Abstract
Mismatch repair contributes to genetic stability, and inactivation of the mammalian pathway leads to tumor development. Mismatch correction occurs by an excision-repair mechanism and has been shown to depend on the 5′ to 3′ hydrolytic activity exonuclease 1 (Exo1) in eukaryotic cells. However, genetic and biochemical studies have indicated that one or more Exo1-independent modes of mismatch repair also exist. We have analyzed repair of nicked circular heteroduplex DNA in extracts of Exo1-deficient mouse embryo fibroblast cells. Exo1-independent repair under these conditions is MutLα-dependent and requires functional integrity of the MutLα endonuclease metal-binding motif. In contrast to the Exo1-dependent reaction, we have been unable to detect a gapped excision intermediate in Exo1-deficient extracts when repair DNA synthesis is blocked. A possible explanation for this finding has been provided by analysis of a purified system comprised of MutSα, MutLα, replication factor C, proliferating cell nuclear antigen, replication protein A, and DNA polymerase δ that supports Exo1-independent repair in vitro. Repair in this system depends on MutLα incision of the nicked heteroduplex strand and dNTP-dependent synthesis-driven displacement of a DNA segment spanning the mismatch. Such a mechanism may account, at least in part, for the Exo1-independent repair that occurs in eukaryotic cells, and hence the modest cancer predisposition of Exo1-deficient mammalian cells.
Keywords: cancer, DNA polymerase, DNA repair, strand displacment
Mismatch repair is an ubiquitous DNA repair pathway that promotes genetic stability by correcting DNA replication errors, processing recombination intermediates, and initiating checkpoint and apoptotic responses to certain types of DNA damage (1–3). The best understood function of mismatch repair is its role in replication fidelity, although the signals that direct correction to the newly-synthesized DNA strand have not been identified in any eukaryote. However, a strand-specific nick or gap, which may reside either 3′ or 5′ to the mismatch, is sufficient to direct mismatch repair in extracts of mammalian cells (4, 5). The nature of this reaction has been partially clarified by study of several purified systems (6–10).
Although understanding of the reaction is at an early stage, purified human proteins have been used to reconstitute a minimal in vitro system that supports 5′ and 3′ nick-directed mismatch repair in a reaction requiring MutSα (MSH2·MSH6 heterodimer), MutLα (MLH1·PMS2 heterodimer), the proliferating cell nuclear antigen (PCNA) replication clamp, the clamp loader replication factor C (RFC), the ssDNA binding protein replication protein A (RPA), exonuclease 1 (Exo1), and DNA polymerase δ (9). Surprisingly, the 5′ to 3′ double-strand hydrolytic activity of human Exo1 is sufficient to support repair directed by either a 3′ or 5′ strand break (7, 9). This paradoxical observation was clarified by the demonstration that unlike Escherichia coli MutL, eukaryotic MutLα harbors a latent endonuclease that is activated in a manner that depends on a preexisting strand break, a mismatch, MutSα, PCNA, RFC, and ATP (10, 11). Incision by activated MutLα is strongly biased to the discontinuous strand of the heteroduplex and tends to occur on the distal side of the mismatch relative to the original strand break. For a 3′ heteroduplex, incision in this fashion results in the introduction of a 5′ strand break. Multiply-nicked molecules produced in this manner are substrates for MutSα-activated Exo1 (6, 10), which hydrolyzes the DNA segment spanning the mispair.
Although Exo1 has been implicated in eukaryotic mismatch repair (12–14), Exo1 null alleles confer relatively modest mutability in yeast and mice, and Exo1-deficient mouse cell extracts display significant levels of residual repair activity (12, 14). The simplest explanation for these findings would be existence of one or more alternate hydrolytic activities that can substitute for Exo1. However, a screen for yeast mutants that enhance the mutability of an Exo1-deficient strain has failed to identify such an activity, yielding mutations in genes that were previously implicated in the reaction (15). In this study we have reexamined the residual repair that occurs in Exo1-deficient mouse cell extracts and demonstrated Exo1-independent repair in a purified system comprised of MutSα, MutLα, RFC, PCNA, RPA, and DNA polymerase δ. Repair in both cases occurs in the absence of a detectable excision intermediate, and in the purified system is mediated by DNA synthesis-driven strand displacement. Taken together, these findings suggest that strand displacement synthesis may account for the Exo1-independent mode of mismatch repair that occurs in eukaryotic cells.
Results
Mismatch Repair in Exo1-Deficient Mouse Cells.
In vitro analysis of extracts derived from Exo1-deficient mouse ES cells has indicated significant residual mismatch repair, particularly on heteroduplexes containing small insertion/deletion mismatches (14). Using heteroduplexes containing a G–T mismatch, we have confirmed residual mismatch rectification activity in whole-cell extracts of this Exo1−/− ES cell line at 20–25% the level observed in the presence of Exo1. To determine whether this residual activity is bona fide mismatch repair, we have examined extracts derived from MLH1−/− Exo1−/− mouse embryo fibroblast (MEF) cells.
