Summary
We describe a mechanism by which nascent RNA can inhibit transcriptional pausing. PutL RNA of bacteriophage HK022 suppresses transcription termination at downstream terminators and pausing within a nearby U-rich sequence. Using in vitro transcription assays and footprinting techniques, we demonstrate that this pausing results from backtracking of RNA polymerase, and that binding of nascent putL RNA to the enzyme limits backtracking by restricting re-entry of the transcript into the RNA exit channel. The restriction is local and relaxes as the transcript elongates. Our results suggest that putL RNA binds to the surface of RNA polymerase close to the RNA exit channel, a region that includes amino acid residues important for antitermination. Although binding is essential for antipausing and antitermination, these two activities of put differ: antipausing is limited to the immediate vicinity of the putL site, but antitermination is not. We propose that RNA anchoring to the elongation complex is a widespread mechanism of pause regulation.
Keywords: E. Coli, RNA polymerase, RNA binding, Phage HK022, Transcript elongation, Antitermination
Introduction
After RNA polymerase (RNAP) binds to a promoter and synthesizes a short transcript, it normally remains bound to the same template and continues to elongate the same RNA chain until it reaches a terminator. At this point, the enzyme frequently releases the template and nascent RNA. The simplest class of terminator is called "intrinsic", because its only essential component is a sequence embedded in the nascent transcript. Intrinsic terminators consist of a short U-rich stretch preceded by a sequence that forms a stable RNA hairpin [reviewed in (Richardson and Greenblatt, 1996)]. Termination occurs within the U-rich stretch, 6–8 nt from the base of the hairpin. Embedded RNA signals can also have the opposite effect, reducing the probability of disruption of the elongation complex (EC) when it encounters terminators. "Intrinsic antiterminators" were discovered and characterized in bacteriophage HK022, where they increase the expression of many virus genes (Clerget et al., 1995). The antiterminator transcripts are encoded by the phage putL and putR sites, which are located downstream of the early promoters PL and PR, respectively. They fold into two stem-loops, and folding is required for activity (Fig. 1A)(King et al., 1996; Banik-Maiti et al., 1997). Ribonuclease protection experiments indicate that the transcripts bind the ECs that made them, and that a mutation of RNAP that prevents put antitermination also prevents binding (Sen et al., 2001). This conclusion is supported by the observation that antitermination-defective mutations in putL or RNAP destabilize the putL transcript in vivo (Sloan et al., 2007). Modification of an EC by putL RNA not only suppresses termination, it also increases the rate of transcript elongation (King et al., 1996; R. Robins, R.A. King, and I. Molyneux, unpublished experiments. et al., 2008)
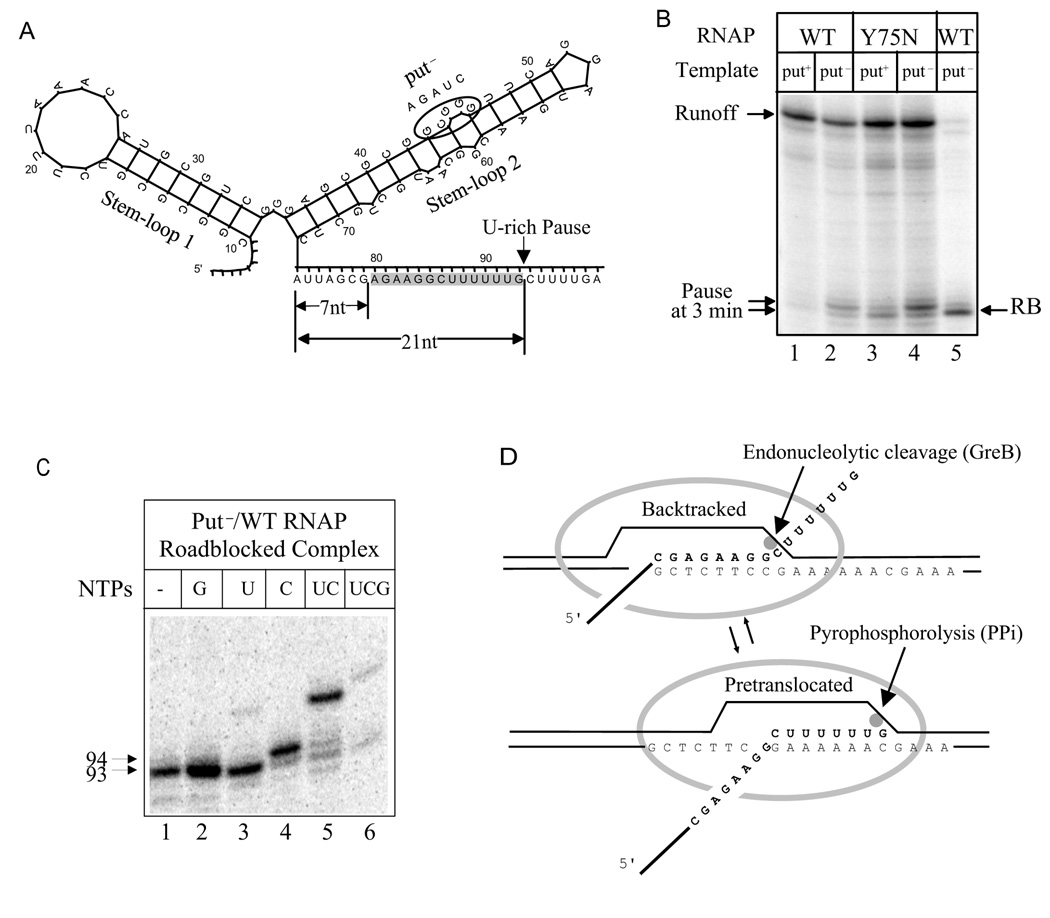
Fig. 1. PutL-mediated suppression of a U-rich pause.
A. A diagram of folded putL RNA and the U-rich pause. The HK022 PL-proximal transcript, which includes the two stem-loops that comprise putL RNA and the downstream U-rich pause, is shown. The RNA segment within an EC paused at position G93 is indicated by nucleotides shown with a gray background. The sequence of the antitermination-defective putL mutation used in most experiments is shown [mutant G of (King et al., 1996)]. B. The effect of antitermination-defective mutations in putL and RNAP on pause intensity. Lanes 1–4: The template encoding WT or mutant putL was transcribed by WT or mutant RNAP for 3 min in the presence of 100 µM ATP, UTP, GTP and 25 µM CTP. Lane 5: An EC roadblocked by Lac repressor at the pause site was obtained as described in Methods. C. Mapping the pause sites by RNAP walking. An EC roadblocked at the pause site (lane 1) was incubated in the presence of the indicated NTPs after the roadblock was removed with IPTG (lanes 2–6). D. Backtracked and pretranslocated states of an EC stalled at position G93. The grey oval represents RNAP and the grey dot, the active center.
Estimates of transcript elongation rates in vivo vary within the range of 28 to 90 nt/sec, depending on the gene, growth conditions, and method of measurement [see (Vogel and Jensen, 1994; Bremer and Dennis, 1996) and references therein]. Elongation rates observed in vitro vary between 4 and 14 nt/sec (Wang et al., 1998; Adelman et al., 2002). The lower rate in vitro can be explained by the lack of accelerating factors in highly purified systems. In addition, the rate of elongation is not uniform throughout a given DNA sequence: ECs have a high probability of pausing for variable amounts of time at particular positions, and such pauses can have important physiological functions [reviewed in (Landick, 2006)]. Several general types of sequence-dependent pauses have been characterized. One type is associated with the formation of an RNA stem-loop immediately upstream of the EC (Levin and Chamberlin, 1987; Chan and Landick, 1993). A second is associated with a retrograde movement of the EC known as backtracking (Komissarova and Kashlev, 1997b; Palangat and Landick, 2001; Nudler et al., 1997) (see below). A third depends on binding of a DNA sequence downstream of the transcription start by the σ subunit of RNAP (Ring et al., 1996). Finally, a fourth type of sequence-dependent pause that is associated neither with RNA stem-loops nor backtracking has recently been described (Herbert et al., 2006).
In earlier work, we showed that putL suppressed pausing at a U-rich sequence located close to and downstream of the putL site (King et al., 1996). Here we show that this pause is associated with backtracking of the EC, and that put-dependent modification of the EC suppresses backtracking. However, in contrast to the antitermination activity of putL, which is not strongly dependent on the distance of the terminator from the putL site (Sloan et al., 2007), the pause was no longer suppressed when it was moved away from putL. Thus, the mechanism of suppression of the U-rich pause differs from that of antitermination.
