Abstract
Several pathogenic bacteria produce adenylyl cyclase toxins, such as the edema factor (EF) of Bacillus anthracis. These disturb cellular metabolism by catalyzing production of excessive amounts of the regulatory molecule cAMP. Here, a structure-based method, where a 3D- pharmacophore that fit the active site of EF was constructed from fragments, was used to identify non-nucleotide inhibitors of EF. A library of small molecule fragments was docked to the EF- active site in existing crystal structures and those with the highest HINT scores were assembled into a 3D-pharmacophore. About 10,000 compounds, from over 2.7 million compounds in the ZINC database, had a similar molecular framework. These were ranked according to their docking scores, using methodology that was shown to achieve maximum accuracy (i.e., how well the docked position matched the experimentally determined site for ATP analogues in crystal structures of the complex). Finally, 19 diverse compounds with the best AutoDock binding/docking scores were assayed in a cell based assay for their ability to reduce cAMP secretion induced by EF. Four of the test compounds, from different structural groups, inhibited in the low micromolar range. One of these has a core structure common to phosphatase inhibitors previously identified by high-throughput assays of a diversity library. Thus, the fragment based pharmacophore identified a small number of diverse compounds for assay, and greatly enhanced the selection process of advanced lead compounds for combinatorial design.
Keywords: HINT program, 3D-pharmacophore, structure-based screening, non-nucleotide ATP analogues, bacterial toxin inhibitor, adenylyl cyclase inhibitor, fragment docking
1. Introduction
Many pathogenic bacteria, regardless of their cellular morphology and grouping, produce toxins with similar functions that are often plasmid encoded. For example, Bacillus anthracis, a Gram-positive, spore-forming, rod-shaped bacterium, produces two types of factors that enhance its lethality, a polysaccharide capsule1 and two protein toxins, lethal toxin (LT) and edema toxin (ET). Both toxins are lethal when injected into mice, and they suppress the functions of macrophages, polymorphoneutrophils, and lymphocytes. One component of both toxins is protective antigen (PA), which enables the cell entry of the enzymatic toxin components lethal factor (LF) and edema factor (EF)2. LF contains metalloprotease activity that is specific for the MAP kinase-kinases; inhibitors have been identified by many paths, including high throughput screening3. One inhibitor of LF has been shown to be an effective adjunct to antibiotic therapy in animal studies4. This inhibitor does not affect the activity of EF, which is an adenylyl cyclase analogous to that produced by Bordetella pertussis (the causative agent of whooping cough).5–7 These “adenylyl cyclase” toxins8, 9 catalyze the production of cAMP from ATP10–13. High levels of cAMP perturb the water homeostasis of the cell leading to abnormalities in the intracellular signaling pathways and stimulation of the chloride channel14–16. This contributes to edema (and widening) of the mediastinum located between the lobes of the lungs of patients with inhalation anthrax. Patients with cutaneous anthrax often display tissue edema near the lesion. Inhibitors that would bind to EF and prevent its enzymatic activity could reduce the severity of infections by B. anthracis and other bacteria that produce similar toxins.
The active site residues of anthrax EF, have been identified by several crystal structures of the toxin alone or complexed with substrate analogues and small molecule inhibitors8, 9, 17–19. Since the active site of the mammalian AC is distinct from that of the toxin, we sought to design inhibitors that bind specifically to anthrax EF. Previous authors have identified nucleotide-like inhibitors of adenylyl cyclases, starting from ATP20–23 or by molecular docking of large libraries24. Our approach was to identify discrete fragments with tight binding to the active site, and assemble these into a flexible 3D-pharmacophore that could be used to screen databases of known compounds for those that would fit the active site.
Fragment-based drug design is an emerging lead discovery approach to construct highly potent inhibitors. There are many variations of this approach for molecular drug design 25–37, all of which begin by determining the binding energy of compounds of low molecular complexity, to identify those with the highest “ligand efficiency” (ΔG of binding per heavy atom 28, 29, 33). Both experimental and computational approaches can be taken to screen fragment libraries. For the former, biophysical methods such as X-ray crystallography26, NMR spectroscopy25, 27, 30, and surface plasmon resonance38 have been used to design and synthesize high-affinity ligands, based on fragments with good binding properties25, 31, 32, 35–37. Some compounds identified using fragment-based approaches have entered clinical trials36, and fragment based discovery can identify quality leads for targets where HTS has not succeeded 31, 32, 39.
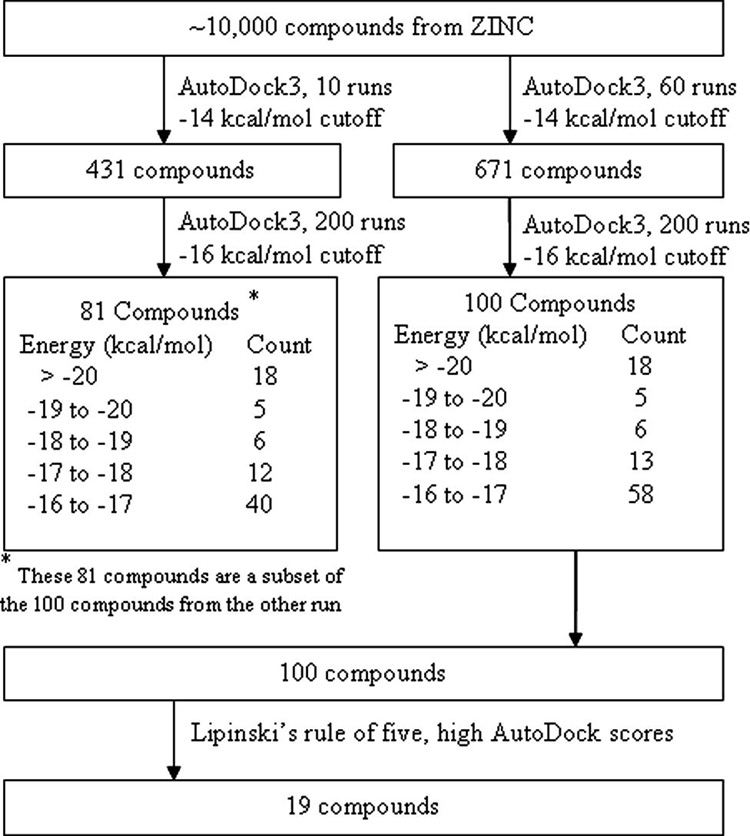
Computational methods40–42, such as computational solvent mapping (CS-Map) 40, 42, have also been developed to enhance ligand efficiency of the starting fragments, by identifying hot spots, or regions of protein binding sites that are major contributors to the binding energy in silico. Our approach was based, in the same fashion, on identifying fragments with high binding energy to discrete areas of the EF active site (Figure 1), as determined by their Hydropathic INTeractions, or HINT, program43–45 scores. The fragments were then converted to molecular frameworks (3D-pharmacophores), using distance constraints based on inter-fragment distances in the active site. Instead of relying on synthesis in the early stages of the project, we used these pharmacophores to screen large compound libraries for small molecules with the desired arrangement of fragments. In initial tests, compounds were selected from the NCI database, and screened for those with docking scores better than those for known inhibitors. We then went on to identify compounds with even better docking scores in the larger ZINC database46. From an initial list of about 10,000 compounds that matched the pharmacophores, AutoDock scores were used to select 19 compounds that were assayed for their ability to inhibit the EF-induced secretion of cAMP from mammalian cells. Three of these compounds inhibited cAMP production in the range of 2–8 µM, and were thus promising lead compounds for combinatorial design. This selection was related to our ability to account for the metal ion charge during the docking, as has been described separately47, and to identify tight binding fragments with the HINT program.
Figure 1.