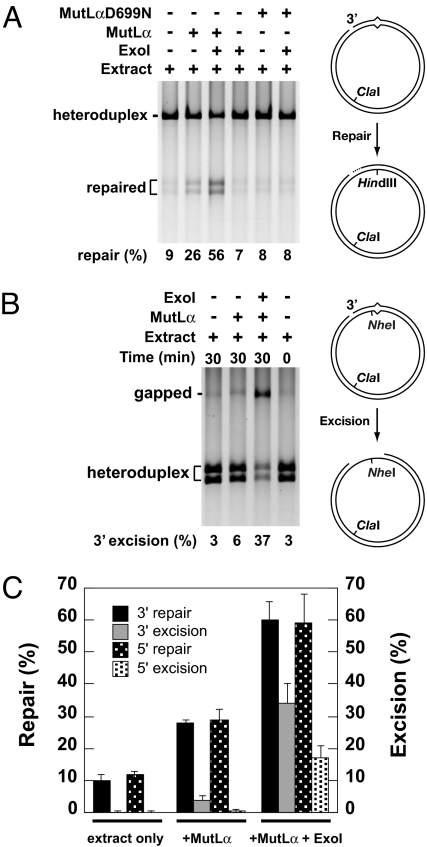
As summarized in Fig. 1A, Fig. S1A, and Table 1, extracts derived from MLH1−/− Exo1−/− cells supported rectification of 7–10% of G–T heteroduplex molecules in a 30-min reaction. Supplementation of the MLH1−/− Exo1−/− MEF cell extracts with near homogeneous human MutLα and Exo1 resulted in robust mismatch repair, at a level 5–6 times that observed with extract alone, an enhancement that was not observed upon supplementation with Exo1 and MutLαD699N, a mutant form of MutLα defective in endonuclease function (Fig. 1A, compare lanes 3 and 6). Supplementation of MLH1- and Exo1-deficient extracts with MutLα alone also potentiated repair ≈3-fold over background levels (Fig. 1A, Fig. S1A, and Table 1), an effect that also depends on the functional integrity of MutLα (Fig. 1A, compare lanes 2 and 5). Repair in extracts supplemented with MutLα alone was inhibited 70% by 300 μM aphidicolin but insensitive to 100–400 μM ddTTP, suggesting involvement of DNA polymerase δ and/or ε but rendering a short patch repair mechanism involving polymerase β highly unlikely. After correction for background rectification, the level of repair observed in the presence of MutLα alone was 35–40% of that observed upon supplementation with both MutLα and Exo1. These findings substantiate occurrence of an Exo1-independent but MutLα-dependent mode of mismatch repair in mouse cell extracts and confirm a previous observation that human MutLα can complement mouse cell extracts deficient in MLH1 and PMS2 (16). They also indicate that human Exo1 can substitute for mouse Exo1 in such extracts.
Fig. 1.
Exo1-independent 3′ directed mismatch repair extracts of MEFs. Mismatch repair and mismatch-provoked excision in extracts of Mlh1−/− Exo1−/− MEF cells were determined as described in Materials and Methods. Reactions were supplemented with MutLα and Exo1 as indicated or with MutLαD699N, which contains an amino acid substitution in the PMS2 metal binding motif that renders the MutLα endonuclease nonfunctional (10). (A) Mismatch repair, which restores a HindIII site, was scored by cleavage with HindIII and ClaI. (B) Mismatch-provoked excision under conditions of repair DNA synthesis block was scored by cleavage with NheI and ClaI. Excision renders the NheI site 5 bp from the mismatch resistant to hydrolysis. (C) 3′ and 5′ Directed mismatch repair and mismatch-provoked excision in MEF cell extracts. Results shown are the average of 4 independent experiments like those in A and B and Fig. S1. Excision values are corrected for 0 time background. Error bars correspond to 1 SD.
Table 1.
Gap formation in MEF cell extracts as scored by oligonucleotide hydbridization assay
| Components | Exp. 1 |
Exp. 2 |
||
|---|---|---|---|---|
| Mismatch repair, % | Excision-gap signal, arbitrary units | Mismatch repair, % | Excision-gap signal, arbitrary units | |
| Mlh1−/−Exo1−/− extract | 7–8 | 4–5 | 7–8 | 7 ± 4 |
| Extract (0 time) | ND | 3–4 | ND | 3–7 |
| Extract + Exo1 | 4–7 | 4–5 | ND | 5 ± 3 |
| Extract + MutLα | 23–24 | 5–5 | 30–33 | 9 ± 2 |
| Extract + MutLα + Exo1 | 58–69 | 20–26 | 54–60 | 23 ± 4 |
Reactions containing 3′ G–T heteroduplex DNA and 120 μg (Exp. 1) or 180 μg (Exp. 2) of Mlh1−/− Exo1−/− MEF cell extract were performed as described in Fig. 1 in the absence (to score repair) or presence of 50 μM aphidicolin (to score excision). Gaps resulting from excision were visualized by hybridization of a 32P-oligonucleotide expected to hybridize to the exposed viral DNA strand within the region spanning the original location of the mismatch (see Materials and Methods). Hybridization results (Excision-gap signal) are expressed as specific activities based on radiolabel and DNA quantification. Results are shown as the range or mean (±1 SD) obtained from 2 or at least 3 independent measurements, respectively. For purposes of comparison, hybridization values obtained in Exp. 1 were normalized to those of Exp. 2 by using the mean value obtained in the presence of MutLα and ExoI as the base. ND, not done.