Results
PutL suppression of backtracking
A transcriptional pause occurs within a U-rich sequence located 16–25 nt downstream of the base of stem 2 of the putL site (the "U-rich pause")(Fig. 1A)(King et al., 1996). Pausing was barely detectable when wild type template and RNAP were used, but strong when antitermination was prevented by a multiple base pair substitution that changes stem-loop 2 (put− of Fig. 1A), or by an RNAP mutation that prevents putL binding [β'-Y75N; (Sen et al., 2001)](Fig. 1B). We located the pause more precisely by adding incomplete sets of NTPs to an immobilized EC that had been roadblocked at the pause position with Lac repressor (see below)(Fig. 1C). There are two major pause positions, G93 and C94, located 21 and 22 nt, respectively, downstream of the base of stem-loop 2 of putL. G93 is more intense than C94 under our transcription conditions (low CTP). We measured the efficiency of pausing at this site under conditions when putL was active and when it was not. The strength of a pause can be estimated by two parameters: the fraction of ECs that pause and the time required to resume elongation (measured by the half-life of the paused ECs) (Landick et al., 1996). The β'-Y75N mutation increased the fraction of paused ECs from less than 8% to 60% and the half-life of these ECs from about 0.2 min to 3.0 min (Suppl. fig. 1). To understand how putL suppresses the pause, we analyzed pause- and put-associated changes in the EC structure.
An EC envelops about 14 nucleotides of nascent RNA. Eight to nine nucleotides adjacent to the growing point at the 3' end are hybridized to the template strand of DNA, and the remainder lies within an exit channel in RNAP (Komissarova and Kashlev, 1998; Korzheva et al., 2000). About 12 bp of DNA are melted to form the "transcription bubble" (Zaychikov et al., 1995). At certain sequences, the EC is prone to "backtracking", a retrograde movement during which nucleotides at the 3' end of the transcript are melted from the template DNA strand and extruded from RNAP, and compensating amounts of upstream RNA and DNA re-enter the EC (Fig. 1D). The length of the RNA-DNA hybrid is maintained during backtracking. Backtracking is favored by thermodynamically weak RNA-DNA hybrid at the forward position and stronger hybrid at the backtracked position (Palangat et al., 1998; Shaevitz et al., 2003). Since the 3' end of the transcript is distant from the active center of a backtracked EC, elongation cannot resume unless RNAP slides forward to re-engage the 3' end with the active center, or the extruded RNA is cleaved to form a new and appropriately positioned 3' end (Komissarova and Kashlev, 1997b; Nudler et al., 1997). Backtracking is associated with a class of sequencedependent transcriptional pauses (below) and increases their duration.
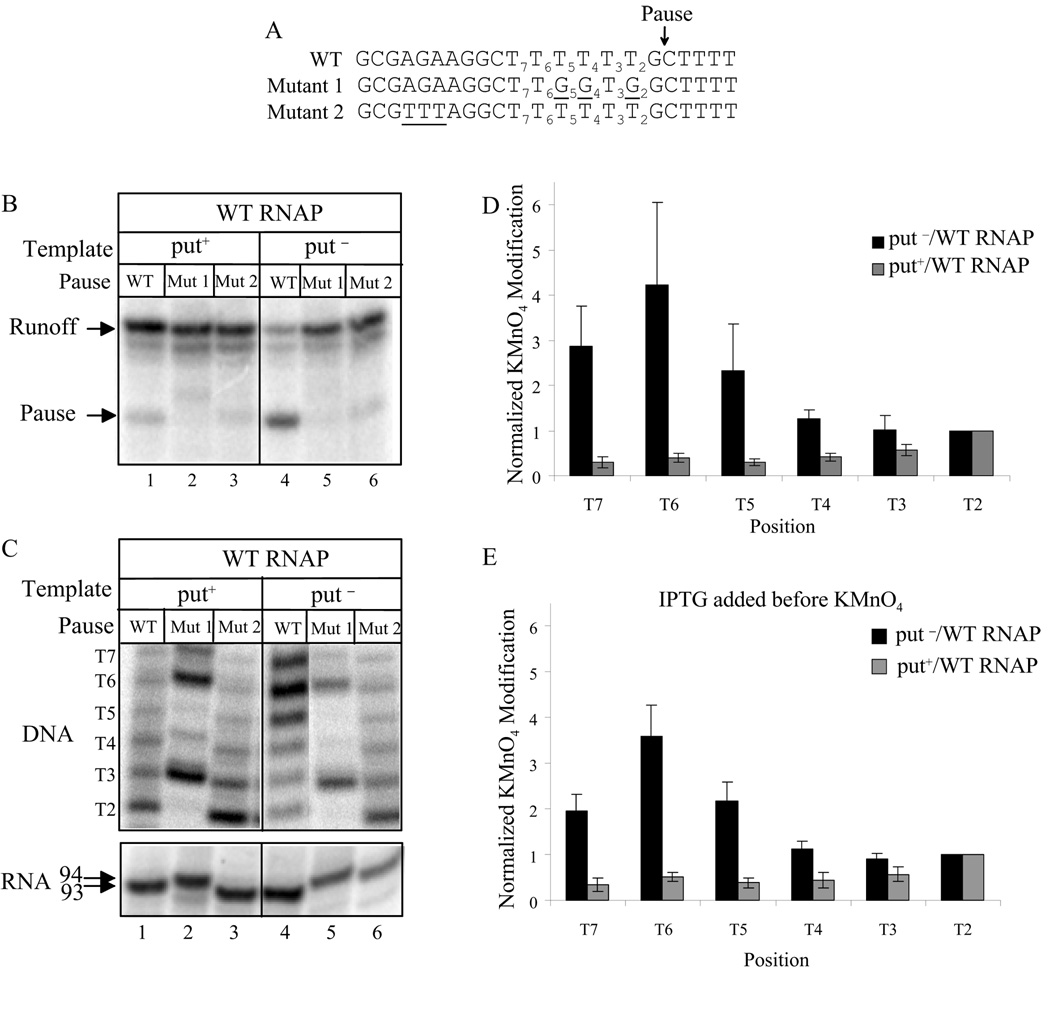
There are six thermodynamically weak rU-dA base pairs in the RNA-DNA hybrid of an EC paused at position G93, and backtracking should increase hybrid stability (Fig. 1D). To see if the U-rich pause is enhanced by backtracking, we made mutant templates that strengthened the RNA-DNA hybrid at positions 2, 4, and 5 nt upstream of G93 (Mutant 1) or weakened the hybrid at (potentially backtracked) positions 12, 13, and 14 upstream of G93 (Mutant 2) (Fig. 2A). Both mutants greatly diminished the intensity of the pause when putL action was prevented (Fig. 2B, lanes 5 and 6). These results are expected if the U-rich pause is associated with and enhanced by backtracking and raise the possibility that putL suppresses the U-rich pause by inhibiting backtracking.
Fig. 2. PutL suppresses backtracking at the U-rich pause site.
A. The sequences of the nontemplate DNA strands of the wild type (WT) U-rich pause and those of mutants 1 and 2 are shown. The sequence numbering is relative to the site of pausing, and changes from the wild type are underlined. The HK022 PL promoter followed by a wild type or mutant putL site is upstream of the pause. B. The templates were transcribed and sampled at 3 minutes as described. C. (Top) KMnO4 cleavage patterns of immobilized ECs that were roadblocked at the pause by Lac repressor and washed free of unincorporated NTPs. The DNA was labeled at the 3' end of the non-template strand. The templates are the same as in panel B, and the thymines are numbered as in panel A. Recall that T2, T4, and T5 are missing in mutant 1 (lanes 2 and 5), so little or no cleavage was seen at these positions. (Bottom) Labeled RNA made in transcription reactions identical to those used for KMnO4 cleavage. The reduced mobility of RNA in lanes 2 and 5 can be explained either as a direct effect of the sequence change or by a sequencedependent change in the position of the roadblocked EC. D. Quantitation of the cleavage patterns shown in panel C, lanes 1 and 4. The amount and recovery of paused ECs differed between the two templates. To compensate for this, the intensities of the bands formed by cleavage at T2 were set to 1.0 for each template, and the intensities at other positions are relative to those at T2. E. As in panel D, except repressor was removed from the roadblocked and washed ECs by exposure to 1 mM IPTG for 10 min before KMnO4 treatment.
To examine this possibility, we used KMnO4 footprinting to locate the position of the transcription bubble in put-modified and unmodified ECs that were artificially stalled at position G93 by a Lac repressor roadblock (Fig. 1B, lane 5). We previously showed that similar stalled ECs could resume transcription after removal of the roadblock, and that restarted ECs retained the put-mediated modification (Sen et al., 2001; King et al., 2003). KMnO4 attack potentiates DNA strand cleavage at unpaired T residues, and such residues may exist within the non-template strand of the transcription bubble. We found that residues T5, T6, and T7 were cleaved less readily than residues T2, T3, and T4 in the non-template strand of stalled put-modified ECs (residue G1 is the pause position, 93 nt from the 5' end)(Fig. 2C, lane 1 and 2D, grey bars). By contrast, when we inactivated putL by mutation we found that residues T5, T6, and T7 were cleaved more readily than residues T2, T3, and T4 (Fig. 2C, lane 4, and 2D, black bars).