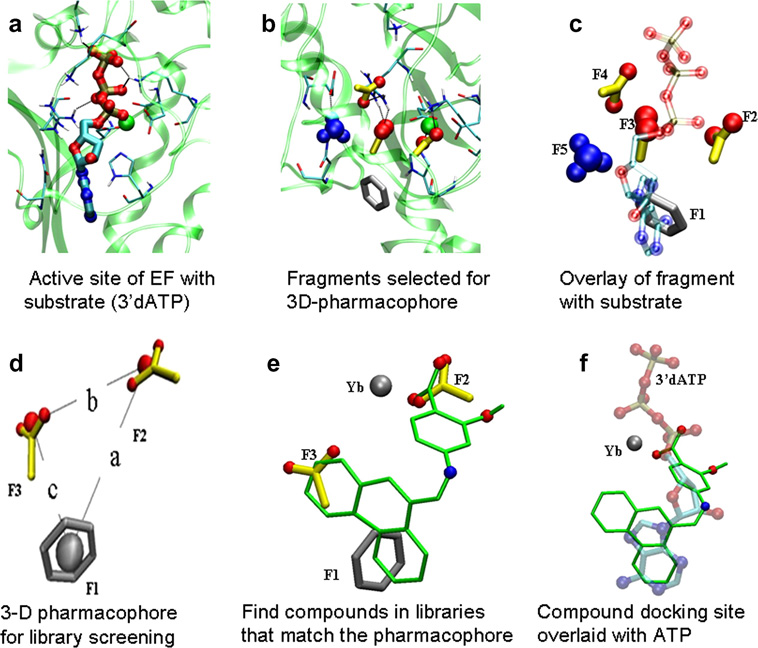
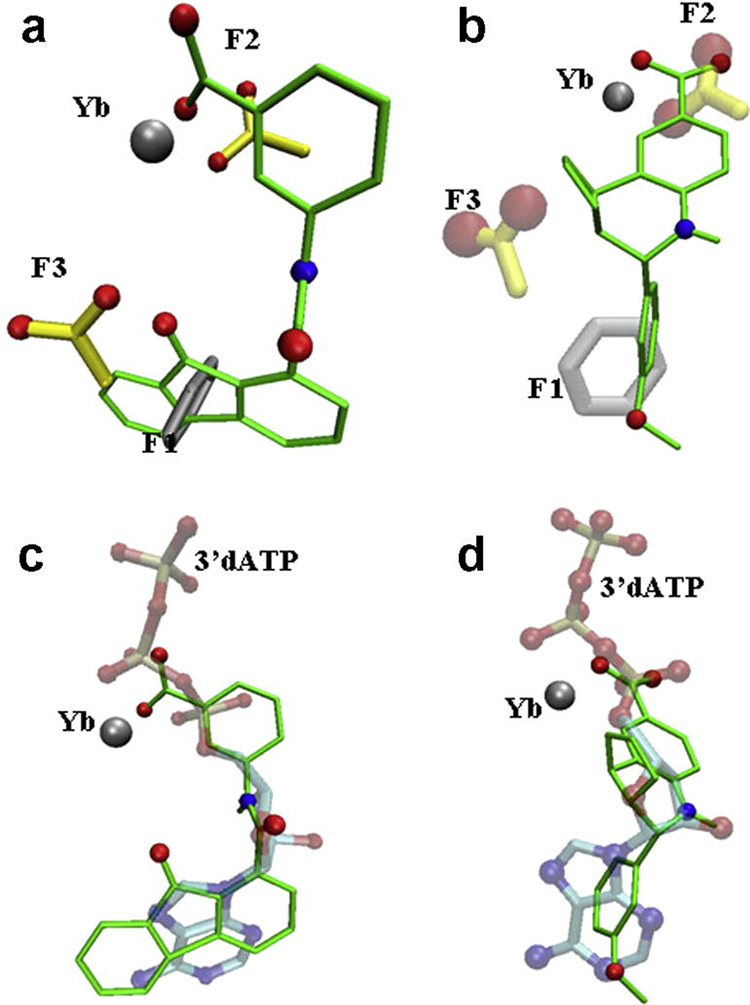
Overview of the design of a 3D-pharmacophore for compound selection. The experimentally determined configuration of a substrate analogue, 3’dATP is shown for orientation purposes in a,c, and f, but fragments were selected based only on force field considerations and HINT scores. a) Active site of EF (PDB code: 1K90) with the position of the substrate 3’dATP bound in the active site; side chains of EF within 5 Ǻ are illustrated; b) The position of the 5 fragments in the active site (F1: phenyl; F2, F3, and F4: carboxyl; F5: ammonium groups) with the best HINT scores. c) Overlay of the position of F1-5 with a). Note that there is a good correlation between the fragments and the substrate analogue, but that the fragments occupy areas of the active site peripheral to the substrate analogue. d) A sample 3D-pharmacophore, consisting of fragments (F1-3) constrained by their distances in the active site, which can be used for database searching with UNITY; e, f) our best ranked (lowest docking energy) compound. DC1, shown in its best docking position, overlays the initial pharmacophore fragments (e) and the position of the substrate analogue 3’d-ATP in the crystal structure of EF (f).
2. Results
2.1. Overall procedure
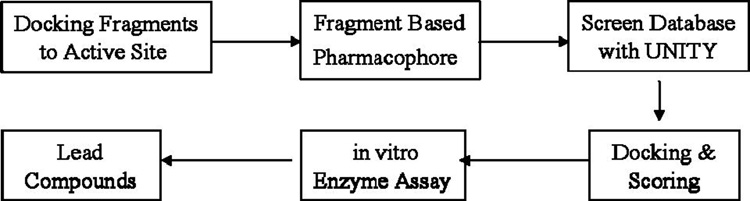
The basic steps in our procedure can be summarized:
Pharmacophore development →Unity search/2D searches of the NCI and ZINC libraries→ Docking→ MW/logP filter →19 compounds for screening→obtained 3 compounds that were active in the cell based assay for further testing.
A pictorial overview is shown in Figure 1. We began searching in the smaller NCI library, which contains many druglike molecules, and then expanded our search to the ZINC database.
2.2. Pharmacophore design
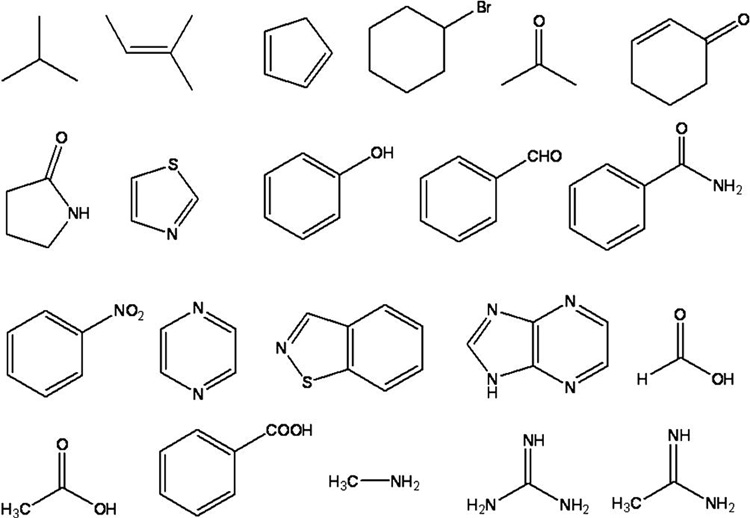
A fragment library (Figure 2) search and docking were conducted as described in Experimental, and 5 fragments (F1-5) were selected that had: a) HINT scores greater than 700 for the indicated positions in the active site and b) no inter-fragment steric hindrance. The 3 fragments with the best binding energies were used to define 3D-pharmacophores for UNITY searches of the NCI database (Figure 1b–d). The fragments to some extent overlay the position of the substrate analogue, 3’dATP, in the active site (Fig. 1c). The fragment, F1, a phenyl ring, is located exactly over the center of the purine moiety of the ATP analogue in the crystal structure, while fragments F2, F3, and F4 are carboxyl groups that lie near the phosphate groups. Fragment F2 is within hydrogen bond distance of the metal ion and Lys346 in the EF active site, while F3 and F4 interact with Arg329. Fragment F5 contains an ammonium group which interacts with Glu588.
Figure 2.
Examples of fragments tested for docking into the EF active site.
2.3. Database screening and molecular docking
As an initial test of our approach, we screened the NCI library, which contains about 250,000 compounds, using the UNITY program and our initial 3 fragment pharmacophore (Figure 1d), with varying tolerances on our distance constraints. A total of 82 compounds that matched the pharmacophores within the constraint distances were obtained, using UNITY. We confirmed, by molecular docking with FlexX, that many of these compounds had docking energies similar to our controls (ATP and analogues thereof). Substructures of the compounds with the best docking values were also selected, based on their hydrogen bond formation to areas of the active site (Figure 3). To search a wider range of chemical space than is available in the NCI Database, we used these substructures, and variations of the initial pharmacophore fragments, to obtain a list of compounds from the much larger ZINC database. Approximately 10,000 compounds were identified, using the search tools provided at that website, that fit our pharmacophores.
Figure 3.
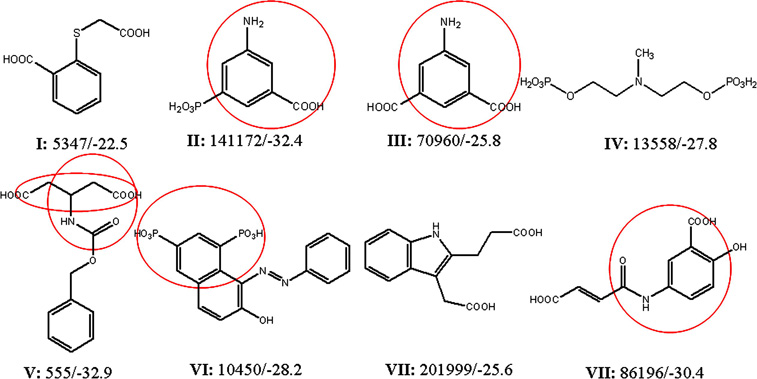
Compounds from the NCI database with the best docking scores, obtained by screening with the initial 3-D pharmacophores and the UNITY program. The NCI code/FlexX docking score is given. Areas of these molecules that formed particularly strong hydrogen bonding within the active site for the docked conformers are circled.
Screening of ~10,000 compounds with AutoDock
Our initial studies indicated that both proper adjustment of the charge on the metal ion in the target and allowing sufficient docking iterations to achieve the lowest possible binding energy were essential to obtain accurate docking.47 We thus used a tiered docking, as shown in Figure 4, to first eliminate all compounds with very low binding scores by rapid comparison, using only a few iterations, and then using more definitive, longer time scale, docking conditions to select those with the best possible binding scores. The properties and structures of the 100 compounds with the most favorable AutoDock scores (i.e., those with estimated binding energy less than or equal to −16 kcal/mol) were compared. These compounds were then screened based on rotatable bonds. If the binding energies were between −16 and −17.0/kcal/mol, the maximum rotatable bonds were set to 8. If the binding energies were between −17.0 and −18.0/kcal/mol, the maximum rotatable bonds were set to 9. If the binding energies were lower than −18.0/kcal/mol, the maximum rotatable bonds were set to 10. That is because the unfavorable energy caused by entropy loss of fixing a rotatable bond (TΔSrot) is between −3.5 and −5 kJ/mol (0.83 ~ 1.2kcal/mol)48, 49, while AutoDock3 binding energy did not included this item. The remaining compounds were screened based on the Lipinski’s rule of five (H-bond donors are not over 5; molecular weight is not over 500; cLogP is not over 5 and H-bond acceptors are not over 10). If two or more parameters of a compound are out of range, the compound is removed. By these rules, 48 compounds remained. Among these compounds, 7, 2, 5, 6 and 28 compounds are in energy range <−20, −20 to −19, −19 to −18, −18 to −17 and −17 to −16 kcal/mol respectively. Finally, 19 compounds, that were indeed commercially available, with the lowest molecular weights and logP values, were selected for assay._These rules agree approximately with Ghose’s rules50 as well. As these compounds also had reasonable docking energies with mammalian AC, we cannot at this time determine selectivity from these numbers. Table 1 shows the AutoDock scores for the 19 compounds, as well as controls for previously selected inhibitors, ATP and analogues thereof (Table 1).