Previous studies have shown that Exo1-dependent mismatch repair proceeds via an excision intermediate that can be visualized as a gap when repair DNA synthesis is blocked (6, 8, 17, 18). To clarify the nature of the mismatch repair events described above, we have evaluated the formation of excision tracts on G–T heteroduplexes incubated in extracts derived from MLH1−/− Exo1−/− cells. As judged by 2 independent assay methods (Fig. 1B, Fig. S1B, and Table 1), excision intermediates were readily evident when extracts were supplemented with both MutLα and Exo1, but were detected only at trace levels in extracts alone or in extracts supplemented with only MutLα. Results of multiple independent experiments documenting these effects are summarized in Fig. 1C. As can be seen, repair in the presence of MutLα but in the absence of Exo1 occurs without a detectable excision intermediate. Furthermore, the extent of mismatch repair observed upon supplementation of MLH1−/− Exo1−/− cell extracts with both MutLα and Exo1 significantly exceeded the level of detectable excision intermediate.
Exo1-Independent Mismatch Repair in a Purified System.
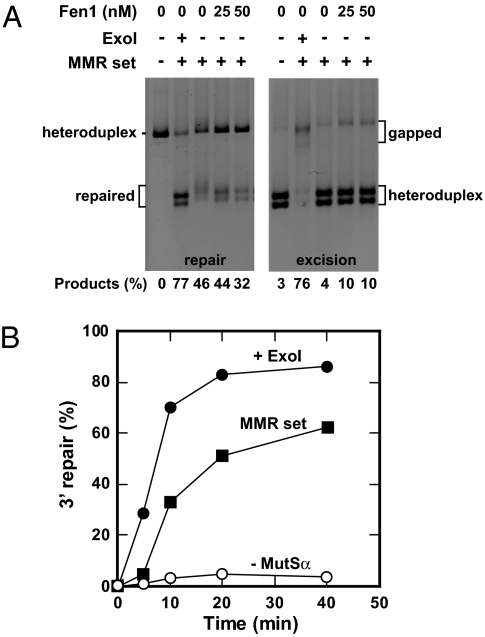
Collectively, these findings suggest that mismatch repair in mammalian cell extracts occurs by an Exo1-dependent mechanism that involves readily detectable excision tracts and at least 1 other pathway that is either linked to ongoing DNA synthesis or involves excision tracts much shorter than those of the Exo1-dependent reaction. However, the latter possibility seems unlikely in view of insensitivity of the reaction to inhibition by ddTTP. DNA polymerase δ, which has been implicated in the DNA synthesis step of mismatch repair (8, 9, 19), is proficient in strand displacement DNA synthesis, a reaction that plays an important role in Okazaki fragment maturation (20). Because strand displacement synthesis could account for the unexpected nature of the Exo1-independent mismatch repair events described above, we have used the reconstituted human repair system to test this possibility. We have previously shown that a 7-component system comprised of MutSα, MutLα, RFC, PCNA, Exo1, RPA, and DNA polymerase δ supports efficient 5′ and 3′ directed mismatch repair, with only low levels of repair observed in the absence of Exo1 during the 5- to 8-min reactions used in these experiments (9). However, the 6-component system comprised of MutSα, MutLα, RFC, PCNA, RPA, and DNA polymerase δ is in fact capable of supporting substantial Exo1-independent mismatch repair provided that reaction time is increased. This effect is illustrated for 30-min incubations in Fig. 2A Left (compare lanes 2 and 3). As judged by conversion of a heteroduplex containing a G–T mismatch within a HindIII recognition site to an endonuclease-sensitive form, the 6-component system supports repair at ≈60% the efficiency of that observed in the presence of Exo1 during a 30-min reaction. In contrast to the Exo1-dependent reaction, repair observed in the absence of the exonuclease occurs with a pronounced lag (Fig. 2B).
Fig. 2.