The upstream skewing of the distribution of cleavages in the putL mutant ECs is expected if these ECs had backtracked, reforming T:A base-pairs at the downstream positions and moving the transcription bubble upstream from the pause site. The modest downstream skewing of the distribution of cleavages in the put-modified ECs can be explained by assuming that all of these T residues are within the transcription bubble of the EC located at the pause, but that there are small differences in their accessibility or intrinsic susceptibility to KMnO4 attack. When pausing was inhibited by mutations 1 or 2 in the pause region, the KMnO4 cleavage pattern was consistent with the downstream location of the transcription bubble, and put-modification had little or no effect on the distribution (Fig. 2C, lanes 2, 3, 5, and 6). This is very clear for mutation 2 but less so for mutation 1, probably because the unreactive G residues at positions 2, 4, and 5 alter the reactivity of the adjacent T3 and T6. This can occur because of changes in base stacking. Nevertheless, even here it appears that cleavage of T3 ≥ T6 > T7, consistent with the downstream location of the bubble.
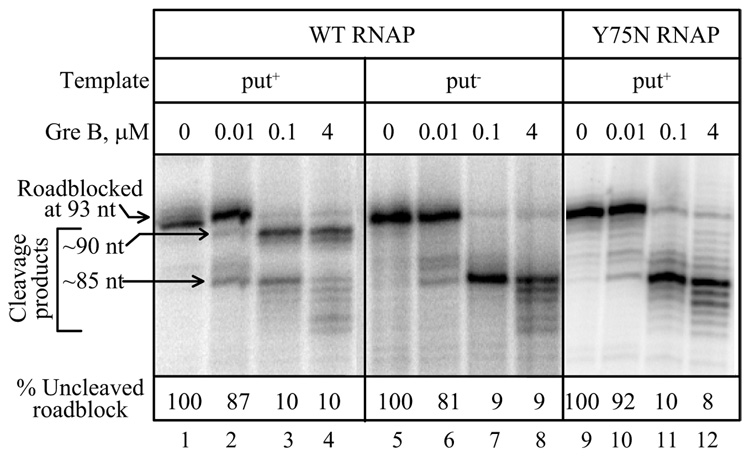
The results of GreB-mediated RNA cleavage of similar stalled ECs are consistent with the patterns of KMnO4 cleavage. GreB is an E. coli protein that promotes hydrolytic cleavage within the 3' proximal region of the transcript in backtracked ECs. Cleavage occurs at the active center of RNAP (Orlova et al., 1995; Sosunov et al., 2003; Laptenko et al., 2003), so the size of the products indicates the extent of backtracking. We found that both put-modified and unmodified roadblocked ECs were cleaved by GreB, but the products differed markedly (Fig. 3). Cleavage of a stalled put-modified EC by low concentrations of GreB gave a major 5'-terminal product that was 2 to 3 nt shorter than the uncleaved RNA, and a minor product that was shorter by 7 to 8 nt (Fig. 3, lanes 1–4). The minor product was further cleaved when the highest concentration of GreB was used. The 7–8 nt but not the 2–3 nt cleavage product was seen after GreB treatment of stalled ECs formed with antitermination-defective putL or RNAP mutants (Fig. 3, lanes 5–12). These observations argue that the active center of halted ECs can be located further downstream when putL is active than when it is not, consistent with the results of KMnO4 mapping of the transcription bubble. Low amounts of cleavage at the -8 position in the put-modified EC (Fig. 3, Suppl. figs. 2 and 3) could be due to absence of the modification in a fraction of the population. Replacement of the 7–8 nt product by products cleaved even further upstream upon addition of the highest concentration of GreB can be explained by cleavage that occurs after backtracking of the unmodified ECs to more upstream and rarely occupied positions.
Fig. 3.
GreB cleavage of put-modified and unmodified ECs. Immobilized ECs were roadblocked at the U-rich pause site and washed as described. GreB was added at the indicated concentrations for 3 min, and the products were analyzed by gel electrophoresis and autoradiography. The amount of uncleaved EC was calculated as a fraction of roadblocked EC. The slight offset of the RNA bands in the first two lanes was not reproducible, and both represent G93 RNAs.
An alternative interpretation of the effect of put on the GreB cleavage pattern is that put modification changes the active site in such a way as to alter GreB cleavage. This interpretation appears unlikely because it fails to explain the KMnO4 results and because we would expect such alteration to affect the probability rather than the location of cleavages. Another potential problem of interpretation arises from the observation that a transcriptional roadblock can induce backtracking at sequences where it does not normally occur [(Epshtein et al., 2007) and references cited therein]. However, the following experiments argue strongly that backtracking occurs at the U-rich pause site even in the absence of a roadblock. First, the KMnO4 and GreB cleavage patterns of arrested ECs were not significantly changed when Lac repressor was removed by preincubation of the ECs with IPTG in the absence of NTPs (Fig. 2E and Suppl. fig. 2A). Second, two mutants that decrease the stability of RNA:DNA hybrid in backtracked relative to active ECs at the U-rich pause site suppressed pausing during unobstructed transcription (Fig. 2B). Finally, we have shown that GreA and GreB suppress pausing during unobstructed transcription (Suppl. fig. 2).
It is worth noting that backtracked ECs probably oscillate among several positions. KMnO4 mapping gives us a snapshot of this population while determination of the 5' products of GreB cleavage might indicate the location of the most extreme backtracked position. This is because cleavage can occur repeatedly from the 3' end as RNAP backtracks (Lee et al., 1994). To map the location of backtracked ECs more precisely, we determined the sizes of the 3'- proximal products of GreB cleavage of paused (i.e., not roadblocked) ECs. These products must be generated by the initial cleavage event. We found that the main product of cleavage of put− transcripts during unobstructed transcription was 8 nt long with smaller amounts of 9 and 10 nt products (Suppl. fig. 3). This finding also confirms that the EC backtracks at the pause site during continuous elongation.
Two other experiments support the hypothesis that put limits backtracking. First, we found that an unmodified EC resumed transcription more slowly than a comparable put-modified EC when repressor was removed and NTPs added (Suppl. fig. 4A). Second, the initial rate of pyrophosphorolysis (the reversal of chain elongation) was higher for put-modified than unmodified ECs (Suppl. fig. 4B). Backtracked ECs are unable to catalyze either forward elongation or pyrophosphorolysis until the 3' end re-engages the active site. Delayed re-engagement in unmodified ECs is consistent with the hypothesis that put limits backtracking.
Antitermination and antipausing activities of putL
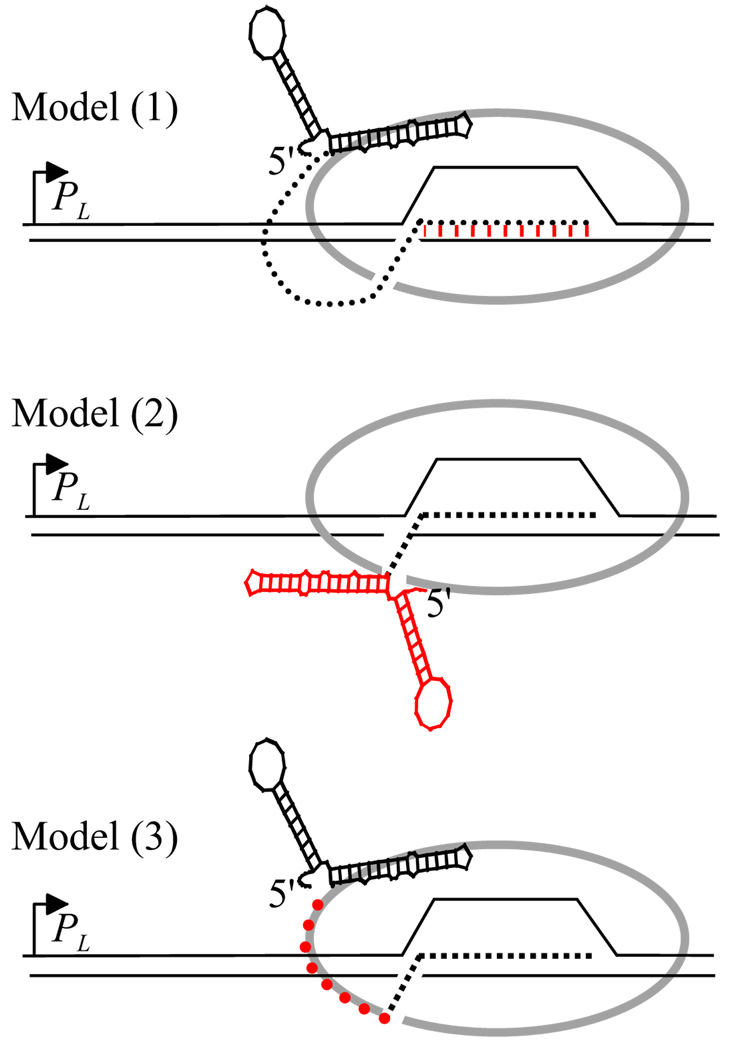
We consider three models of the relationship between antitermination and antipausing (Fig. 4). In the first, the modification conferred by putL on the EC suppresses pausing and termination by the same (unknown) mechanism. In Fig. 4 we suggest that binding of putL RNA to the EC alters the RNA:DNA hybrid, but this is only one of several possibilities. Another is a put-mediated alteration in the active site that increases elongation rate. In model 2, backtracking, and therefore the U-rich pause, are suppressed by folding of putL RNA into a structure that inhibits re-entry of nascent RNA into the RNA exit channel of RNAP [see (Komissarova and Kashlev, 1997b; Reeder and Hawley, 1996)]. In this model the putL mutation (Fig. 1A) and β'-Y75N would alter or delay RNA folding. Binding of putL RNA to the EC is required only for antitermination. In the third model, binding of putL RNA to the EC is required for both antipausing and antitermination, but the mechanisms are different. Backtracking, and therefore the U-rich pause, are suppressed because the anchored RNA is unable to re-enter the exit channel. Terminators are suppressed because bound putL RNA confers a persistent although unknown modification on the EC, a modification that does not prevent backtracking. Note that in model 3, unlike model 2, RNA folding does not by itself prevent backtracking. The evidence presented next contradicts the first two models and is consistent with the third.
