating familial relationship data in an SS-ANOVA model as described by our methodology not only improves the predictive performance of existing models of pigmentary abnormality risk, but also partly describes how these three sources of data relate in a predictive model. Refining this statistical methodology to further understand the interaction of these data sources would be of both technical and scientific interest.
Discussion
Throughout our experiments and simulations we have used genetic marker data in a very simple manner by including single markers for each gene in an additive model. A more realistic model should include multiple markers per gene and would include interaction terms between these markers. Although we have data on two additional markers for each of the two genes included in our case study (CFH and ARMS2) for a total of six markers (three per gene), we chose to use the additive model on only two markers since, for this cohort, this model showed the same predictive ability as models including all six markers with interaction terms. Furthermore, due to some missing entries in the genetic marker data, including multiple markers reduced the sample size.
Along the same lines, we currently use a very simple inheritance model to define pedigree dissimilarity. Including, for example, dissimilarities between unrelated subjects should prove advantageous. A simple example would be including a spousal relationship when defining dissimilarity because this would be capturing some shared environmental factors. Extensions to this methodology that include more complex marker models and multiple or more complex dissimilarity measures are fertile ground for future work.
Other methods for including graph-based data in predictive models have been proposed recently, especially in the Machine Learning community. They range from semi-supervised methods that regularize a predictive model by applying smoothness penalties over the graph (30), to discriminative graphical models (31), and methods closer to ours that define kernels from graph relationships (32).
There are issues in the risk-modeling setting with general pedigrees, where relationship graphs encode relationships between subsets of a study cohort that are usually not explicitly addressed in the general graph-based setting. Most important is the assumption that, although graph structure has some influence in the risk model, it is not necessarily an overwhelming influence. Thus, a model that produces relative weights between components of the model, one being graph relationships, is required. That is the motivation for using the SS-ANOVA framework in this article. Although graph regularization methods have a parameter that controls the influence of the graph structure in the predictive model, it is not directly comparable to the influence of other model components, e.g., genetic data or environmental covariates. However, graphical model techniques define a probabilistic model over the graph to define the predictive model. This gives the graph relationships too much influence over the predictive model in the sense that it imposes conditional independence properties over subjects determined by the relationship graph that might not be valid for the other data sources, e.g., environmental covariates.
Materials and Methods
The model in Eq. 5 included genotype data for the Y402H region of the complement factor H (CFH) gene and for SNP rs10490924 in the LOC387715 (ARMS2) gene. A variable for each SNP is coded according to the subject genotype for that SNP as (11,12,22). For identifiability, the 11 level of each SNP is modeled by the intercept μ, while an indicator variable is included for each of the other two levels. This results in each level (other than the 11 level) having its own model coefficient. Functions f1 and f2 are cubic splines, while f12 uses the tensor product construction (4). The remaining covariates are modeled as linear terms with Ij as indicator functions. All continuous variables were scaled to lie in the interval [0, 1].
The cohort used were female subjects of the BDES baseline study for which we had full data for genetic markers, environmental covariates, and pedigrees. The cohort was further restricted to those from pedigrees containing two or more subjects within the cohort (n = 684). This resulted in 175 pedigrees in the dataset, with sizes ranging from 2 to 103 subjects. More than a third of the subjects were in pedigrees with 8 or more observations. We chose to only include female subjects in this study to make our model a direct extension of that in Lin et al. (2), which used only female subjects in their cohort and included exposure to hormone replacement therapy as a covariate. The cross-validation folds used to measure AUC were created such that for every subject in each test fold, at least one other member of their pedigree was included in the labeled training set. Pedigree kernels were built on all members of the study cohort with hyperparameters chosen independently for each fold by using GACV on the labeled training set. Cosine diagnostics are defined on the vector of fitted “pseudo”-gaussian responses f̂ for the entire cohort. The π diagnostic decomposes the norm of f̂ according to the additive terms of the SS-ANOVA decomposition assigning a relative weight to each term in the model.
The penalized likelihood problem of Eq. 3 was solved by the quasi-Newton method implemented in the gss R package (33) using the function gssanova0 with slight modifications to address some numerical instabilities. The RKE semidefinite problem of Eq. 4 was solved by using the CSDP library (34) with input dissimilarities given by Definition 1. A number of additional edges between unrelated individuals encoding the “infinite” dissimilarity were added randomly to the graph. The dissimilarity encoded by these edges was arbitrarily chosen to be the sum of all dissimilarities in the entire cohort, whereas the number of additional edges was chosen such that each subject had an edge to at least 25 other subjects in the cohort (including all members of the same pedigree). The kernel matrix obtained from RKE was then truncated to those leading eigenvalues that account for 90% of the matrix trace to create a “pseudo”-attribute embedding. A third-order Matérn kernel was then built over the points in the resulting embedding (see SI Appendix). Pedigree dissimilarities were derived from kinship coefficients calculated using the kinship R package (35).