Exo1-independent mismatch repair in a purified human system. (A) Reactions containing 3′ G–T heteroduplex DNA, MutSα, MutLα, RFC, PCNA, RPA, DNA polymerase δ, and ATP (MMR set; see Materials and Methods) were supplemented with Exo1 or Fen1 as indicated. Gels show repair (Left) or excision (Right) in the presence or absence of the 4 dNTPs respectively, using the methods outlined in Fig. 1. Excision on otherwise identical A·T homoduplex DNA was ≤5% under all conditions. (B) Kinetics of mismatch repair in the presence of MutSα, MutLα, RFC, PCNA, RPA, DNA polymerase δ, and ATP (MMR set) plus the 4 dNTPs, in the presence of the MMR set-dNTP mix from which MutSα was omitted or in the presence of the MMR set, dNTPs and Exo1.
Mismatch repair in the purified system lacking Exo1 differs in several ways from the reaction that occurs in the presence of the exonuclease. Exo1-dependent repair of a 3′ heteroduplex requires MutSα, MutLα, RFC, PCNA, and DNA polymerase δ and is stimulated by RPA (9); the requirements for Exo1-independent repair of a 3′ heteroduplex are similar (Fig. 2B and Table 2). Unlike the 3′ directed reaction, Exo1-dependent repair of a 5′ heteroduplex can occur in the absence of MutLα, an effect attributable to MutSα-dependent activation of 5′ to 3′ hydrolysis by Exo1 (6, 8, 9). By contrast, MutLα is required for efficient repair of a 5′ heteroduplex in the absence of Exo1 (Table 2), and MutLαD699N cannot substitute in this regard, suggesting that function of MutLα as an endonuclease is required for this effect. Exo1-dependent and -independent reactions also differ with respect to production of an excision intermediate. The omission of dNTPs leads to production of gapped intermediates in the Exo1-dependent system, but these species are not observed in the absence of the exonuclease (Fig 2A Right, compare lanes 2 and 3), reminiscent of the results obtained with MEF cell extracts in the absence of Exo1 described above. Last, the HindIII-sensitive repair products produced in the absence of Exo1 display a retarded mobility as compared with those produced in the presence of the exonuclease (Fig. 2A Left, compare lanes 2 and 3). In view of the known ability of DNA polymerase δ to strand displace, this observation suggests that 5′ ssDNA flaps are produced during Exo1-independent repair. The finding that this anomalous mobility was largely resolved by treatment of isolated repair products with the Fen1 flap endonuclease or Fen1 supplementation of Exo1-independent repair reactions (Fig. 2A Left, compare lanes 3–5) is consistent with this idea.
Table 2.
Requirements for reconstituted Exo1-independent mismatch repair
| Repair system | 3′ G–T heteroduplex repair, % | 5′ G–T heteroduplex repair, % |
|---|---|---|
| Complete | 53 | 51 |
| − MutSα | 7 | 7 |
| − MutLα | 7 | 8 |
| − RPA | 20 | 18 |
| − PCNA | <3 | <3 |
| − RFC | <3 | <3 |
| − DNA pol δ | <3 | <3 |
| − dNTPs | <3 | <3 |
| − MutLα + MutLαD699N | 3 | 8 |
Complete reactions contained 3′ or 5′ G–T heteroduplex DNA, MutSα, MutLα, RFC, PCNA, RPA, DNA polymerase δ, ATP, and the 4 dNTPs (see Materials and Methods), with omissions as indicated. When present, 10 nM MutLαD699N was substituted for 10 nM MutLα.
Mismatch- and Synthesis-Dependent Displacement of DNA Segments from Heteroduplex DNA.
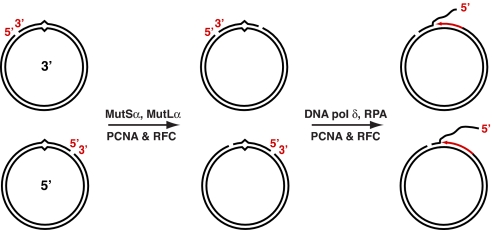
The results summarized above suggested the model for Exo1-independent heteroduplex repair shown in Fig. 3. In this mechanism, activation of the latent MutLα endonuclease leads to strand breaks bracketing the mismatch. These multiply-nicked molecules serve as substrates for synthesis by DNA polymerase δ, resulting in concerted displacement of a DNA segment spanning the mismatch and heteroduplex repair. Such a mechanism would account for the features of the Exo1-independent reaction described above, including the dependence of 5′ heteroduplex repair on MutLα.
Fig. 3.
Model for Exo1-independent mismatch repair. Results presented here are consistent with the 2-stage mechanism shown. MutLα endonuclease, which is activated on a nicked DNA in a mismatch-, MutSα-, RFC, and PCNA-dependent manner, introduces additional breaks into the incised DNA strand in a manner biased to the distal side of the mispair (10). The multiply-nicked product serves as substrate for DNA polymerase δ, which is capable of synthesis-driven strand displacement (20), resulting in coordinate displacement of a DNA segment spanning the mismatch and heteroduplex repair.