Fig. 4.
Models of put-mediated pause suppression. Each diagram shows RNAP (grey ovals) that has initiated at the HK022 PL promoter, translocated through the putL site and stopped at the U-rich pause. The end of the RNA exit channel is indicated by a break in the grey oval, RNA by dotted lines (except for put) and DNA by thin lines. In model (1), putL binds to and modifies RNAP. The modification suppresses backtracking and termination by the same mechanism. For purposes of illustration we show the modification as an alteration in the RNA:DNA hybrid (red hatching), although in fact the nature of the modification is unknown (see text). In model (2), the secondary structure of putL (in red) prevents the re-entry of RNA that is immediately upstream of the EC into the exit channel. Binding of putL RNA to RNAP is needed to suppress termination but not pausing. In model (3), putL RNA binds close to the exit channel as or shortly after it emerges from the enzyme, and the short size of the polyribonucleotide chain between the end of anchored putL RNA and the end of the channel (in red) prevents RNA re-entry. Further elongation of the transcript relieves this restriction. The bound putL RNA suppresses termination by an unknown mechanism.
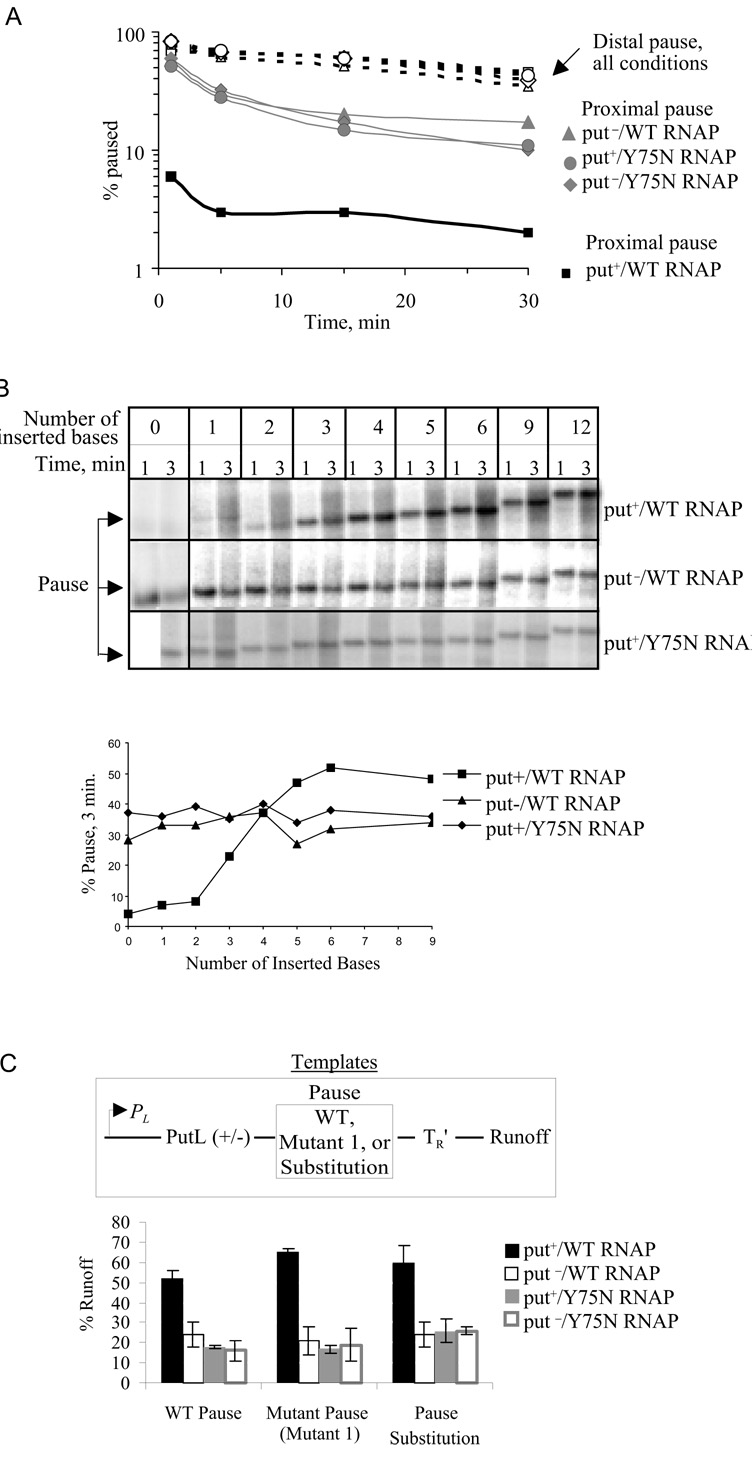
The efficiency of put-mediated antitermination is relatively insensitive to the distance between the putL site and the terminator in vivo (Sloan et al., 2007) and in vitro (King et al., 1996). If the antipause and antitermination activities of putL are the result of the same modification of the EC (model 1), pause suppression should be similarly independent of distance. By contrast, models 2 and 3 predict that antipause activity should decrease with increasing distance between the putL site and the pause, since the effects of both secondary structure and RNA anchoring on backtracking will be local (see Fig. 4 legend). We increased the distance between the pause and the base of stem-loop 2 from 21 to 48 nt and measured the effect of putL on pausing at this distal location. Contrary to model 1, putL no longer suppressed pausing at the distal site (Fig. 5A, dashed lines). Indeed, the intensity of the pause increased dramatically with even a small increase in the put-pause distance: 3 extra nt significantly reduced the effect of putL, and 4 nt abolished it (Fig. 5B).
Fig. 5.
The effect of pause displacement on pause suppression. A. Both classes of templates contained PL-putL (wild type or mutant G), and this was followed either by the U-rich pause in its normal position ("proximal pause"), or by the same pause displaced by insertion of 27nt ("distal pause"). Four transcription conditions were used: putL+ template with WT RNAP (squares), putL− template with WT RNAP (triangles), putL+ template with Y75N RNAP (circles), and putL− template with Y75N RNAP (diamonds). Templates were transcribed for various times, and the fraction of paused ECs was plotted as function of time. B. (Top) Templates with 1 to 12 bp inserted immediately downstream of the end of putL were transcribed and sampled 1 and 3 min after the initiation of transcription. (Bottom) Quantitation of the amount of transcript paused at 3 min. C. The effect of the U-rich pause on terminator readthrough. Templates containing WT or mutant putL sites followed by WT or altered pause sites and the TR′ terminator were transcribed for 3 min, and the fraction of runoff ECs was calculated (bottom).