Figure 4.
Flowchart illustrating the scheme used to select the final set of 19 compounds from approximately 10,000 initial hits from the ZINC database.
Table 1.
AutoDock results for known ligands and test compounds used in this study. Our computationally selected compounds with significant activity in the bioassay (see Fig. 1,Fig. 6,Fig. 7) are bold.
| Compound | BEEF1 (kcal/mol) |
BEAC2 (kcal/mol) |
|---|---|---|
| ATP | −19.0 | −13.0 |
| 3’dATP | −18.7 | −13.8 |
| 2’3’ddATP | −18.7 | −13.8 |
| PGE2-imidazole | −14.7 | −12.1 |
| Selected compounds | ||
| DC1(ZINC132715) | −20.6 | −17.8 |
| DC2 | −20.6 | −16.1 |
| DC3 | −20.4 | −17.8 |
| DC4 | −20.1 | −15.5 |
| DC5 (ZINC75209) | −19.2 | −17.5 |
| DC6 | −18.8 | −14.1 |
| DC7 (ZINC94352) | −18.6 | −15.4 |
| DC8 (ZINC75022) | −18.1 | −16.2 |
| DC9 | −17.1 | −13.7 |
| DC10 | −17.0 | −14.6 |
| DC11 | −16.8 | −14.1 |
| DC12 | −16.7 | −13.3 |
| DC13 | −16.7 | −13.5 |
| DC14 | −16.6 | −14.3 |
| DC15 | −16.5 | −14.2 |
| DC16 | −16.4 | −16.1 |
| DC17 | −16.3 | −13.6 |
| DC18 | −16.2 | −14.7 |
| DC19 | −16.0 | −13.5 |
| Soelaiman7 (25 µM)3 | −10.7 | −10.7 |
| Soelaiman3 (60 µM)3 | −11.4 | −10.8 |
| Soelaiman2 (70 µM)3 | −11.0 | −10.3 |
AutoDock binding energies for anthrax EF.
AutoDock binding energies for mammalian AC
These compounds, selected by docking a large compound library, had the best IC50 values (in parentheses) for EF, according to Soelaiman et al.24
2.5. Three compounds have better activity than previously identified EF inhibitors
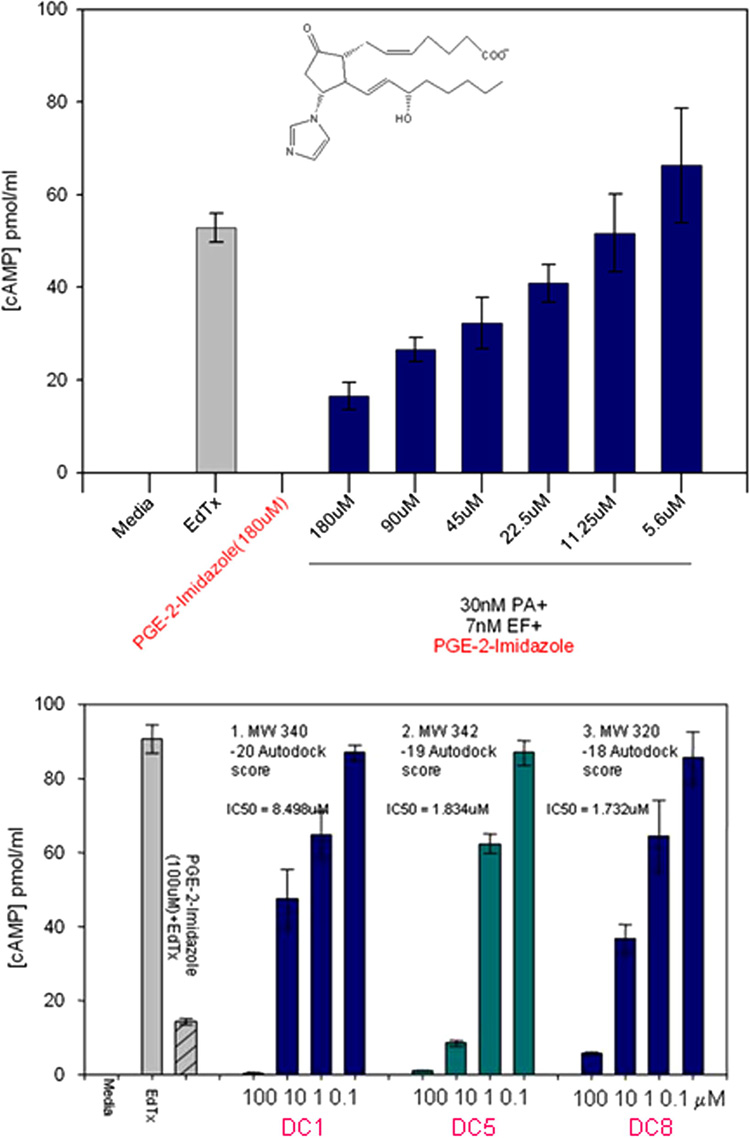
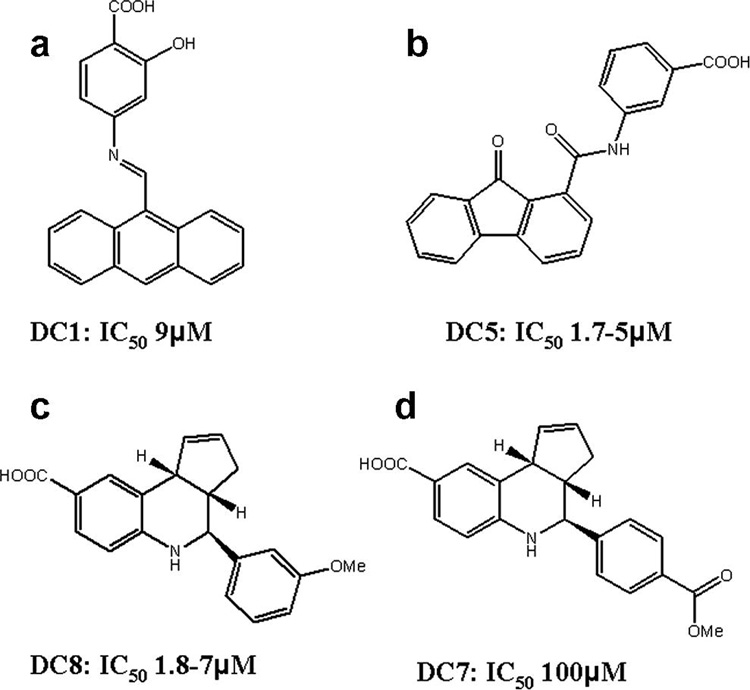
The inhibitory activities of the compounds on preventing the secretion of cAMP by cultured cells were compared to that of a previously known inhibitor, PGE2-imidazole16, which inhibits the EF-induced production of cAMP in cells in the range of 100 µM (Figure 5a). Their docking and IC50 values also compared favorably to those reported for other inhibitors of EF, which were identified by docking in a previous study24 (last three lines of Table 1). Three compounds from our list, with quite different molecular structures, had IC50 values in the low µmolar range (Fig. 5b, Fig. 6): 3-[(9-oxo-9H-fluorene-1-carbonyl)-amino]-benzoic acid (DC5 in Table 1; ZINC #75209; 1.7–5 µM), 4-(3-methoxy-phenyl)-3a,4,5,9b-tetrahydro-3H-cyclopenta[c]quinoline-8-carboxylic acid (DC8; ZINC #75022; 1.8–7 µM), and 4-[(anthracen-9-ylmethylene)-amino]-2-hydroxybenzoic acid (DC1; ZINC #132715; 9 µM). Despite their diverse structures, none of which resemble ATP, the docking positions of all 3 of these compounds were close to that of ATP in crystal structures of EF complexes (Figure 1f, Figure 6c,d) and with the initial pharmacophore (Figure 1e and Figure 6 a,b). Common substructures in all three compounds include a multiring, planar aromatic structure, which docks near the position of the (planar) purine of ATP (and centers on the phenyl fragment F1 position), and a carboxyl group (that corresponds approximately to the carboxyl fragment F2) that interacts in the docking with the metal ion and positively charged side chains in EF.
Figure 5.
Bioassay results. a) PGE2-imidazole16 inhibits cAMP production induced by Edema Toxin (EdTx). RAW 264.7 cells were incubated with 30 nM PA and 7 nM EF for 4 hours, in the absence (EdTx) or presence of various concentrations of PGE2-imidazole. Controls (no PA or EF) were media alone or plus 180 µM PGE2-imidazole. IBMX (50 µM, phosphodiesterase inhibitor) was added to each well before assaying the concentration of cAMP in the medium, as described previously 47. Data is representative of several experiments, where each sample was done in triplicate. b) Bioassay of the 3 best compounds (DC1,5, and 8; from the 19 selected with low AutoDock scores) inhibited EF with IC50 between 1.7–8.5 µM; all 3 were more effective inhibitors than 100 µM PGE2-imidazole (lane 3). Controls (no PA or EF) were media alone (lane 1) or plus 100 µM of each compound tested (lanes 4,9,14). Assays shown were done on one day together, IC50 values given in Figure 6 are calculated from 2 other assays with differing amounts of the compounds.
Figure 6.