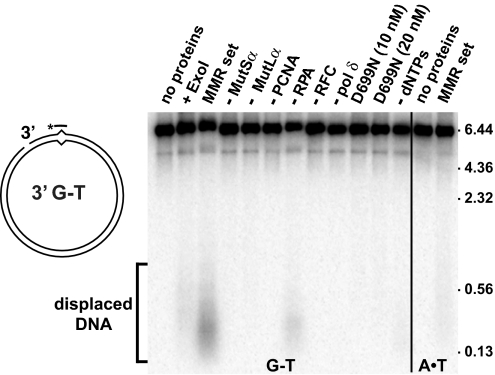
Production of displaced segments of ssDNA in the reconstituted Exo1-independent system was evaluated by nondenaturing gel electrophoresis, followed by Southern analysis with a probe complementary to particular sites within the nicked heteroduplex strand. Strand displacement products spanning the mismatch were produced from both 3′ (Fig. 4) and 5′ (Fig. S2) heteroduplexes by the reconstituted Exo1-independent repair system. Production of this material, which ranged in size from ≈70 to 300 nt, peaking at 100–150 nt, was mismatch-dependent (Fig. 4, compare lanes 3 and 14) and required MutSα, MutLα, PCNA, RFC, DNA polymerase δ, and the presence of dNTPs (Fig. 4, lanes 4–6, 8, 9, and 12). Based on the latter 2 requirements, we infer that strand displacement depends on DNA synthesis. Omission of RPA reduced efficiency of the reaction by ≈70% (Fig. 4, lane 7), and displacement products were not produced by reactions in which wild-type MutLα was replaced with endonuclease-defective MutLαD699N (Fig. 4, lanes 10 and 11). Supplementation of strand displacement reactions with Exo1 dramatically reduced the yield of displaced DNA segments (Fig. 4 and Fig. S2, compare lanes 2 and 3). This finding suggests that excision and concomitant gap formation by Exo1 is the default pathway for mismatch removal in the exonuclease-supplemented purified system and that strand displacement synthesis becomes highly active only in the absence of Exo1.
Fig. 4.
Mismatch-dependent, synthesis-driven displacement of DNA segments from a 3′ G–T heteroduplex. Repair reactions as in Fig. 2 contained 3′ G–T heteroduplex DNA, MutSα, MutLα, RFC, PCNA, RPA, DNA polymerase δ, and ATP (MMR set) plus the 4 dNTPs, with individual additions/omissions as indicated (lanes 2, 4–9 and 12). Results obtained with otherwise identical A·T homoduplex DNAs are shown in lanes 13 and 14. For the reactions shown in lanes 10 and 11, MutLαD699N was substituted for wild-type MutLα. The reactions were analyzed by Southern hybridization assay as detailed in Materials and Methods with a 32P-labeled oligonucleotide [d(gctttcgagtctagaaattcggct)] complementary to that portion of the incised DNA strand spanning the mismatch as a probe. Sizes shown on the right (kB) are based on duplex DNA markers. Comparative electrophoresis of double- and single-stranded markers indicated that displaced DNA segments in lane 3 range in size from ≈70 to 300 nt.
Although endonucleolytic incision of a nicked heteroduplex by purified MutSα, MutLα, PCNA, and RFC is biased to the vicinity the mismatch, cleavage can nevertheless occur throughout the incised strand of a nicked circular heteroduplex, an effect that is attenuated by Exo1 and perhaps by 1 or more additional factors present in cell extracts (10). We therefore tested for displacement of DNA segments distant from the mismatch. As shown in Fig. S3, displacement of such segments does occur, suggesting that DNA synthesis may initiate from multiple sites of MutLα incision along the discontinuous heteroduplex strand.
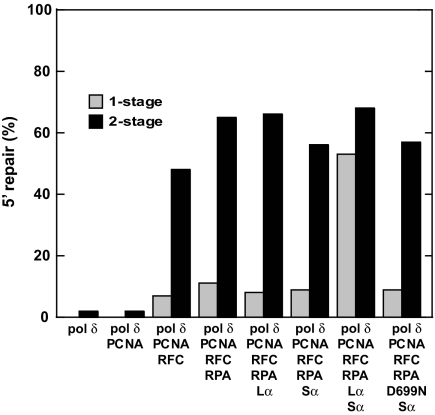
The multiprotein dependence of the strand displacement reaction shown in Fig. 4 and Fig. S2 could reflect required interactions of MutSα and/or MutLα with DNA polymerase δ or its accessory factors. Alternatively, these requirements could indicate that the multiply-nicked heteroduplex molecules produced by activated MutLα are better substrates for strand displacement synthesis by polymerase δ. A 2-stage reaction protocol was used to distinguish between these possibilities. In these experiments, a 5′ heteroduplex was subjected to preincision by incubation with MutSα, MutLα, PCNA, RFC, and RPA. After deproteinization, DNA products were incubated with polymerase δ in the absence or presence of other components of Exo1-independent repair system. As shown in Fig. 5, polymerase δ, PCNA, and RFC in the second-stage reaction were sufficient to support efficient mismatch repair, and the additional presence of RPA provided further modest stimulation. The efficiency of repair observed with these 4 components in the second stage was comparable with that observed in single-stage reactions that contained all 6 activities required for strand displacement-mediated repair. As can be seen, the presence of MutSα and MutLα in the second stage of the 2-stage protocol did not alter the extent of repair, indicating that preincision by MutLα activates the heteroduplex for repair by strand displacement; possible mechanisms for this activation will be considered below. The level of repair observed in 2-stage reactions, where the second incubation contained DNA polymerase δ, PCNA, RFC, and RPA, was ≈6 times that observed in single-stage reactions that contained only these components (Fig. 5). Repair in the latter case presumably resulted from mismatch-independent strand displacement synthesis initiating at the original nick in the 5′ heteroduplex, located 128 bp from the mismatch.