An alternative interpretation of these results is that the U-rich pause site in its normal location is required for put-dependent modification of the EC. If so, translocation or modification of the pause site will prevent antitermination. We previously showed that the pause is not required for antitermination in vivo (King et al., 1996), and, indeed, we do not know if it has any biological function for HK022. To see if the pause improves antitermination in vitro, we measured the efficiency of termination on a template in which the pause site in its normal location was inactivated by mutation (Mutant 1; Fig. 2A), and on a second template that lacked the U-rich pause ("substitution"). PutL suppressed termination on both templates (Fig. 5C). We conclude that antitermination does not depend on the U-rich pause site, and therefore that suppression of the U-rich pause is the result of a localized rather than a distance-independent effect of putL, contrary to model 1 but predicted by models 2 and 3.
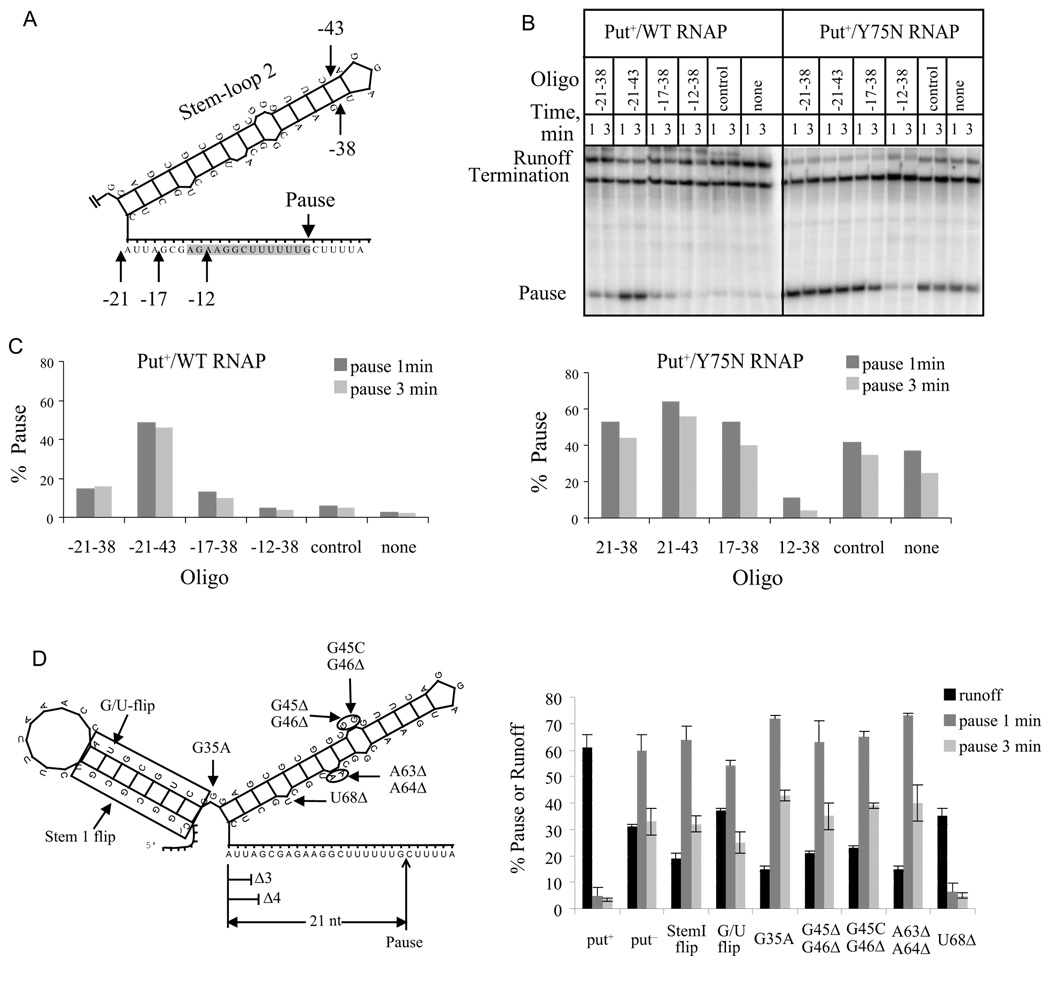
If the secondary structure of putL is sufficient to prevent backtracking (model 2), it is likely that an RNA-DNA hybrid whose structure mimics that of stem 2 will also suppress the pause (Komissarova and Kashlev, 1997a; Komissarova and Kashlev, 1997b). We therefore transcribed put+ and put− templates in the presence of various antisense oligonucleotides. Contrary to the prediction of model 2, two oligonucleotides whose 5' ends are complementary to position -21, the downstream end of stem-loop 2, and a third whose 5' end is complementary to position -17 relative to the pause site did not inhibit pausing in reactions containing β'-Y75N RNAP (Figs. 6B and 6C) or a put− template (data not shown). In fact, these oligonucleotides enhanced pausing, perhaps because they inhibited residual putL activity by hybridizing to putL RNA. In agreement, these three oligonucleotides also increased pausing when they were added to a transcription reaction containing wild type RNAP and a putL+ template. By contrast, a fourth oligonucleotide, whose 5' end was complementary to position -12, immediately upstream of the paused EC, did suppress the pause, consistent with previous data (Komissarova and Kashlev, 1997b).
Fig. 6.
Effect of antisense oligonucleotide and putL mutations on pausing. The templates contained the PL promoter followed by WT or mutant putL sites followed by the U-rich pause followed by the TR′ terminator. A. The location of the 5' (-21, -17, and -12) and 3' (-43 and -38) ends of the antisense oligos are shown on stem-loop 2 of putL RNA. The numbers show position relative to G93. Stem-loop 1 is not shown, but is present in the templates of panels B and C. B. Transcription reactions were carried out using WT or Y75N RNAP in the absence or presence of the indicated oligonucleotide (100 µM). C. The fraction of paused transcripts obtained with WT (left) or Y75N RNAPs (right). The larger effect of oligo -21–43 compared to oligo -21–38 is probably because the former hybridizes more rapidly to putL RNA. D. (Left) A diagram of folded putL RNA showing mutations used in this experiment and that of Fig. 7A. (Right) Transcription was performed with WT RNAP, and the fraction of paused and runoff ECs calculated.
These results suggest that the formation of stem-loop 2 of putL is not sufficient to suppress backtracking at the U-rich sequence. However, it is possible that a simple RNA-DNA hybrid does not accurately mimic the bulkier structure of folded putL RNA. Therefore, we investigated the antipause activity of seven more putL mutants, which, like the mutant we used up to now (mutant G), are predicted to retain a secondary structure that is similar or identical to that of the wild type, yet are deficient in antitermination (Fig. 6D) (Banik-Maiti et al., 1997)(S. Sloan and R.A.W., unpublished experiments). We decided to investigate many mutants rather than rely on just one to reduce the probability that any lack of effect of the mutants on the pause was due to formation of an alternative secondary structure that was not predicted by the RNA folding program (Zuker, 2003). We also confirmed the secondary structure prediction for mutant G35A by determining its sensitivity to structure-specific RNases (Suppl. fig. 5). Six of these new mutants lost both antipause and antitermination activities. These results are consistent with the conclusion that folding of putL RNA is not sufficient to suppress backtracking. An additional step is necessary, and we propose that this step is binding of putL RNA to the EC. Such binding has been demonstrated in vivo and in vitro. Indeed, one of the putL mutations that failed to suppress pausing (stem 1 flip) also reduced binding to the EC in vitro (Sen et al., 2001), and another (mutant G) reduced binding in vivo (Sloan et al., 2007). In addition, the β'-Y75N mutation of RNAP reduced putL RNA binding in vivo and in vitro. Note that one of the putL mutants, U68Δ, lost antitermination but retained antipause activity. We consider this mutant further below (see Discussion).