We also tested an alternative to RKE where the pedigree dissimilarity is treated as a distance and a kernel, e.g., a Matérn kernel, is defined directly over it. However, since the pedigree dissimilarity does not satisfy the definitions of a distance, the resulting kernel might not be positive semidefinite. In our implementation, we computed the projection under Frobenius norm of the resulting kernel matrix to the cone of positive semidefinite matrices, by setting the negative eigenvalues of the matrix to zero. Since solving the RKE problem is computationally expensive, this is an attractive alternative due to its computational efficiency. However, the RKE problem gives a sound and principled way of generating a kernel encoding relationships, while this alternative method is ad hoc. Although this efficient alternative might perform well in some cases, we expect the RKE method to be more robust and work better in the general case. Thus, gains in efficiency must be weighted against possible losses in the general applicability of this alternative method.
Supplementary Material
Acknowledgments.
This work was partially supported by National Institutes of Health (NIH) GrantEY09946, National Science Foundation Grant DMS-0604572 and Office of Naval Research Grant N0014-06-0095 (to H.C.B. and G.W.), NIH Grant EY06594 (to K.L., R.K. and B.K.), the Research to Prevent Blindness Senior Scientific Investigator Awards (to R.K. and B.K.), and NIH Grant 5R01 EY018510 (to S.I.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902906106/DCSupplemental.
Hereafter, we will use the term pigmentary abnormalities when referring to retinal pigmentary abnormalities.
References
- 1.Wahba G, Wang Y, Gu C, Klein R, Klein B. Smoothing spline ANOVA for exponential families, with application to the Wisconsin Epidemiological Study of Diabetic Retinopathy. Ann Stat. 1995;23:1865–1895. [Google Scholar]
- 2.Lin X, et al. Smoothing spline ANOVA models for large data sets with Bernoulli observations and the randomized GACV. Ann Stat. 2000;28:1570–1600. [Google Scholar]
- 3.Xiang D, Wahba G. A generalized approximate cross validation for smoothing splines with non-Gaussian data. Stat Sin. 1996;6:675–692. [Google Scholar]
- 4.Gu C. Smoothing Spline Anova Models. New York: Springer; 2002. [Google Scholar]
- 5.Klein BE, Klein R, Jensen SC, Ritter LL. Are sex hormones associated with age-related maculopathy in women? The Beaver Dam Eye Study. Trans Am Ophthalmol Soc. 1994;92:289–297. [PMC free article] [PubMed] [Google Scholar]
- 6.Klein R, Klein BE, Linton KL. Prevalence of age-related maculopathy. The Beaver Dam Eye Study. Ophthalmology. 1992;99:933–943. doi: 10.1016/s0161-6420(92)31871-8. [DOI] [PubMed] [Google Scholar]
- 7.Thompson C, et al. Complement Factor H and Hemicentin-1 in age-related macular degeneration and renal phenotypes. Hum Mol Genet. 2007;16:2135–2148. doi: 10.1093/hmg/ddm164. [DOI] [PubMed] [Google Scholar]
- 8.Thompson C, et al. Genetics of pigment changes and geographic atrophy. Invest Ophthalmol Visual Sci. 2007;48:3005–3013. doi: 10.1167/iovs.06-1325. [DOI] [PubMed] [Google Scholar]
- 9.Magnusson K, et al. CFH Y402H confers similar risk of soft drusen and both forms of advanced AMD. PLoS Med. 2006;3:e5. doi: 10.1371/journal.pmed.0030005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li M, Atmaca-Sonmez P, et al. CFH haplotypes without the Y402H coding variant show strong association with susceptibility to age-related macular degeneration. Nat Genet. 2006;38:1049–1054. doi: 10.1038/ng1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Klein R, et al. Complement Factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanda A, et al. A variant of mitochondrial protein LOC387715/ARMS2, not HTRA1, is strongly associated with age-related macular degeneration. Proc Natl Acad Sci USA. 2007;104:16227–16232. doi: 10.1073/pnas.0703933104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haines J, et al. Complement Factor H variant increases the risk of age-related macular degeneration. Science. 2005;308:419–421. doi: 10.1126/science.1110359. [DOI] [PubMed] [Google Scholar]