The structures of the active inhibitors (Figure 5b) and their IC50 values (average or range of several assays); see Table 1 for docking energies.
None of the three best compounds resembled any known drug or metabolite. However, DC8 has some similarity to a phosphatase inhibitor family identified experimentally by assaying a diversity library of 10,000 compounds51. Substituting the 3-methoxy group (-OMe) on the phenyl of DC-8 to 4-methoxycarbonyl (-CO2Me) (DC7) greatly reduced activity (Figure 6). The docked conformations of DC7 and DC8 are very similar, with no real difference in the binding energy (Table 1). These results indicate that, despite the obvious accuracy of the force field calculations used in the different docking methods (discussed in more detail elsewhere47), the inhibitory potential of our compounds is not indicated solely by docking energies. This result is consistent with those of other groups52. Other factors that contribute to a compound’s ability to inhibit include its solubility53, ability to penetrate into the cell or the active site of the target protein, and binding to alternate sites on the same or other proteins.
Figure 1e, 1f, and Figure 7 illustrate how well the lowest energy docking conformation of the three active compounds corresponds to both the initial pharmacophore and that of the ATP analogue in the crystal structure of EF. Combinatorial design of these compounds, for enhanced pharmaceutical properties, is ongoing to optimize this fit to the active site.
Figure 7.
Comparison between the docked conformations of inhibitors DC5 and DC8 in the active site of EF, overlaying the position of the pharmacophore fragments (a, b) or and that of 3’dATP in the crystal structure (c,d).
3. Discussion
There are several ways to begin screening for compounds in libraries that could be inhibitors of a given protein. The most common begin with the known substrate, or another lead compound identified experimentally. The major problem with these approaches is that they are inherently limiting in the compound space that can be explored. Here, rather than rely on the ubiquitous reaction substrate, ATP, as our pharmacophore for in silico compound selection, we directly interrogated the active site to determine, de novo, an optimal configuration of binding fragments (Figures 1) using the HINT program. Fragment docking avoids many of the artifacts that may arise when determining binding energies of large compounds for protein active sites, as there are few possible conformations and solvent interactions are better defined. We then used the configuration of a subset of these fragments to search databases, using the UNITY program, beginning with the NIH/NCI library of druglike compounds (Figure 3) and expanding to the multipurpose catalogue of commercially available compounds, ZINC. The compounds from that search were then ranked according to their calculated binding affinity for EF, with the AutoDock program (Figure 4 and Table 1). Our use of pharmacophore based in silico screening methods enabled us to identify novel lead compounds by querying the existing databases, using a reasonable amount of CPU. As the resulting compound list yielded several candidates with significant inhibitory activity (Figure 5–Figure 7), they reduced the need for a large number of cell culture assays. The procedure thus proved to be both cost- and resource-efficient. A further advantage of the fragment based approach to pharmacophore design is that it did not restrict us to nucleotide based analogues, as a pharmacophore based only on the bound ligand in the crystal structure would have.
In addition to our fragments, we also used the compounds initially identified from the NCI library, with good docking values to identify additional modes of interaction with active site groups. Hydrogen bond interactions are very important in ligand-receptor interactions and are important in drug design. Fragment based methods, that begin with discrete molecules with few rotatable bonds, allow one to identify areas within the target that can be targeted to form hydrogen bonds with ligands. Our fragment based pharmacophore is different from the traditional ones in that we emphasize the atom types of the hydrogen bond forming atoms (Figure 2; Figure 1b). By using a fragment based pharmacophore, it is possible to make every hydrogen bond between the ligand and receptor contribute significantly to the binding energy, and thus enhance ligand efficiency.
To choose fragments which could form the most favorable hydrogen bonds with atoms in the active site of the receptor, we used the HINT scoring function to calculate the binding energy of individual fragments to the receptor. The HINT scoring function takes into account the desolvation energy of hydrogen bond donors (HD) or acceptors (HA) and calculates the ligand-receptor binding energy by using an empirical scoring function. It estimates the free energy of binding based on experimental measurements of octanol/water transfer free energies (logPo/w), ie, the free energy for solute transfer between the two solvents, for small organic molecules.43–45, 54..This measure, which is widely used as a measure of aqueous solubility, can also be used to evaluate structure-activity relationships.55
The main aim of our fragment based pharmacophore method, in which the atom types (especially the atom types of hydrogen bond forming groups) were specified, is to identify fragments with high ligand efficiency, and at most one rotatable bond. Similar approaches are incorporated into other design programs, including GANDI (Genetic Algorithm-based de Novo Design of Inhibitors), 56, LUDI57, GROW58, GroupBuild59 and FlexDovo60. GANDI automatically docks a 40,000 fragment library into into a target active site with SEED61 and links the prodocked fragments. FlexNovo uses a sequential growth strategy. based on the FlexX57, 62–65 docking software for the docking calculation of all fragments and makes use of an incremental algorithm and underlying chemical models in a sequential growth process.
In our fragment identification step, we focus on the atom types of fragments, especially the atom types of the hydrogen bond forming groups. The linkers between fragments were assigned according to the placement in the active site, but generous tolerances were used in applying these distance constraints, to obtain a more flexible pharmacophore.
Reducing CPU in library screening
Preselecting compounds, compared to random docking of the whole library to the active site, greatly reduced the computational time required for our compound search. We were able to concentrate on molecules that had a good chance to bind within the active site. We found in initial trials that the default “rapid screening” conditions (10 iterations) with AutoDock did not give accurate results for docking known inhibitors. While 60 iterations (about 6 min CPU/compound, more for those with many rotatable bonds66) were adequate to discriminate good compounds from the bulk of the list, longer docking times were needed to distinguish the very best compounds, as energy levels did not approach their true minimum until about 200 iterations (up to 30 min CPU per compound). We have since found (data not shown) that the GOLD program67, 68 also gives more accurate results if longer iterations are used.
As we needed to dock “only” 10,000 compounds, and used very long docking times for only the most likely compounds, our final ranking order was based on the lowest energy conformation one could obtain by docking. We could also compare the dockings in several different PDB files of the active sites of EF and the related Pertussis toxin, to control for how the hydrogen bonding pattern in the active site was affected by variations due to crystal conditions or the composition of the complex. The advantages in this tiered docking, compared to high speed docking of a larger number of unselected compounds, is indicated by the fact that we obtained 3 possible lead inhibitors from a small diverse starting list.
Types of compounds resulting from the search
Compounds identified in this study differ considerably from P-site inhibitors (adenine nucleotides with a 3’-O phosphate or polyphosphate groups) that have been shown to inhibit mammalian AC20, 21, 69–71, but have no effect on the catalytic activity of EF72. The compounds that populated our lowest binding energy class were quite diverse, and would not have been obtained had we only begun with direct analogues of the substrate. All three of the active compounds have two ring systems, connected by a flexible linker. The three most active compounds are similar in that each compound has a free carboxyl group, which docks in a positively charged region of the active site formed from residues highly conserved in EF and related adenylyl cyclase toxins, and a largely planar, aryl hydrocarbon ring system with some internal flexibility, that overlays the area where the adenine ring binds.
Pharmacokinetic factors will eventually determine the suitability of these compounds for use in therapy. Neither DC5 nor DC8 revealed significant toxicity in initial animal testing, and have progressed to more rigorous assays for suitability as an adjunct treatment for inhalation anthrax and further combinatorial optimization. Our efforts are now concentrated on obtaining derivatives of our lead compounds with improved pharmaceutical properties, such as solubility, stability, oral uptake, and bioavailability.
4. Conclusions
The fragment based, 3D-pharmacophore and in silico screening enabled us to identify novel inhibitors of EF from compound databases, using a reasonable amount of CPU. As the small list of compounds for assay yielded several candidates with significant inhibitory activity, lab assays were kept to a minimum (an important consideration when dealing with assays that involve toxins and expensive reagents). The major advantage of the flexible, fragment based pharmacophore design is that it did not restrict us to obvious analogues of the ubiquitous biological substrate, ATP, but allowed us to explore a much larger chemical space. This means that the inhibitors should be less likely to cause side reactions by binding to other enzymes that bind adenine nucleotides.
These results demonstrate that pharmacophore based docking methods can be used to widen the search for lead compounds to inhibit bacterial toxins, and to fully use the diversity now available in web-based compound databases. Besides its usefulness in cases like ours, the methodology could also be easily adapted to identifying potential substrates of proteins of unknown function, such as those revealed by the structural genomics initiatives. 73,74, 75, or to cryptic, conserved sites in known enzymes76.
5. Experimental
5.1. Protein Data Bank Structures
Structure of anthrax EF
The Protein Data Bank (PDB) structure, 1K908, (resolution 2.75 Å, r-value 0.225) was used. Here, anthrax EF, with the PA binding domain removed, is complexed with calmodulin and a non-cyclizable nucleotide analogue, 3’-deoxy-ATP (3’dATP). One Yb3+ ion in the active site coordinates carboxyl groups of residues Asp491, Asp493 and His577 (Yb- N: 2.78Å) and an oxygen atom of the α-phosphate group of the 3’dATP ligand (Figure 1a). For all the dockings using 1K90, Yb3+ was replaced with the more physiological Mg2+ at the same position, which allowed better comparison of the energy data47. As noted in our previous work, EF is active in the presence of Yb3+, which is easier to see in electron density maps than the smaller, but physiological, metal Mg2+. Metal binding sites for both metals could appear similar but have discrete differences at the protonation stage.47, 77 We found that we could obtain similar docking results for crystal structures that contained either the one bound Yb3+, or another with 2 Mg2+ at the active site if we corrected for the protonation state of the ligand. However, dockings to the 1K90 structure were most accurate (i.e., correlated better with the position of the ligand seen in the crystal structure), so only those results are shown here.47 All water molecules and 3’dATP were removed. The protein model was used for docking and ligand-receptor binding energy calculation without minimization. When using the HINT program for calculating fragment-protein interaction energies and using FlexX for docking, all hydrogen atoms were added. When using AutoDock3 for docking, only polar hydrogen atoms are added to protein. This structure also has a substrate analogue bound, thus accounting for changes in sidechain orientations induced by binding of suitable ligands78.