Fig. 5.
Exo1-independent mismatch repair in a 2-stage reaction. Single-stage reactions (gray bars) contained 5′ G–T heteroduplex DNA, ATP, the 4 dNTPs, and the protein components as indicated on the abscissa (see Materials and Methods; D699N indicates substitution of MutLα by MutLαD699N). Repair was scored by cleavage with HindIII and ClaI (Fig. 1). No detectable repair was observed in the presence of only polymerase δ or polymerase δ plus PCNA. In 2-stage reactions (black bars), the 5′ G–T heteroduplex was preincised by 30-min incubation with MutSα, MutLα, RFC, PCNA, RPA, the 4 dNTPs, and ATP, followed by deproteinization (see Materials and Methods). Isolated DNA was then incubated for 30 min with ATP, the 4 dNTPs, and the indicated protein components. Repair was scored with HindIII and ClaI as above.
Discussion
Using extracts derived from MLH1−/− Exo1−/− MEF cells, we have confirmed occurrence of Exo1-independent mismatch repair and have shown this reaction to be largely aphidicolin-sensitive and dependent on functional MutLα. Surprisingly, this reaction occurs by a mechanism that does not involve the long excision tracts that are characteristic of the Exo1-dependent reaction. Analysis of a minimal purified system that supports human mismatch repair in vitro has demonstrated that this system also supports an Exo1-independent mode of repair, and this reaction occurs without production of an excision tract spanning the mismatch. Repair in the purified system is accompanied by production of DNA flaps and the displacement of ssDNA segments from the heteroduplex, with displacement dependent on MutLα incision of the nicked heteroduplex strand and synthesis by DNA polymerase δ. Based on these observations, we have concluded that Exo1-independent repair that occurs in the reconstituted system occurs by a mechanism involving polymerase δ-dependent synthesis-driven displacement of a DNA segment spanning the mismatch as illustrated in Fig. 3. As mentioned above, strand displacement synthesis by DNA polymerase δ is believed to play an important role in Okazaki fragment maturation (20).
Given the similarity of the Exo1-independent reactions that occur in MEF cell extracts and the purified system, repair by strand displacement provides a plausible mechanism for at least some of the mismatch rectification events that occur in Exo1-deficient cells. It is important to note in this regard that the activity concentrations used in the purified system described here are comparable to those present in 100 μg of mammalian nuclear extract, which is typically used to score mismatch repair in vitro (4, 7, 17). The idea that strand displacement repair may account for some mismatch rectification events that occur in the cell is also consistent with the findings of Amin et al. (15), who identified mutations that synthetically enhance the modest mutability of a Sacccharomyces cerevisiae exo1Δ allele. With the exception of a mutation in ribonucleotide reductase, all of the exo1-dependent mutations identified in this study alter proteins previously implicated in mismatch repair, namely MLH1, PMS1 (corresponds to mammalian PMS2), MSH2, MSH3, POL30 (PCNA), and POL32. The pol32 defect is of particular interest because this gene encodes a nonessential 40-kDa subunit of yeast polymerase δ that plays an important role in the production of long flaps via extended strand displacement synthesis by the enzyme (20). As a further test of the strand displacement mechanism for mismatch repair, we have attempted to identify strand displacement products produced upon incubation of heteroduduplex substrates in Exo1-deficient MEF cell extracts. However, these experiments have yielded negative results, a failure that could be caused by nucleolytic processing in the extract, e.g., degradation of strand displacement intermediates by Fen1 or DNA2 exo/endonucleases during the flap stage of the reaction, or single-strand specific exonucleases after displacement from the helix.