- 14.Baird P, et al. Analysis of the Y402H variant of the Complement Factor H gene in age-related macular degeneration. Invest Ophthalmol Visual Sci. 2006;47:4194–4198. doi: 10.1167/iovs.05-1285. [DOI] [PubMed] [Google Scholar]
- 15.Edwards A, et al. Complement factor H polymorphism and age-related macular degeneration. Science. 2005;308:421–424. doi: 10.1126/science.1110189. [DOI] [PubMed] [Google Scholar]
- 16.Fisher S, et al. Meta-analysis of genome scans of age-related macular degeneration. Hum Mol Genet. 2005;14:2257–2264. doi: 10.1093/hmg/ddi230. [DOI] [PubMed] [Google Scholar]
- 17.Fritsche L, et al. Age-related macular degeneration is associated with an unstable ARMS2 (LOC387715) mRNA. Nat Genet. 2008;40:892–896. doi: 10.1038/ng.170. [DOI] [PubMed] [Google Scholar]
- 18.Hageman G, et al. A common haplotype in the complement regulatory gene factor H (HF1/CFH) predisposes individuals to age-related macular degeneration. Proc Natl Acad Sci USA. 2005;102:7227–7232. doi: 10.1073/pnas.0501536102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klein R, Peto T, Bird A, Vannewkirk M. The epidemiology of age-related macular degeneration. Am J Ophthalmol. 2004;137:486–495. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 20.Lu F, Keles S, Wright S, Wahba G. Framework for kernel regularization with application to protein clustering. Proc Natl Acad Sci USA. 2005;102:12332–12337. doi: 10.1073/pnas.0505411102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimeldorf G, Wahba G. Some results on tchebycheffian spline functions. J Math Anal Appl. 1971;33:82–95. [Google Scholar]
- 22.Malécot G. Les Mathématiques de l'Hérédité. Paris: Masson; 1948. [Google Scholar]
- 23.Klein R, Klein B, Linton K, De M, ets D. The Beaver Dam Eye Study: Visual acuity. Ophthalmology. 1991;98:1310–1315. doi: 10.1016/s0161-6420(91)32137-7. [DOI] [PubMed] [Google Scholar]
- 24.Klein R, et al. Fifteen-year cumulative incidence of age-related macular degeneration: The Beaver Dam Eye Study. Ophthalmology. 2007;114:253–262. doi: 10.1016/j.ophtha.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 25.Klein R, Klein B, Tomany S, Meuer S, Huang G. Ten-year incidence and progression of age-related maculopathy: The Beaver Dam eye study. Ophthalmology. 2002;109:1767–1779. doi: 10.1016/s0161-6420(02)01146-6. [DOI] [PubMed] [Google Scholar]
- 26.Klein R, Klein B, Jensen S, Meuer S. The five-year incidence and progression of age-related maculopathy: The Beaver Dam Eye Study. Ophthalmology. 1997;104:7–21. doi: 10.1016/s0161-6420(97)30368-6. [DOI] [PubMed] [Google Scholar]
- 27.Lee K, Klein B, Klein R, Knudtson M. Familial aggregation of retinal vessel caliber in the Beaver Dam Eye Study. Invest Ophthalmol Visual Sci. 2004;45:3929–3933. doi: 10.1167/iovs.04-0462. [DOI] [PubMed] [Google Scholar]
- 28.Fawcett T. An introduction to roc analysis. Pattern Recognit Lett. 2006;27:861–874. [Google Scholar]
- 29.Gu C. Diagnostics for nonparametric regression models with additive terms. J Am Stat Assoc. 1992;87:1051–1058. [Google Scholar]
- 30.Sindhwani V, Niyogi P, Belkin M. Beyond the point cloud: From transductive to semi-supervised learning. ACM Int Conf Proc Ser. 2005;119:824–831. [Google Scholar]
- 31.Chu W, Sindhwani V, Ghahramani Z, Keerthi S. Advances in Neural Information Processing Systems: Proceedings of the 2006 Conference. Cambridge, MA: MIT Press; 2007. Relational learning with gaussian processes. [Google Scholar]
- 32.Smola A, Kondor R. Conference on Learning Theory. Heidelberg, Germany: Springer Verlag; 2003. Kernels and regularization on graphs; pp. 144–158. [Google Scholar]
- 33.Gu C. [Accessed February 13, 2008];gss: General smoothing splines. 2007 R package version 1.0-0. Available at: http://cran.r-project.org/web/packages/gss/index.html. [Google Scholar]
- 34.Borchers B. CSDP: A C library for semidefinite programming. Optimiz Methods Software. 1999;11:613–623. [Google Scholar]
- 35.Atkinson B, Therneau T. [Accessed June 22, 2008];Kinship: Mixed-Effects Cox Models, Sparse Matrices, and Modeling Data from Large Pedigrees. 2007 R package version 1.1.0-18. Available at: http://cran.r-project.org/web/packages/kinship/index.html. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.