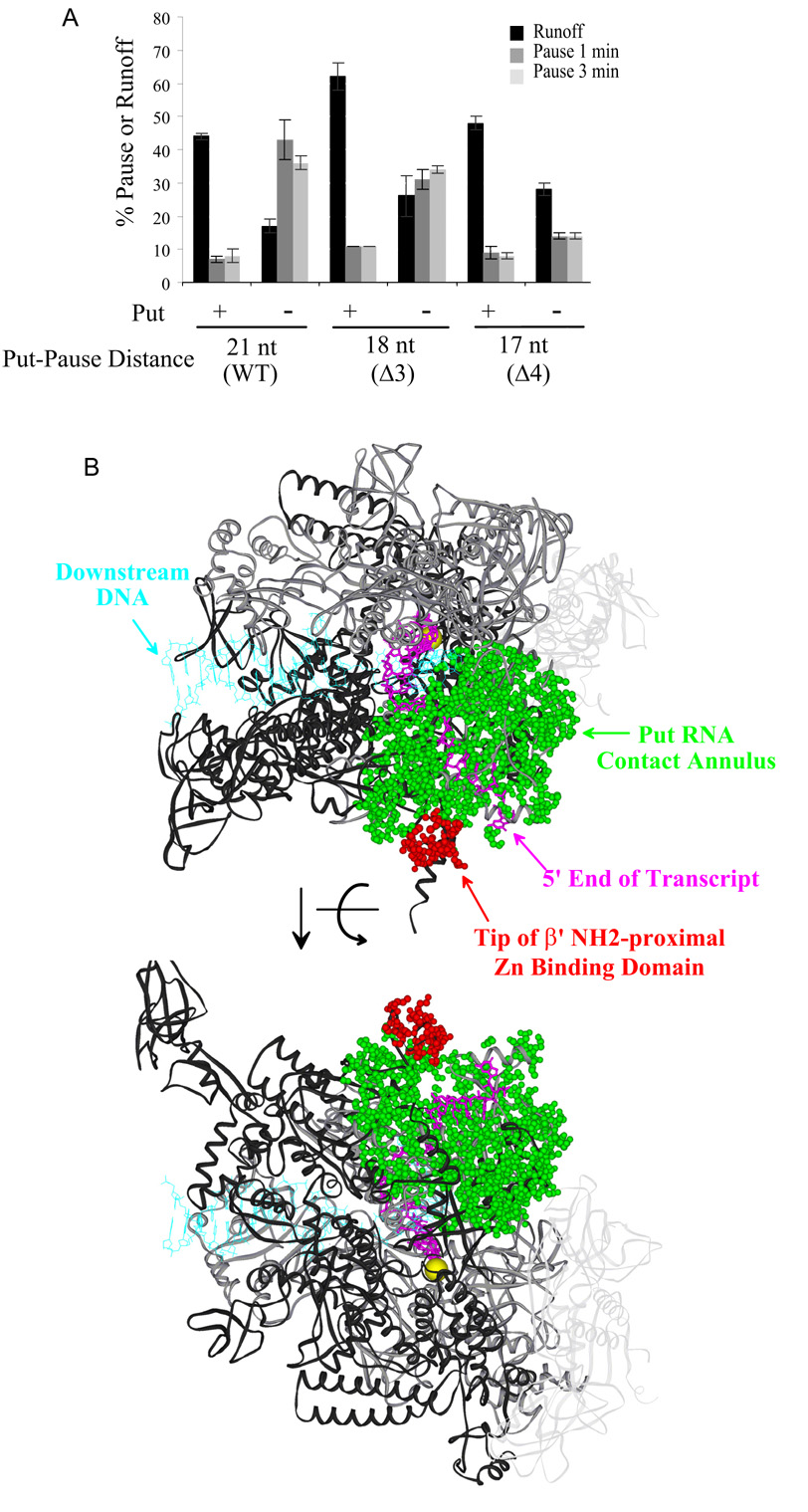
If RNA anchoring suppresses pausing, we can use the distance dependence of this effect as a molecular "tape measure" to estimate where anchoring occurs [see (Rodgers and Schleif, 2008)]. To this end, we determined the effect of progressive shortening of the putL-pause distance on the efficiency of antipausing. A decrease from 21 nt to 18 nt reduced the antipause effect of putL only slightly, suggesting that binding is efficient at this distance (Fig. 7A). However, a decrease to 17 nt (Δ4) reduced pausing on the putL mutant template to the extent that it was nearly the same as that on the put+ template. Thus, it is likely that at a distance of 17 nt, the folded putL RNA is an obstacle to backtracking whether or not it binds the EC. Such a binding-independent effect of put could contribute to pause suppression even when the pause is 21 nt from put, since the pause duration increased on a put− template or with RNAP-Y75N when we increased the distance to 48 nt (Fig. 5A). However, the data of Fig. 5A are consistent with our previous conclusion that binding makes the major contribution to pause suppression when the put-pause distance is 21 nt. We conclude that the EC can bind putL RNA when it is as close as 18 nt from the 3' end of the transcript, or about 2 to 4 nt from the end of the RNA exit channel see (Discussion)
Fig. 7.
Mapping put binding on RNAP. (A) Transcription was performed on either full-size templates or on templates containing a three or four nucleotide deletion immediately downstream of putL (Fig. 6D), and the fraction of paused and runoff ECs was calculated. (B) Two views of the Thermus thermophilus EC that differ by a ~180o rotation around the horizontal axis (Vassylyev et al., 2007). The green spheres indicate atoms potentially contacted by the base of stem-loop 2 of putL RNA (the "contact annulus"), and the red spheres indicate potentially contacted atoms that are within the tip of the β' amino-terminal zinc binding domain (cys70- cys88, E. coli numbering). The contact annulus includes atoms that are on the surface of RNAP and within 20 to 28 Å of position 14 of the transcript, within 15 to 21 Å of position 15 of the transcript, or within 10 to 15 Å of position 16 of the transcript. The transcript (16 nt) is colored magenta, the DNA (downstream and template strand of RNA-DNA hybrid) is colored light blue, and the Mg2+ atom at the active center is a yellow sphere. The ω subunit of RNAP has been omitted, the two α subunits are very light grey ribbons, the β subunit is a darker grey, and the β' subunit is an even darker grey
Discussion
PutL reduced the extent of backtracking by ECs that were artificially stalled at a U-rich pause located just downstream of the putL site. Mutations that increased the stability of RNA:DNA hybrid at the forward relative to the backtracked position prevented pausing at this site during unobstructed transcription. The presence of GreB during transcription reduced the intensity of pausing. This and other observations argue strongly that backtracking enhances the intensity of the U-rich pause during unobstructed transcription, and that putL suppresses the pause by limiting the extent of backtracking.
PutL no longer suppressed the U-rich pause when the distance between them was increased by a few nucleotides. PutL suppression of terminators is not strongly distance-dependent, and this difference implies that antipausing at the U-rich site and antitermination have different mechanisms. We propose that the EC binds putL RNA as it leaves the exit channel (see below), and that this step is common to antipausing and antitermination. Binding initially inhibits backtracking by constraining re-entry of nascent RNA into the exit channel, but continued chain elongation relaxes the constraint. PutL RNA remains bound to the EC as it translocates, and this ensures the persistence of the antiterminating modification. Folding of the putL transcript is required for antitermination (Banik-Maiti et al., 1997; King et al., 1996) and, very likely, for binding to the EC, but putL secondary structure is not sufficient to suppress the pause by itself, probably because it is too far away from RNAP.
Transcriptional pausing is an important component of many regulatory networks and our knowledge about these networks is growing. For example, most recently it was shown that in a large number of Drosophila embryo genes, ECs are stalled near the transcription start site, possibly waiting for activation at later developmental stages (Muse et al., 2007; Zeitlinger et al., 2007)[reviewed in (Core and Lis, 2008)]. Backtracking is a mechanism that accounts for many known pauses [for example (Adelman et al., 2005)], and RNA anchoring to the EC is potentially a general mechanism to regulate backtracking-associated pauses. Moreover, since the effect of anchoring is local, individual pauses can be targeted.
We know of two cases, both involving eukaryotic RNA polymerase II, in which RNA anchoring might have altered the intensity of backtracking-associated arrests and pauses, one immediately downstream from the adenovirus late promoter and the other in the initially transcribed region of HIV-1 (Ujvari et al., 2002; Palangat et al., 1998). In both cases, the nascent transcript affected backtracking. The possibility that formation of a hairpin in the upstream RNA blocked backtracking was considered, but no structure could be predicted. Therefore, the hypothesis that RNA anchoring suppresses backtracking was suggested but never tested (Ujvari et al., 2002).
The phenomenon of binding of nascent RNA to RNAP after the transcript leaves the RNA exit channel has been documented in several studies involving the E. coli enzyme and human RNA polymerase II. In one study, upstream nascent RNA extending from position 30 to 45 from the 3' end is partially protected from digestion by RNase T1 by E. coli RNAP (Milan et al., 1999). More recently, Ujvari and Luse (2006) have shown that the region extending from 26 to 32 nt upstream from the 3' end can be cross-linked to the Rpb7 subunit of polymerase II as well as to a splicing factor associated with the EC. We speculate that the existence of a clear precedent for suppression of backtracking by put RNA anchoring to the EC will lead to the discovery of additional examples of this phenomenon.