5.2. Pharmacophore design
Fragment database
The fragment database consisted of 3-D structures (MOL2 files built in SYBYL) of about 60 small molecules, containing hydrogen bond donor/acceptor or hydrophobic moieties, with at most one rotatable bond (Figure 2). These were either common ionizable molecules, or were selected from the SYBYL fragment database.
HINT score
The Hydropathic INTeractions, or HINT, program43–45 uses experimental solvent partitioning data as a basis for an empirical molecular interaction model that calculates free energy scores that were shown to accurately reproduce experimental measurements of binding43– 45. “Hydropathic” interactions are non-covalent interactions such as hydrogen-bonding, acid-base, Coulombic, and hydrophobic interactions. The HINT calculation is the summation of hydropathic interactions between all atom pairs:
where bij is the interaction score between atoms i and j, S is the solvent accessible surface area, a is the hydrophobic atom constant, Rij and rij are the functions of distance between i and j, and Tij is a logic function with value of 1 and −1, depending on the character of interacting polar atoms. The item rij is an appropriately weighted implementation of the Lennard-Jones 6–12 function adapted from the literatures79−81. It is scaled with an empirical constant of "50/50” without using "vector focusing". In practice, a HINT score difference of 500 corresponds to an energy difference of about 1 kcal/mol.
Fragment based pharmacophore design
The goal was to find a scaffold of fragments with flexible distance constraints that would most completely fill the active site. The optimal binding position of each molecule in the fragment database in the active site of EF (PDB 1K90) was obtained by translating and rotating the fragment, using an algorithm reported previously82, so that the best HINT score for the interaction of the fragment and active site was achieved. The coordinates of the small molecules that had the best HINT score with the receptor were saved. The molecule with the best interaction energy was incorporated into the receptor so as to block that area in the active site. Then the fragment library was redocked into this compound receptor to find secondary binding locations that did not sterically conflict with previously bound fragments. The five fragments with the most favorable HINT scores (Figure 1b) surround the position of 3’dATP in the crystal structure of EF, but also indicate additional possible strong interaction sites peripheral to this (Figure 1c). Different combinations of these five fragments at the indicated relative positions were used to identify several pharmacophores such that a given pharmacophore included three or four fragments (Figure 1d).
5.3. Database screening
NCI database search with UNITY
A UNITY (in SYBYL from Tripos) search was conducted using the pharmacophores obtained as above. All hydrogen atoms in the fragments were removed and distance constraints (i.e., the distances between the heavy atoms of the bound fragments in the configuration shown in Figure 1d) were automatically extracted from a PDB file used to start the 3D UNITY searches. The 3D-pharmacophore search was done in the NCI-2000 database, containing ~250,000 structures, as integrated in SYBYL. The hits obtained were docked into the EF active site using FlexX and those with the lowest FlexX Scores were selected (Figure 3). The interactions of these compounds were then analyzed, and their major interactions with the metal and the active site, such as hydrogen bonding, hydrophobic, and metal coordination interactions identified (circled in figure 3).
ZINC database screening
ZINC83 is a web database of over 2.7 million commercially available compounds for virtual screening. A variety of 2-D fragments, based on our initial fragments and other interactions identified from the larger compounds from the NCI database, were used to search the ZINC database (http://blaster.docking.org/zinc/choose.shtml), to identify about 10,000 compounds that matched our initial 3D-pharmacophore. These were ranked using AutoDock and FlexX scores for binding to our target enzymes, as described below.
5.4. Docking
FlexX
FlexX57, 62–65, 84 was used to dock with EF the compounds obtained from UNITY search of the NCI database. Default docking settings were used, except that the number of conformations was set to 100. Formal charges of the metals were assigned by the SYBYL program. Compounds were selected that had a better (lower) docking score than that obtained for ATP and our assay control inhibitor PGE2-imidazole.
AutoDock
The compounds obtained from the ZINC database screening were docked with EF using AutoDock version 3.0.5_(which proved more accurate47 and was easier to implement in parallel for multiple dockings than FlexX). Autodock4 gave similar results but not identical docking scores. AutoDock85, 86 (http://www.scripps.edu/mb/olson/doc/autodock) provides three different algorithms for docking: simulated annealing (SA), genetic algorithm (GA) and “Lamarckian” genetic algorithm (LGA). We docked 3’dATP to the protein model 1K90 with LGA, SA and GA respectively. The docking results showed that the docking conformation using LGA was in better agreement with experimental conformation than using SA or using GA. Also we found in our initial that the LGA gave better results than SA or GA in rapidly discriminating compounds with potentially useful docking energies from those that did not, and others have found that both LGA and GA are much more efficient and robust than SA86. Thus, LGA was used in these studies._For initial screening, default parameters (10 iterations) were used (Figure 4). This was increased to 60 iterations and the results compared. For final scoring, the number of iterations was set to 200 and population size to 100. The ligand and solvent molecules were removed from the crystal structure to obtain the docking grid and the active site was defined using AutoGrid. The grid size was set to 90 × 90 × 90 points with grid spacing of 0.375 Å. The grid box was centered on the center of the ligand from the corresponding crystal structure complexes. The Amber force field potentials were used for the Mg2+ ions as defined in the AutoDock program. We assigned a partial charge of +0.8 as we found that if the formal charge was set to +1.2, the interaction of the ligand and the carboxyl group of the ligand was overestimated and led to very short O-Mg distances. The conformation with the lowest binding energy was used to analyze ligand placement in the active site.
5.5. Bioassay
Compounds for assay
PGE2-imidazole, used as the positive control in the biological assays, was synthesized as previously described16 and stored frozen. PGE2-imidazole was first identified as an inhibitor of mammalian adenylyl cyclase activity following cholera toxin stimulation16 but has also been shown to be an effective inhibitor of anthrax EF in the µM range (Figure 5a). Other compounds were obtained from the National Cancer Institute (NCI) or purchased from Asinex, Ryan Scientific, Sigma-Aldrich, or TimTec. Compounds were dissolved in cell culture medium or DMSO, and where necessary the pH was adjusted to neutrality with small amounts of NaOH or HCl.
Cell culture
Murine monocyte/macrophage cells (RAW 264.7) were propagated in T75 flasks containing Dulbecco’s Modified Eagle’s Medium (DMEM) (Mediatech, Inc., Herndon, VA) at 37°C with 5% CO2. The culture media contained 10% fetal bovine serum (FBS), 100 µg/ml penicillin/streptomycin and L-glutamine.
Cell Assay
Cells were plated 1 × 106 cells per well in DMEM containing 10% FBS, 100 µg/ml penicillin/streptomycin and L-glutamine with isobutylmethylxanthine (IBMX) (50µM) IBMX in 48 well tissue culture plates and allowed to adhere overnight at 37°C in 5% CO2. PGE2- imidazole, PA (2.5µg/ml) and EF (0.625 µg/ml) were diluted with assay media containing DMEM (without phenol red) with 100 µg/ml penicillin/streptomycin and L-glutamine. Media was aspirated from the cells and replaced with the varying concentrations of PGE2-imidazole (180 µM, 90 µM, 45 µM, 22.5 µM, 11.25 µM and 5.6µM) along with PA and EF. The plates were then incubated for 4 hours at 37°C in 5% CO2. Following incubation, the culture supernatants were removed (extracellular cAMP) and transferred to a new 48 well plate for cAMP determination.
cAMP determination
The extracellular cAMP concentration in the culture supernatants was measured with a cAMP-specific ELISA from Assay Designs, Inc. (Ann Arbor, Michigan) per manufacturer directions. Previous assays of the toxin effects have shown that the extracellular levels were more reliable than the intracellular levels of cAMP. A recent report describing the ribonucleotide efflux mechanism offers an explanation for this87. The increase in cAMP secreted by mammalian cells affected by adenylyl cyclase toxins could play a role in quorum sensing that enables bacterial colonization of the intestine.88–90
Estimation of cytotoxic effects
All compounds were tested for cytotoxicity, and any that elicited a cytotoxic response within the concentration range tested (up to 100 µM) were discarded. Cytotoxicity was measured by visual observation of the control cells (compound without PA/EF added) and quantitatively by lactate dehydrogenase (LDH) enzyme release from a murine monocyte-macrophage cell line (RAW 264.7; American Type Culture Collection, Manassas, VA)91 or the MTT assay, which is a colorimetric test based on the uptake of 3-(4, 5- dimethylthiazolyl-2)-2,5-diphenyltetrazolium bromide (MTT) by proliferating cells (cytotoxic compounds reduce the MTT taken by cells as the drug concentration is increased)92. For the LDH assay, the RAW 264.7 cells are propagated in Dulbecco’s modified essential medium supplemented with 10% fetal bovine serum, 100 µg/ml penicillin-streptomycin, and 2 mM L-glutamine (Mediatech, Inc.,Herndon, VA) at 37°C with 5% CO2 using tissue culture flasks. Subsequently, thecells are plated in 96-well flat-bottom tissue culture plates (Corning) at adensity of 1 × 106 cells/ml and incubated overnight at 37°C in 5% CO2. Themonolayers are washed twice with Dulbecco’s modified essential medium devoidof serum or phenol red. LeTx-mediated cytotoxicity is measured as afunction of the amount of LDH enzyme released from the macrophages into thecell culture supernatants. Various dilutions of compounds are incubated for 4 h at 37°C in 5% CO2. LDH release into the culture supernatant of the macrophage cells is measured using the CytoTox 96 nonradioactive cytotoxicity assay kit (Promega, Madison, WI) and quantitated by measuring wavelength absorbance at 490 nm. An increase in color of the culture medioium is an indication of cytotoxicity.