We have found that preincision of a nicked 5′ heteroduplex with activated MuLα dramatically activates the substrate for strand displacement repair relative to that observed with the untreated substrate (Fig. 5). The native heteroduplex (5′ nick to mismatch distance of 128 bp) might be expected to support strand displacement synthesis, and in fact, limited mismatch rectification was observed when this DNA was challenged with polymerase δ, RFC, and PCNA (± RPA). Preincision with activated MutLα led to 6-fold enhancement of the repair observed upon subsequent incubation with the DNA biosynthetic activities, but the basis of this effect is not clear. Although MutLα incision of this 5′ heteroduplex is biased to the distal side of the mispair relative to the location of the original strand break, limited incision does occur between the 2 DNA sites (10). Incision in the latter manner would reduce the distance over which displacement synthesis would have to occur to result in repair. Although such effects might account for the enhancement observed, it is difficult to reconcile this idea with the finding that DNA segments displaced from the heteroduplex during Exo1-independent repair range in size from ≈70 to 300 nt (Fig. 4). An alternate possibility is that multiply-nicked molecules are simply better substrates for strand displacement synthesis. For example, such molecules may be better substrates for loading of polymerase accessory proteins like PCNA than are molecules with only 1 DNA strand discontinuity.
Materials and Methods
Cell Lines, Cell Culture, Extracts, and Proteins.
Cell lines and methods for cell culture, extract preparation, and protein isolation are described in SI Text and Figs. S4 and S5.
Mismatch Repair Substrates and Reactions.
3′ G–T heteroduplex, 3′ A·T homoduplex, 5′ G–T heteroduplex, and 5′ A·T homoduplex DNAs were prepared as described (7, 17). 3′ Heteroduplex DNA contained a single-strand break 141 bp 3′ to the mismatch (shorter path in the circular molecule), whereas the nick-mismatch distance in the 5′ heteroduplex was 128 bp.
Mismatch repair reactions were performed by a modification of the described method (10). Unless indicated otherwise, reconstituted mismatch repair reactions (40 μL) contained 20 mM Hepes-NaOH (pH 7.6), 140 mM KCl, 5 mM MgCl2, 2 mM ATP, 2 mM DTT, 0.2 mg/mL BSA, 5% (vol/vol) glycerol, 1.2 nM DNA (0.2 μg of nicked 5′ or 3′ DNA), and 0.2 mM each dATP, dGTP, dCTP, and dTTP. MutSα (12.5 nM), MutLα (10 nM), RFC (9 nM), PCNA (30 nM), RPA (100 nM), Exo1b (2.5 nM), and DNA polymerase δ (1.1 nM), were present as indicated. Reconstituted mismatch-provoked excision reactions were performed in the same manner, but dNTPs were omitted. Mismatch repair in mouse cell extracts was determined in 40-μL reactions containing 120 μg of extract protein under conditions described above except that KCl concentration was 100 mM, ATP was 3 mM, glycerol was 1.5% (vol/vol), and heteroduplex DNA was 0.6 nM (0.1 μg). Extract reactions were supplemented with 20 nM MutLα, 20 nM MutLαD699N, and/or 2.5 nM Exo1b as indicated. Mismatch-provoked excision in cell extracts was performed in a similar manner except that exogenous dNTPs were omitted and aphidicolin was included at 100 μM.
After incubation at 37 °C for 30 min, reactions were terminated by the addition of 30 μL of 0.35% SDS, 0.3 mg/ml Proteinase K, 0.4 M NaCl, 0.3 mg/mL glycogen, and 13 mM EDTA, followed by incubation at 50 °C for 15–30 min. After extraction with phenol/chloroform, isopropanol precipitation, and wash with 70% ethanol, recovered DNA was dissolved in NEB4 buffer (NEB) supplemented with 0.1–0.2 mg/mL BSA and digested with HindIII and ClaI to score mismatch repair or with NheI and ClaI to score excision. Restriction endonuclease digestions were usually for 1–2 h at 37 °C. In the case of the extract-based reactions, restriction digests were supplemented with 27–36 μg/mL RNase A, and cleavage with HindIII and ClaI was for 2 h, whereas hydrolysis with NheI and ClaI was for 16 h. Products were visualized and quantified after native agarose gel electrophoresis (13).
Southern hybridization assay was used to score displacement of DNA segments from DNA substrates, in which case recovered DNAs were not subjected to restriction digestion. After electrophoresis through a nondenaturing 1.1% agarose gel and alkaline transfer to a nylon membrane, products were hybridized with the indicated 32P-labeled oligonucleotide (7, 17). When indicated, a hybridization-based assay (18) was used to score mismatch-provoked excision in MEF cell extracts. In this case recovered DNAs (4–8 fmol) treated with RNase A were hybridized with a 32P-labeled oligonucleotide [20 fmol, d(agccgaatttctagactcgaaagc)] in a 10-μL reaction [20 mM Hepes·NaOH (pH 7.4), 100 mM NaCl, and 10 mM MgCl2] at 40 °C for 2 h and immediately resolved by electrophoresis through a nondenaturing 0.8% agarose gel at 5 V/cm for 90 min. DNA bands were visualized and quantified as above, and the gel was dried on DEAE paper. Probe hybridization was visualized and quantified by using Molecular Dynamics or GE Healthcare phosphorimagers.