Pausing at the U-rich site occurs when the 3' end of the nascent transcript is 21 nt from the downstream end of stem-loop 2 of putL RNA. The GreB cleavage pattern (Fig. 3) shows that a put-modified EC at this position can still backtrack by 2 or 3 nt, but no more. This suggests that the polynucleotide chain between the end of anchored putL RNA and the outside end of the exit channel is fully extended when the 3' end of the transcript is 18 nt (= 21 - 3) from the base of stem-loop 2 of putL. Consistent with this suggestion, we found that putL still suppressed pausing when the putL-pause distance was reduced to 18 nt, but gave an equivocal result at a 17 nt spacing (Fig. 7A). Thus, it is likely that put RNA binds poorly to the EC when the distance between the 3' end and the base of stem-loop 2 is less than 18 nt. Nuclease protection and other types of experiment suggest that the EC envelops about 14 nt of RNA [(Korzheva et al., 2000) and references cited therein], and the structure of the Thermus thermophilus EC shows that the 3' phosphates of nt -14 and -16 make polar interactions with RNAP (Vassylyev et al., 2007). Therefore, when the 3' end of the transcript is 18 nt from putL, we estimate that there are 2 to 4 nt between the base of anchored putL RNA and the point at which the nascent transcript loses its interactions with RNA polymerase (i.e., the "end" of the RNA exit channel). Estimates of distance between adjacent phosphate groups of a fully extended single-stranded polynucleotide chain range from 5 to 7 Å (Murphy et al., 2004; Woodside et al., 2006). Accordingly, the base of stem-loop 2 of anchored putL RNA should be located between 10 and 28 Å from the end of the exit channel.
The atoms colored green in Fig. 7B show the annulus of potential contacts on the surface of the Thermus thermophilus EC that is generated by these assumptions. It is of interest that many of the amino acid residues that comprise the tip of the β' amino-proximal zinc binding domain, including the residue that corresponds to E. coli Tyr75, are included in this annulus. Other mutations that alter this region also reduced put-dependent antitermination, and the extent of the reduction differed between putL and putR for certain mutations (Sen et al., 2002). These findings led us to conclude that the tip of the zinc binding domain was part of a put RNA binding site, and the work presented here strengthens this conclusion.
This work revealed an unusual putL mutation, U68Δ, that lost antitermination activity but retained antipausing activity in vitro. This mutant, unlike any of the others used here, does support antitermination in vivo (Banik-Maiti et al., 1997)(S. Sloan and R.A.W., unpublished results), which suggests that modification of the EC is not drastically compromised. It is possible that U68Δ weakens putL RNA binding to RNAP to the extent that it exists at the pause site but is lost by the time the enzyme reaches the terminator unless a cellular factor is present. Alternatively, the mutation could delay putL RNA folding by creating a competing secondary structure unless a cellular factor is present. One such structure, pairing of C67GCU with A76GCG, would sterically prevent backtracking at the neighboring pause site (suggested by a reviewer). Even if the RNA subsequently refolded in the “correct”, thermodynamically favorable secondary structure, binding would be inhibited if it had to occur co-transcriptionally. Clearly, more work is required to understand the difference between this mutation and the others we tested.
Antitermination, unlike antipausing, is relatively independent of the distance between putL and terminators (Sloan et al., 2007; King et al., 1996), and therefore reflects a different and as yet unknown property of the put RNA-RNAP complex. The failure of putL to suppress the U-rich pause when the putL-pause spacing was increased to 25 bp or more (Fig. 5B) argues that antitermination does not result from selective stabilization of rU-dA base pairs, since such stabilization should also suppress backtracking at any distance. However, it remains possible that put-modification of the EC generally stabilizes RNA:DNA hybrid regardless of composition. A second possibility is that put inhibits the formation or action of terminator hairpins, as has been suggested for the phage λQ antiterminator (Yarnell and Roberts, 1999; Shankar et al., 2007). The location of the put binding site on RNAP, near the RNA exit channel, makes this possibility attractive. Consistent with this hypothesis, put decreased the probability of a hairpin-dependent pause about two-fold (Suppl. fig. 1). We note that the RNAP site that interacts with the p7 antiterminator protein of X. oryzae bacteriophage Xp10 was recently mapped close to the put interaction site that we suggest here (Yuzenkova et al., 2008). Put modification also increases the average elongation rate about two-fold in vivo, and this increase persists for long distances (W. Robins, R.A.K, and I. Molineux, unpublished work). Although faster elongation can reduce termination (McDowell et al., 1994; Jin et al., 1992; Shankar et al., 2007), it is not yet clear how much it contributes to put-mediated antitermination. In any event, our work argues that faster elongation must result from something other than suppression of backtracking. It could result from suppression of pauses that are not associated with backtracking [e.g., hairpin-associated pauses or "ubiquitous" pauses (Herbert et al., 2006)] or from a general increase in the rate of addition of nucleotides at many or all steps.
Methods
Materials
Wild-type RNAP was purchased from Epicenter. His-tagged RNAP, Lac repressor, and GreB were gifts of Drs. M. Kashlev, S. Adhya, and S. Borukhov, respectively. RNAP β'-Y75N was purified as described (Hager et al., 1990). Ultrapure NTPs and [α- 32P]-CTP were purchased from GE Healthcare Bio-Sciences (Piscataway, NJ), DNA oligonucleotides from Integrated DNA Technologies, streptavidin coated magnetic beads from Promega, Ni-NTA agarose form Qiagen, and Klenow fragment from New England Biolabs.
Templates
The transcription templates were PCR amplified products of DNA molecules carrying the PL promoter of HK022 and 98 nt from transcription start point followed by a 22 bp symmetrical lac operator site (King et al., 2003). PutL mutations are shown in figures or described in the text. The templates used for KMnO4 footprinting had an AgeI restriction site located immediately downstream from the lac operator. The templates used for the experiments of FIG. 5B, 5C, Fig 6 and Fig 7A contained the TR′ terminator inserted 3 bp downstream from the lac operator. The 5' end of the non-template strand was biotinylated in all templates.
In the distal pause template of Fig. 5A, a 27 bp fragment was inserted between G79 and A80 of Fig. 1A. The pause substitution template of Fig. 5C contained the same 27 bp insertion, and, in addition, the segment after position C86 of Fig. 1A was replaced with a segment containing the λ TR' terminator. The templates used in Fig. 5B contained one of the following insertions after position C72 of Fig. 1A: A, AT, ATT, ATTC, ATTCG, ATTCGA, ATTCGAATT, or CGAATTCGAATT. Full sequences of the templates are available on request.
Measuring the efficiency of pausing and termination
Procedures used for in vitro transcription and the fractionation and quantitation of products are similar to those described previously (Sen et al., 2001; King et al., 2003)(details in Supplementary Data). The concentrations of ATP, GTP and UTP were 100 µM and that of CTP was 5 µM, unless otherwise noted. The percent of paused ECs (%P) was calculated as %P = [P/(P + T + RO)] × 100, where T = ECs terminated at TR' (when present), and RO = Runoff. The percent of ECs that failed to terminate at TR′ was calculated as %RO = [RO/(T + RO)] × 100.
Roadblocked ECs
Immobilized washed ECs that were roadblocked at the pause site by Lac repressor were obtained on biotinylated templates linked to streptavidin-coated magnetic or agarose beads as described (King et al., 2003). In some experiments, Ni-NTA agarose was used to immobilize His-tagged RNAP. In some experiments, a subset of NTPs was added to generate an EC stalled at G16, washed, and chased with NTPs (details Supplementary Data). The 3' end of the transcript in roadblocked ECs was 5 nt upstream from the lac operator.
GreB-induced cleavage and pyrophosphorolysis
Aliquots of roadblocked ECs immobilized on streptavidin-coated magnetic beads were incubated with the indicated concentrations of GreB for 3 min or with 20 mM sodium pyrophosphate for the indicated time periods at 37°C.
Walking RNAP along the template
Roadblocked ECs were obtained using Histidine-tagged RNAP and immobilized on Ni-NTA agarose beads (Quiagen). NTPs were removed from roadblocked ECs by 5 cycles of centrifugation and resuspension in 1 ml TB. Two mM IPTG was added to dissociate Lac repressor from the template. After 2 min, NTPs or the NTP subset indicated in Fig. 1C (5 µM each) were added for 5 min, and the reactions were stopped and analyzed.
KMnO4 footprinting of roadblocked ECs
ECs labeled at the 3' end of the non-template DNA strand were immobilized on Ni-NTA beads and treated with KMnO4 as described in Supplementary data. Cleavage of modified residues was measured by polyacrylamide gel electrophoresis as described (Komissarova and Kashlev, 1997b).