For the MTT assay, a kit purchased from the American Type Culture Collection (Manassas, VA) was used. J774A.1 murine monocyte/macrophage-like cells (ATCC) were plated at 5 × 105 cells/ml and grown to 60 to 80% confluence at 37°C overnight in 5% CO2. Twofold dilutions of each compound were added to the cells and incubated for 4 h. After incubation, 10 µl/well of yellow tetrazolium MTT salt was added to the cells and left for 2 h. The salt was reduced by metabolically active cells. The resulting intracellular purple formazan was solubilized overnight in detergent reagent (ATCC catalog no. 30-1010K) provided in the MTT assay kit. The reaction product was measured at 570 nm and quantified. Reduction in color is an indication of cytotoxicity.
ACKNOWLEDGMENTS
Funding for this project was provided by grants from the NIH (5UO1-AI053858-03), the US Army (DAMD17-02-1-0699) and MISSION PHARMACAL COMPANY, San Antonio, TX.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Drysdale M, Heninger S, Hutt J, Chen YH, Lyons CR, Koehler TM. Capsule synthesis by Bacillus anthracis is required for dissemination in murine inhalation anthrax. EMBO J. 2005;24(1):221–227. doi: 10.1038/sj.emboj.7600495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrami L, Reig N, van der Goot FG. Anthrax toxin: the long and winding road that leads to the kill. Trends Microbiol. 2005;13(2):72–78. doi: 10.1016/j.tim.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 3.Nguyen TL, Panchal RG, Topol IA, Lane D, Kenny T, Burnett JC, Hermone AR, McGrath C, Burt SK, Gussio R, Bavari S. A theoretical study of anthrax lethal factor inhibition by a set of novel carbamimidolyl-aryl-vinyl-carboxamidines: A possible mechanism involving zinc-ligation by amidine. J of Mol Struct-Theochem. 2007;821(1–3):139–144. [Google Scholar]
- 4.Xiong YS, Wiltsie J, Woods A, Guo J, Pivnichny JV, Tang W, Bansal A, Cummings RT, Cunningham BR, Friedlander AM, Douglas CM, Salowe SP, Zaller DM, Scolnick EM, Schmatz DM, Bartizal K, Hermes JD, MacCoss M, Chapman KT. The discovery of a potent and selective lethal factor inhibitor for adjunct therapy of anthrax infection. Bioorganic & Medicinal Chemistry Letters. 2006;16(4):964–968. doi: 10.1016/j.bmcl.2005.10.088. [DOI] [PubMed] [Google Scholar]
- 5.Munier H, Bouhss A, Krin E, Danchin A, Gilles AM, Glaser P, Barzu O. The role of histidine 63 in the catalytic mechanism of Bordetella pertussis adenylate cyclase. J. Biol. Chem. 1992;267(14):9816–9820. [PubMed] [Google Scholar]
- 6.Hewlett EL, Underhill LH, Cook GH, Manclark CR, Wolff J. A protein activator for the adenylate cyclase of Bordetella pertussis. J. Biol. Chem. 1979;254(13):5602–5605. [PubMed] [Google Scholar]
- 7.Hewlett EL, Urban MA, Manclark CR, Wolff J. Extracytoplasmic adenylate cyclase of Bordetella pertussis. Proc. Natl. Acad. Sci. USA. 1976;73(6):1926–1930. doi: 10.1073/pnas.73.6.1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Drum CL, Yan SZ, Bard J, Shen YQ, Lu D, Soelaiman S, Grabarek Z, Bohm A, Tang WJ. Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature. 2002;415(6870):396–402. doi: 10.1038/415396a. [DOI] [PubMed] [Google Scholar]
- 9.Shen Y, Zhukovskaya NL, Guo Q, Florian J, Tang WJ. Calcium-independent calmodulin binding and two-metal-ion catalytic mechanism of anthrax edema factor. EMBO J. 2005;24(5):929–941. doi: 10.1038/sj.emboj.7600574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leppla S. Anthrax toxin edema factor: a bacterial adenylate cyclase that increases cyclic AMP concentrations of eukaryotic cells. Proc. Natl. Acad. Sci. USA. 1982;79(10):3162–3166. doi: 10.1073/pnas.79.10.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Rooij J, Zwartkruis FJ, Verheijen MH, Cool RH, Nijman SM, Wittinghofer A, Bos JL. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature. 1998;396(6710):474–477. doi: 10.1038/24884. [DOI] [PubMed] [Google Scholar]
- 12.Lacy DB, Collier RJ. Structure and function of anthrax toxin. Curr. Top. Microbiol. Immunol. 2002;271:61–85. doi: 10.1007/978-3-662-05767-4_4. [DOI] [PubMed] [Google Scholar]
- 13.Lacy DB, Mourez M, Fouassier A, Collier RJ. Mapping the anthrax protective antigen binding site on the lethal and edema factors. J. Biol. Chem. 2002;277(4):3006–3010. doi: 10.1074/jbc.M109997200. [DOI] [PubMed] [Google Scholar]
- 14.Ahuja N, Kumar P, Bhatnagar R. The adenylate cyclase toxins. Crit. Rev. Microbiol. 2004;30(3):187–196. doi: 10.1080/10408410490468795. [DOI] [PubMed] [Google Scholar]
- 15.Ascenzi P, Visca P, Ippolito G, Spallarossa A, Bolognesi M, Montecucco C. Anthrax toxin: a tripartite lethal combination. FEBS Lett. 2002;531(3):384–388. doi: 10.1016/s0014-5793(02)03609-8. [DOI] [PubMed] [Google Scholar]
- 16.Peterson JW, King D, Ezell EL, Rogers M, Gessell D, Hoffpauer J, Reuss L, Chopra AK, Gorenstein D. Cholera toxin-induced PGE(2) activity is reduced by chemical reaction with L-histidine. Biochim. Biophys Acta. 2001;1537(1):27–41. doi: 10.1016/s0925-4439(01)00052-7. [DOI] [PubMed] [Google Scholar]
- 17.Shen Y, Guo Q, Zhukovskaya NL, Drum CL, Bohm A, Tang WJ. Structure of anthrax edema factor-calmodulin-adenosine 5′-(alpha,beta-methylene)-triphosphate complex reveals an alternative mode of ATP binding to the catalytic site. Biochem. Biophys. Res. Commun. 2004;317(2):309–314. doi: 10.1016/j.bbrc.2004.03.046. [DOI] [PubMed] [Google Scholar]
- 18.Shen Y, Zhukovskaya NL, Zimmer MI, Soelaiman S, Bergson P, Wang CR, Gibbs CS, Tang WJ. Selective inhibition of anthrax edema factor by adefovir, a drug for chronic hepatitis B virus infection. Proc. Natl. Acad. Sci. U S A. 2004;101(9):3242–3247. doi: 10.1073/pnas.0306552101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo Q, Shen Y, Zhukovskaya NL, Florian J, Tang WJ. Structural and kinetic analyses of the interaction of anthrax adenylyl cyclase toxin with reaction products cAMP and pyrophosphate. J. Biol. Chem. 2004;279(28):29427–29435. doi: 10.1074/jbc.M402689200. [DOI] [PubMed] [Google Scholar]
- 20.Johnson RA, Desaubry L, Bianchi G, Shoshani I, Lyons E, Jr, Taussig R, Watson PA, Cali JJ, Krupinski J, Pieroni JP, Iyengar R. Isozyme-dependent sensitivity of adenylyl cyclases to P-site-mediated inhibition by adenine nucleosides and nucleoside 3′-polyphosphates. J. Biol. Chem. 1997;272(14):8962–8966. doi: 10.1074/jbc.272.14.8962. [DOI] [PubMed] [Google Scholar]
- 21.Tesmer JJ, Sunahara RK, Johnson RA, Gosselin G, Gilman AG, Sprang SR. Two-metal-Ion catalysis in adenylyl cyclase. Science. 1999;285(5428):756–760. doi: 10.1126/science.285.5428.756. [DOI] [PubMed] [Google Scholar]
- 22.Wang JL, Guo JX, Zhang QY, Wu JJQ, Seifert R, Lushington GH. A conformational transition in the adenylyl cyclase catalytic site yields different binding modes for ribosyl-modified and unmodified nucleotide inhibitors. Bioorganic & Medicinal Chemistry. 2007;15(8):2993–3002. doi: 10.1016/j.bmc.2007.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gottle M, Dove S, Steindel P, Shen Y, Tang W-J, Geduhn J, Konig B, Seifert R. Molecular Analysis of the Interaction of Bordetella pertussis Adenylyl Cyclase with Fluorescent Nucleotides. Mol Pharmacol. 2007;72(3):526–535. doi: 10.1124/mol.107.034413. [DOI] [PubMed] [Google Scholar]
- 24.Soelaiman S, Wei BQ, Bergson P, Lee YS, Shen Y, Mrksich M, Shoichet BK, Tang WJ. Structure-based inhibitor discovery against adenylyl cyclase toxins from pathogenic bacteria that cause anthrax and whooping cough. J. Biol. Chem. 2003;278(28):25990–25997. doi: 10.1074/jbc.M301232200. [DOI] [PubMed] [Google Scholar]
- 25.Shuker SB, Hajduk PJ, Meadows RP, Fesik SW. Discovering high-affinity ligands for proteins: SAR by NMR. Science. 1996;274(5292):1531–1534. doi: 10.1126/science.274.5292.1531. [DOI] [PubMed] [Google Scholar]
- 26.Blundell TL, Jhoti H, Abell C. High-throughput crystallography for lead discovery in drug design. Nat. Rev. Drug Discov. 2002;1(1):45–54. doi: 10.1038/nrd706. [DOI] [PubMed] [Google Scholar]
- 27.Carr R, Jhoti H. Structure-based screening of low-affinity compounds. Drug Discov Today. 2002;7(9):522–527. doi: 10.1016/s1359-6446(02)02245-6. [DOI] [PubMed] [Google Scholar]