Supplementary Material
Acknowledgments.
We thank Mike O'Donnell, Alan Tomkinson, and Roger Woodgate for contributions to the design and costs of the RFC expression constructs; Ellen Fanning (Vanderbilt University) and Guo-Min Li (University of Kentucky) for DNA polymerase δ baculovirus constructs; Binghui Shen (City of Hope) for the Fen1 expression plasmid; and Leonid Dzantiev for insightful discussions. This work was supported in part by National Institutes of Health Grants GM45190 (to P.M.) and CA93484 (to W.E.) and start-up funds from the Southern Illinois University School of Medicine (to F.A.K.). P.M. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0903654106/DCSupplemental.
References
- 1.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 2.Modrich P. Mechanisms in eukaryotic mismatch repair. J Biol Chem. 2006;281:30305–30309. doi: 10.1074/jbc.R600022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98. doi: 10.1038/cr.2007.115. [DOI] [PubMed] [Google Scholar]
- 4.Holmes J, Clark S, Modrich P. Strand-specific mismatch correction in nuclear extracts of human and Drosophila melanogaster cell lines. Proc Natl Acad Sci USA. 1990;87:5837–5841. doi: 10.1073/pnas.87.15.5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thomas DC, Roberts JD, Kunkel TA. Heteroduplex repair in extracts of human HeLa cells. J Biol Chem. 1991;266:3744–3751. [PubMed] [Google Scholar]
- 6.Genschel J, Modrich P. Mechanism of 5′-directed excision in human mismatch repair. Mol Cell. 2003;12:1077–1086. doi: 10.1016/s1097-2765(03)00428-3. [DOI] [PubMed] [Google Scholar]
- 7.Dzantiev L, et al. A defined human system that supports bidirectional mismatch-provoked excision. Mol Cell. 2004;15:31–41. doi: 10.1016/j.molcel.2004.06.016. [DOI] [PubMed] [Google Scholar]
- 8.Zhang Y, et al. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
- 9.Constantin N, Dzantiev L, Kadyrov FA, Modrich P. Human mismatch repair: Reconstitution of a nick-directed bidirectional reaction. J Biol Chem. 2005;280:39752–39761. doi: 10.1074/jbc.M509701200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLα in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 11.Kadyrov FA, et al. Saccharomyces cerevisiae MutLα is a mismatch repair endonuclease. J Biol Chem. 2007;282:37181–37190. doi: 10.1074/jbc.M707617200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tishkoff DX, et al. Identification and characterization of Saccharomyces cerevisiae EXO1, a gene encoding an exonuclease that interacts with MSH2. Proc Natl Acad Sci USA. 1997;94:7487–7492. doi: 10.1073/pnas.94.14.7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Genschel J, Bazemore LR, Modrich P. Human exonuclease I is required for 5′ and 3′ mismatch repair. J Biol Chem. 2002;277:13302–13311. doi: 10.1074/jbc.M111854200. [DOI] [PubMed] [Google Scholar]
- 14.Wei K, et al. Inactivation of exonuclease 1 in mice results in DNA mismatch repair defects, increased cancer susceptibility, and male and female sterility. Genes Dev. 2003;17:603–614. doi: 10.1101/gad.1060603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amin NS, Nguyen MN, Oh S, Kolodner RD. Exo1-dependent mutator mutations: Model system for studying functional interactions in mismatch repair. Mol Cell Biol. 2001;21:5142–5155. doi: 10.1128/MCB.21.15.5142-5155.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tomer G, Buermeyer AB, Nguyen MM, Liskay RM. Contribution of human mlh1 and pms2 ATPase activities to DNA mismatch repair. J Biol Chem. 2002;277:21801–21809. doi: 10.1074/jbc.M111342200. [DOI] [PubMed] [Google Scholar]
- 17.Fang W-h, Modrich P. Human strand-specific mismatch repair occurs by a bidirectional mechanism similar to that of the bacterial reaction. J Biol Chem. 1993;268:11838–11844. [PubMed] [Google Scholar]
- 18.Wang H, Hays JB. Mismatch repair in human nuclear extracts. Quantitative analyses of excision of nicked circular mismatched DNA substrates, constructed by a new technique employing synthetic oligonucleotides. J Biol Chem. 2002;277:26136–26142. doi: 10.1074/jbc.M200357200. [DOI] [PubMed] [Google Scholar]
- 19.Longley MJ, Pierce AJ, Modrich P. DNA polymerase δ is required for human mismatch repair in vitro. J Biol Chem. 1997;272:10917–10921. doi: 10.1074/jbc.272.16.10917. [DOI] [PubMed] [Google Scholar]
- 20.Stith CM, Sterling J, Resnick MA, Gordenin DA, Burgers PM. Flexibility of eukaryotic Okazaki fragment maturation through regulated strand Displacement Synthesis. J Biol Chem. 2008;283:34129–34140. doi: 10.1074/jbc.M806668200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.