Supplementary Material
Acknowledgments
We are grateful to Gali Prag for help in the preparation of Fig. 7B, and to Sankar Adhya, Sergei Borukhov, Mikhail Kashlev, and Arkady Mustaev for materials and helpful advice. This work was supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Adelman K, La Porta A, Santangelo TJ, Lis JT, Roberts JW, Wang MD. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc. Natl. Acad. Sci. U. S. A. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell. 2005;17:103–112. doi: 10.1016/j.molcel.2004.11.028. [DOI] [PubMed] [Google Scholar]
- Banik-Maiti S, King RA, Weisberg RA. The antiterminator RNA of phage HK022. J. Mol. Biol. 1997;272:677–687. doi: 10.1006/jmbi.1997.1251. [DOI] [PubMed] [Google Scholar]
- Bremer H, Dennis PP. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt FC, Curtiss RC III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia Coli and Salmonella: Cellular and Molecular Biology. Washington, D.C.: ASM Press; 1996. pp. 1553–1569. [Google Scholar]
- Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J. Mol. Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- Clerget M, Jin DJ, Weisberg RA. A zinc binding region in the β" subunit of RNA polymerase is involved in antitermination of early transcription of phage HK022. J. Mol. Biol. 1995;248:768–780. doi: 10.1006/jmbi.1995.0259. [DOI] [PubMed] [Google Scholar]
- Core LJ, Lis JT. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science. 2008;319:1791–1792. doi: 10.1126/science.1150843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol. Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- Hager DA, Jin DJ, Burgess RR. Use of Mono Q high-resolution ion-exchange chromatography to obtain highly pure and active Escherichia coli RNA polymerase. Biochemistry. 1990;29:7890–7894. doi: 10.1021/bi00486a016. [DOI] [PubMed] [Google Scholar]
- Herbert KM, La PA, Wong BJ, Mooney RA, Neuman KC, Landick R, Block SM. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin DJ, Burgess RR, Richardson JP, Gross CA. Termination efficiency at rhodependent terminators depends on kinetic coupling between RNA polymerase and rho. Proc. Natl. Acad. Sci. U. S. A. 1992;89:1453–1457. doi: 10.1073/pnas.89.4.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RA, Banik-Maiti S, Jin DJ, Weisberg RA. Transcripts that increase the processivity and elongation rate of RNA polymerase. Cell. 1996;87:893–903. doi: 10.1016/s0092-8674(00)81996-0. [DOI] [PubMed] [Google Scholar]
- King RA, Sen R, Weisberg RA. Using a lac repressor roadblock to analyze the E. coli transcription elongation complex. Methods Enzymol. 2003;371:207–218. doi: 10.1016/S0076-6879(03)71015-5. [DOI] [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. RNA polymerase switches between inactivated and activated states By translocating back and forth along the DNA and the RNA. J. Biol. Chem. 1997a;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3' end of the RNA intact and extruded. Proc. Natl. Acad. Sci. U. S. A. 1997b;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komissarova N, Kashlev M. Functional topography of nascent RNA in elongation intermediates of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 1998;95:14699–14704. doi: 10.1073/pnas.95.25.14699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- Landick R. The regulatory roles and mechanism of transcriptional pausing. Biochem. Soc. Trans. 2006;34:1062–1066. doi: 10.1042/BST0341062. [DOI] [PubMed] [Google Scholar]
- Landick R, Wang D, Chan CL. Quantitative analysis of transcriptional pausing by Escherichia coli RNA polymerase: his leader pause site as paradigm. Methods Enzymol. 1996;274:334–353. doi: 10.1016/s0076-6879(96)74029-6. 334–353. [DOI] [PubMed] [Google Scholar]
- Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;22:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DN, Feng G, Landick R. GreA-induced transcript cleavage is accompanied by reverse translocation to a different transcription complex conformation. J. Biol. Chem. 1994;269:22295–22303. [PubMed] [Google Scholar]
- Levin JR, Chamberlin MJ. Mapping and characterization of transcriptional pause sites in the early genetic region of bacteriophage T7. J. Mol. Biol. 1987;196:61–84. doi: 10.1016/0022-2836(87)90511-0. [DOI] [PubMed] [Google Scholar]
- McDowell JC, Roberts JW, Jin DJ, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- Milan S, D'Ari L, Chamberlin MJ. Structural Analysis of Ternary Complexes of Escherichia coli RNA Polymerase: Ribonuclease Footprinting of the Nascent RNA in Complexes. Biochemistry. 1999;38:218–225. doi: 10.1021/bi9818422. [DOI] [PubMed] [Google Scholar]
- Murphy MC, Rasnik I, Cheng W, Lohman TM, Ha T. Probing single-stranded DNA conformational flexibility using fluorescence spectroscopy. Biophys. J. 2004;86:2530–2537. doi: 10.1016/S0006-3495(04)74308-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palangat M, Landick R. Roles of RNA:DNA hybrid stability, RNA structure, and active site conformation in pausing by human RNA polymerase II. J. Mol. Biol. 2001;311:265–382. doi: 10.1006/jmbi.2001.4842. [DOI] [PubMed] [Google Scholar]
- Palangat M, Meier TI, Keene RG, Landick R. Transcriptional pausing at +62 of the HIV-1 nascent RNA modulates formation of the TAR RNA structure. Mol. Cell. 1998;1:1033–1042. doi: 10.1016/s1097-2765(00)80103-3. [DOI] [PubMed] [Google Scholar]
- Reeder TC, Hawley DK. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- Richardson JP, Greenblatt J. Control of RNA chain elongation and termination. In: Curtiss RC III, Lin ECC, Low KB, Magasanik B, Neidhardt FC, Reznikoff WS, Riley M, Schaechter M, Umbarger HE, editors. Escherichia coli and Salmonella: Cellular and molecular biology. Washington, DC: ASM Press; 1996. pp. 822–848. [Google Scholar]
- Ring BZ, Yarnell WS, Roberts JW. Function of E. coli RNA polymerase sigma factor sigma 70 in promoter-proximal pausing. Cell. 1996;86:485–493. doi: 10.1016/s0092-8674(00)80121-x. [DOI] [PubMed] [Google Scholar]
- Rodgers ME, Schleif R. DNA tape measurements of AraC. Nucleic Acids Res. 2008;36:404–410. doi: 10.1093/nar/gkm1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen R, King RA, Mzhavia N, Madsen PL, Weisberg RA. Sequence-specific interaction of nascent antiterminator RNA with the zinc-finger motif of Escherichia coli RNA polymerase. Mol. Microbiol. 2002;46:215–222. doi: 10.1046/j.1365-2958.2002.03154.x. [DOI] [PubMed] [Google Scholar]
- Sen R, King RA, Weisberg RA. Modification of the properties of elongating RNA polymerase by persistent association with nascent antiterminator RNA. Mol. Cell. 2001;7:993–1001. doi: 10.1016/s1097-2765(01)00243-x. [DOI] [PubMed] [Google Scholar]
- Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankar S, Hatoum A, Roberts JW. A transcription antiterminator constructs a NusA-dependent shield to the emerging transcript. Mol. Cell. 2007;27:914–927. doi: 10.1016/j.molcel.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan S, Rutkai E, King RA, Velikodvorskaya T, Weisberg RA. Protection of antiterminator RNA by the transcript elongation complex. Mol. Microbiol. 2007;63:1197–1208. doi: 10.1111/j.1365-2958.2006.05579.x. [DOI] [PubMed] [Google Scholar]
- Sosunov V, Sosunova E, Mustaev A, Bass I, Nikiforov V, Goldfarb A. Unified two-metal mechanism of RNA synthesis and degradation by RNA polymerase. EMBO J. 2003;22:2234–2244. doi: 10.1093/emboj/cdg193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ujvari A, Luse DS. RNA emerging from the active site of RNA polymerase II interacts with the Rpb7 subunit. Nat. Struct. Mol. Biol. 2006;13:49–54. doi: 10.1038/nsmb1026. [DOI] [PubMed] [Google Scholar]
- Ujvari A, Pal M, Luse DS. RNA polymerase II transcription complexes may become arrested if the nascent RNA is shortened to less than 50 nucleotides. J. Biol. Chem. 2002;277:32527–32537. doi: 10.1074/jbc.M201145200. [DOI] [PubMed] [Google Scholar]
- Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- Vogel U, Jensen KF. The RNA chain elongation rate in Escherichia coli depends on the growth rate. J. Bacteriol. 1994;176:2807–2813. doi: 10.1128/jb.176.10.2807-2813.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang MD, Schnitzer MJ, Yin H, Landick R, Gelles J, Block SM. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. U. S. A. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW. Mechanism of intrinsic transcription termination and antitermination. Science. 1999;284:611–615. doi: 10.1126/science.284.5414.611. [DOI] [PubMed] [Google Scholar]
- Yuzenkova Y, Zenkin N, Severinov K. Mapping of RNA polymerase residues that interact with bacteriophage Xp10 transcription antitermination factor p7. J. Mol. Biol. 2008;375:29–35. doi: 10.1016/j.jmb.2007.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaychikov E, Denissova L, Heumann H. Translocation of the Escherichia coli transcription complex observed in the registers 11 to 20: "jumping" of RNA polymerase and asymmetric expansion and contraction of the "transcription bubble". Proc. Natl. Acad. Sci. U. S. A. 1995;92:1739–1743. doi: 10.1073/pnas.92.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet. 2007;39:1512–1516. doi: 10.1038/ng.2007.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.