- 28.Ciulli A, Abell C. Fragment-based approaches to enzyme inhibition. Current Opinion in Biotechnology. 2007;18(6):489–496. doi: 10.1016/j.copbio.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov Today. 2004;9(10):430–431. doi: 10.1016/S1359-6446(04)03069-7. [DOI] [PubMed] [Google Scholar]
- 30.Lepre CA, Moore JM, Peng JW. Theory and applications of NMR-based screening in pharmaceutical research. Chem. Rev. 2004;104(8):3641–3676. doi: 10.1021/cr030409h. [DOI] [PubMed] [Google Scholar]
- 31.Erlanson DA, McDowell RS, O'Brien T. Fragment-based drug discovery. J. Med. Chem. 2004;47(14):3463–3482. doi: 10.1021/jm040031v. [DOI] [PubMed] [Google Scholar]
- 32.Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nat. Rev. Drug. Discov. 2004;3(8):660–672. doi: 10.1038/nrd1467. [DOI] [PubMed] [Google Scholar]
- 33.Carr RA, Congreve M, Murray CW, Rees DC. Fragment-based lead discovery: leads by design. Drug Discov Today. 2005;10(14):987–992. doi: 10.1016/S1359-6446(05)03511-7. [DOI] [PubMed] [Google Scholar]
- 34.Fattori D. Molecular recognition: the fragment approach in lead generation. Drug Discov. Today. 2004;9(5):229–238. doi: 10.1016/S1359-6446(03)03007-1. [DOI] [PubMed] [Google Scholar]
- 35.Gill AL, Frederickson M, Cleasby A, Woodhead SJ, Carr MG, Woodhead AJ, Walker MT, Congreve MS, Devine LA, Tisi D, O'Reilly M, Seavers LC, Davis DJ, Curry J, Anthony R, Padova A, Murray CW, Carr RA, Jhoti H. Identification of novel p38alpha MAP kinase inhibitors using fragment-based lead generation. J. Med. Chem. 2005;48(2):414–426. doi: 10.1021/jm049575n. [DOI] [PubMed] [Google Scholar]
- 36.Liebeschuetz JW, Jones SD, Morgan PJ, Murray CW, Rimmer AD, Roscoe JM, Waszkowycz B, Welsh PM, Wylie WA, Young SC, Martin H, Mahler J, Brady L, Wilkinson K. PRO_SELECT: combining structure-based drug design and array-based chemistry for rapid lead discovery. 2. The development of a series of highly potent and selective factor Xa inhibitors. J. Med. Chem. 2002;45(6):1221–1232. doi: 10.1021/jm010944e. [DOI] [PubMed] [Google Scholar]
- 37.Card GL, Blasdel L, England BP, Zhang C, Suzuki Y, Gillette S, Fong D, Ibrahim PN, Artis DR, Bollag G, Milburn MV, Kim SH, Schlessinger J, Zhang KA. A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design. Nat. Biotechnol. 2005;23(2):201–207. doi: 10.1038/nbt1059. [DOI] [PubMed] [Google Scholar]
- 38.Lundqvist T. The devil is still in the details--driving early drug discovery forward with biophysical experimental methods. Curr. Opin. Drug Discov. Devel. 2005;8(4):513–519. [PubMed] [Google Scholar]
- 39.Hajduk PJ, Huth JR, Fesik SW. Druggability indices for protein targets derived from NMR-based screening data. J. Med. Chem. 2005;48(7):2518–2525. doi: 10.1021/jm049131r. [DOI] [PubMed] [Google Scholar]
- 40.Silberstein M, Dennis S, Brown L, Kortvelyesi T, Clodfelter K, Vajda S. Identification of substrate binding sites in enzymes by computational solvent mapping. J. Mol. Biol. 2003;332(5):1095–1113. doi: 10.1016/j.jmb.2003.08.019. [DOI] [PubMed] [Google Scholar]
- 41.Joseph-McCarthy D, Tsang SK, Filman DJ, Hogle JM, Karplus M. Use of MCSS to design small targeted libraries: application to picornavirus ligands. J. Am. Chem. Soc. 2001;123(51):12758–12769. doi: 10.1021/ja003972f. [DOI] [PubMed] [Google Scholar]
- 42.Landon MR, L DJ, Yu J, Thiel SC, Vajda S. Identification of hot spots within druggable binding regions by computational solvent mapping of proteins. J Med Chem. 2007;50(6):1231–1240. doi: 10.1021/jm061134b. [DOI] [PubMed] [Google Scholar]
- 43.Fornabaio M, Cozzini P, Mozzarelli A, Abraham DJ, Kellogg GE. Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 2. Computational titration and pH effects in molecular models of neuraminidase-inhibitor complexes. J. Med. Chem. 2003;46(21):4487–4500. doi: 10.1021/jm0302593. [DOI] [PubMed] [Google Scholar]
- 44.Fornabaio M, Spyrakis F, Mozzarelli A, Cozzini P, Abraham DJ, Kellogg GE. Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 3. The free energy contribution of structural water molecules in HIV-1 protease complexes. J. Med. Chem. 2004;47(18):4507–4516. doi: 10.1021/jm030596b. [DOI] [PubMed] [Google Scholar]
- 45.Cozzini P, Fornabaio M, Marabotti A, Abraham DJ, Kellogg GE, Mozzarelli A. Simple, intuitive calculations of free energy of binding for protein-ligand complexes. 1. Models without explicit constrained water. J. Med. Chem. 2002;45(12):2469–2483. doi: 10.1021/jm0200299. [DOI] [PubMed] [Google Scholar]
- 46.Irwin JJ, Shoichet BK. ZINC - A Free Database of Commercially Available Compounds for Virtual Screening. J. Chem. Inf. Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen DL, Menche G, Power TD, Sower L, Peterson JW, Schein CH. Accounting for ligand-bound metal ions in docking small molecules on adenylyl cyclase toxins. Proteins-Structure Function and Bioinformatics. 2007;67(3):593–605. doi: 10.1002/prot.21249. [DOI] [PubMed] [Google Scholar]
- 48.Searle MS, Williams DH, Gerhard U. Partitioning of free energy contributions in the estimation of binding constants: residual motions and consequences for amide-amide hydrogen bond strengths. J. Am. Chem. Soc. 1992;114(27):10697–10704. [Google Scholar]
- 49.Williams DH, Searle MS, Mackay JP, Gerhard U, Maplestone RA. Toward an estimation of binding constants in aqueous solution: studies of associations of vancomycin group antibiotics. Proc. Natl. Acad. Sci. USA. 1993;90(4):1172–1178. doi: 10.1073/pnas.90.4.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ghose AK, Viswanadhan VN, Wendoloski JJ. A knowledge-based approach in designing combinatorial or medicinal chemistry libraries for drug discovery. 1. A qualitative and quantitative characterization of known drug databases. J. Comb. Chem. 1999;1(1):55–68. doi: 10.1021/cc9800071. [DOI] [PubMed] [Google Scholar]
- 51.Brisson M, Nguyen T, Vogt A, Yalowich J, Giorgianni A, Tobi D, Bahar I, Stephenson CRJ, Wipf P, Lazo JS. Discovery and characterization of novel small molecule inhibitors of human Cdc25B dual specificity phosphatase. Molecular Pharmacology. 2004;66(4):824–833. doi: 10.1124/mol.104.001784. [DOI] [PubMed] [Google Scholar]
- 52.Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, Lambert MH, Lindvall M, Nevins N, Semus SF, Senger S, Tedesco G, Wall ID, Woolven JM, Peishoff CE, Head MS. A critical assessment of docking programs and scoring functions. Journal of Medicinal Chemistry. 2006;49(20):5912–5931. doi: 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- 53.Babaoglu K, Simeonov A, Irwin JJ, Nelson ME, Feng B, Thomas CJ, Cancian L, Costi MP, Maltby DA, Jadhav A, Inglese J, Austin CP, Shoichet BK. Comprehensive Mechanistic Analysis of Hits from High-Throughput and Docking Screens against beta-Lactamase. J. Med. Chem. 2008 doi: 10.1021/jm701500e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kellogg GE, Semus SF, Abraham DJ. HINT: a new method of empirical hydrophobic field calculation for CoMFA. J. Comput. Aided Mol. Des. 1991;5(6):545–552. doi: 10.1007/BF00135313. [DOI] [PubMed] [Google Scholar]
- 55.Muñoz-Muriedas J, Perspicace S, Bech N, Guccione S, Orozco M, Luque FJ. Hydrophobic Molecular Similarity from MST Fractional Contributions to the Octanol/water Partition Coefficient. J Comput Aided Mol Des. 2005;19(6):401–419. doi: 10.1007/s10822-005-7928-3. [DOI] [PubMed] [Google Scholar]
- 56.Dey F, Caflisch A. Fragment-based de novo ligand design by multiobjective evolutionary optimization. J. Chem. Inf. Model. 2008;48(3):679–690. doi: 10.1021/ci700424b. [DOI] [PubMed] [Google Scholar]
- 57.Bohm HJ. LUDI: rule-based automatic design of new substituents for enzyme inhibitor leads. Comput. Aided Mol. Des. 1992;6(6):593–606. doi: 10.1007/BF00126217. [DOI] [PubMed] [Google Scholar]
- 58.Moon JB, Howe WJ. Computer design of bioactive molecules: a method for receptor-based de novo ligand design. Proteins. 1991;11(4):314–328. doi: 10.1002/prot.340110409. [DOI] [PubMed] [Google Scholar]
- 59.Rotstein SH, Murcko MA. GroupBuild: a fragment-based method for de novo drug design. J. Med. Chem. 1993;36(12):1700–1710. doi: 10.1021/jm00064a003. [DOI] [PubMed] [Google Scholar]
- 60.Degen J, Rarey M. FlexNovo: structure-based searching in large fragment spaces. ChemMedChem. 2006;1(8):854–868. doi: 10.1002/cmdc.200500102. [DOI] [PubMed] [Google Scholar]
- 61.Majeux N, Scarsi M, Apostolakis J, Ehrhardt C, Caflisch A. Exhaustive docking of molecular fragments with electrostatic solvation. Proteins. 1999;37(1):88–105. [PubMed] [Google Scholar]
- 62.Bohm HJ. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J. Comput. Aided Mol. Des. 1994;8(3):243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 63.Bohm HJ. Computational tools for structure-based ligand design. Prog. Biophys. Mol. Biol. 1996;66(3):197–210. doi: 10.1016/s0079-6107(97)00005-9. [DOI] [PubMed] [Google Scholar]
- 64.Bohm HJ. Prediction of binding constants of protein ligands: a fast method for the prioritization of hits obtained from de novo design or 3D database search programs. J. Comput. Aided Mol. Des. 1998;12(4):309–323. doi: 10.1023/a:1007999920146. [DOI] [PubMed] [Google Scholar]
- 65.Rarey M, Wefing S, Lengauer T. Placement of medium-sized molecular fragments into active sites of proteins. J. Comput. Aided Mol. Des. 1996;10(1):41–54. doi: 10.1007/BF00124464. [DOI] [PubMed] [Google Scholar]
- 66.Erickson JA, Jalaie M, Robertson DH, Lewis RA, Vieth M. Lessons in molecular recognition: the effects of ligand and protein flexibility on molecular docking accuracy. J. Med. Chem. 2004;47(1):45–55. doi: 10.1021/jm030209y. [DOI] [PubMed] [Google Scholar]
- 67.Cole JC, Murray CW, Nissink JWM, Taylor RD, Taylor R. Comparing protein-ligand docking programs is difficult. Proteins-Structure Function and Bioinformatics. 2005;60(3):325–332. doi: 10.1002/prot.20497. [DOI] [PubMed] [Google Scholar]
- 68.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins-Structure Function and Genetics. 2003;52(4):609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 69.Dessauer CW, Gilman AG. The catalytic mechanism of mammalian adenylyl cyclase. Equilibrium binding and kinetic analysis of P-site inhibition. J. Biol. Chem. 1997;272(44):27787–27795. doi: 10.1074/jbc.272.44.27787. [DOI] [PubMed] [Google Scholar]
- 70.Onda T, Hashimoto Y, Nagai M, Kuramochi H, Saito S, Yamazaki H, Toya Y, Sakai I, Homcy CJ, Nishikawa K, Ishikawa Y. Type-specific regulation of adenylyl cyclase. Selective pharmacological stimulation and inhibition of adenylyl cyclase isoforms. J. Biol. Chem. 2001;276(51):47785–47793. doi: 10.1074/jbc.M107233200. [DOI] [PubMed] [Google Scholar]
- 71.Gille A, Lushington GH, Mou TC, Doughty MB, Johnson RA, Seifert R. Differential inhibition of adenylyl cyclase isoforms and soluble guanylyl cyclase by purine and pyrimidine nucleotides. J. Biol. Chem. 2004;279(19):19955–19969. doi: 10.1074/jbc.M312560200. [DOI] [PubMed] [Google Scholar]
- 72.Johnson RA, Shoshani I. Inhibition of Bordetella pertussis and Bacillus anthracis adenylyl cyclases by polyadenylate and "P"-site agonists. J. Biol. Chem. 1990;265(31):19035–19039. [PubMed] [Google Scholar]
- 73.Gerlt JA. A protein structure (or function?) initiative. Structure. 2007;15(11):1353–1356. doi: 10.1016/j.str.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 74.Ivanciuc O, Oezguen N, Mathura V, Schein CH, Xu Y, Braun W. Using property based sequence motifs and 3D modeling to determine structure and functional regions in CASP5 targets. Curr. Med. Chem. 2004;11(5):583–593. doi: 10.2174/0929867043455819. [DOI] [PubMed] [Google Scholar]
- 75.Song L, Kalyanaraman C, Fedorov AA, Fedorov EV, Glasner ME, Brown S, Imker HJ, Babbitt PC, Almo SC, Jacobson MP, Gerlt JA. Prediction and assignment of function for a divergent N-succinyl amino acid racemase. Nat Chem Biol. 2007;3(8):486–491. doi: 10.1038/nchembio.2007.11. [DOI] [PubMed] [Google Scholar]
- 76.Frembgen-Kesner T, Elcock AH. Computational sampling of a cryptic drug binding site in a protein receptor: Explicit solvent molecular dynamics and inhibitor docking to p38 MAP kinase. Journal of Molecular Biology. 2006;359(1):202–214. doi: 10.1016/j.jmb.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 77.Schein CH, Zhou B, Oezguen N, Mathura VS, Braun W. Molego-based definition of the architecture and specificity of metal-binding sites. Proteins-Structure Function and Bioinformatics. 2005;58(1):200–210. doi: 10.1002/prot.20253. [DOI] [PubMed] [Google Scholar]
- 78.Zavodszky MI, Kuhn LA. Side-chain flexibility in protein-ligand binding: the minimal rotation hypothesis. Protein Sci. 2005;14(4):1104–1114. doi: 10.1110/ps.041153605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Levitt M. Molecular dynamics of native protein. I. Computer simulation of trajectories. J. Mol. Biol. 1983;168(3):595–617. doi: 10.1016/s0022-2836(83)80304-0. [DOI] [PubMed] [Google Scholar]
- 80.Levitt M, Meirovitch H. Integrating the equations of motion. J. Mol. Biol. 1983;168(3):617–620. doi: 10.1016/s0022-2836(83)80305-2. [DOI] [PubMed] [Google Scholar]
- 81.Levitt M, Perutz MF. Aromatic rings act as hydrogen bond acceptors. J. Mol. Biol. 1988;201(4):751–754. doi: 10.1016/0022-2836(88)90471-8. [DOI] [PubMed] [Google Scholar]
- 82.Kellogg GE, Chen DL. The Importance of Being Exhaustive. Optimization of Bridging Structural Water Molecules and Water Networks in Models of Biological Systems. Chem. & Biodivers. 2004;1(1):98–105. doi: 10.1002/cbdv.200490016. [DOI] [PubMed] [Google Scholar]
- 83.Irwin JJ, Raushel FM, Shoichet BK. Virtual screening against metalloenzymes for inhibitors and substrates. Biochemistry. 2005;44(37):12316–12328. doi: 10.1021/bi050801k. [DOI] [PubMed] [Google Scholar]
- 84.Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267(3):727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 85.Morris GM, Goodsell DS, Huey R, Olson AJ. Distributed automated docking of flexible ligands to proteins: parallel applications of AutoDock 2.4. J. Comput. Aided Mol. Des. 1996;10(4):293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 86.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19(14):1639–1662. [Google Scholar]
- 87.Lin ZP, Zhu Y-L, Johnson DR, Rice KP, Nottoli T, Hains BC, McGrath J, Waxman SG, Sartorelli AC. Disruption of cAMP and Prostaglandin E2 Transport by Multidrug Resistance Protein 4 Deficiency Alters cAMP-Mediated Signaling and Nociceptive Response. Mol Pharmacol. 2008;73(1):243–251. doi: 10.1124/mol.107.039594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liang W, Pascual-Montano A, Silva AJ, Benitez JA. The cyclic AMP receptor protein modulates quorum sensing, motility and multiple genes that affect intestinal colonization in Vibrio cholerae. Microbiology. 2007;153:2964–2975. doi: 10.1099/mic.0.2007/006668-0. [DOI] [PubMed] [Google Scholar]
- 89.Wang L, Hashimoto Y, Tsao CY, Valdes JJ, Bentley WE. Cyclic AMP (cAMP) and cAMP receptor protein influence both synthesis and uptake of extracellular autoinducer 2 in Escherichia coli. J Bacteriol. 2005;87(6):2066–2076. doi: 10.1128/JB.187.6.2066-2076.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Higgins DA, Pomianek ME, Kraml CM, Taylor RK, Semmelhack MF, Bassler BL. The major Vibrio cholerae autoinducer and its role in virulence factor production. Nature. 2007;450(7171):805–807. doi: 10.1038/nature06284. [DOI] [PubMed] [Google Scholar]
- 91.Peterson JW, Comer JE, Noffsinger DM, Wenglikowski A, Walberg KG, Chatuev BM, Chopra AK, Stanberry LR, Kang AS, Scholz WW, J S. Human monoclonal anti-protective antigen antibody completely protects rabbits and is synergistic with ciprofloxacin in protecting mice and guinea pigs against inhalation anthrax. Infect. Immun. 2006;74:1016–1024. doi: 10.1128/IAI.74.2.1016-1024.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peterson JW, Comer JE, Baze WB, Noffsinger DM, Wenglikowski A, Walberg KG, Hardcastle J, Pawlik J, Bush K, Moen S, Thomas J, Chatuev BM, Sower L, Chopra AK, Stanberry LR, Sawada R, Scholz WW, J S. Human monoclonal antibody (AVP-21D9) to protective antigen reduces the dissemination of Bacillus anthracis Ames strain from the lungs in a rabbit model. Infect. Immun. 2007;75:3414–3424. doi: 10.1128/IAI.00352-07. [DOI] [PMC free article] [PubMed] [Google Scholar]