SUMMARY
H2B ubiquitylation has been implicated in active transcription but is not well understood in mammalian cells. Beyond earlier identification of hBRE1 as the E3 ligase for H2B ubiquitylation in human cells, we now show (i) that hRAD6 serves as the cognate E2 conjugating enzyme, (ii) that hRAD6, through direct interaction with hPAF-bound hBRE1, is recruited to transcribed genes and ubiquitylates chromatinized H2B at lysine 120, (iii) that hPAF-mediated transcription is required for efficient H2B ubiquitylation as a result of hPAF-dependent recruitment of hBRE1-hRAD6 to the Pol II transcription machinery, (iv) that H2B ubiquitylation per se does not affect the level of hPAF-, SII- and p300-dependent transcription and likely functions downstream and (v) that H2B ubiquitylation directly stimulates hSET1-dependent H3K4 di- and tri-methylation. These studies establish the natural H2B ubiquitylation factors in human cells and also detail the mechanistic basis for H2B ubiquitylation and function during transcription.
INTRODUCTION
Chromosomal histones are subject to a variety of well-studied covalent modifications -- including acetylation, methylation, and phosphorylation -- that have been implicated in transcriptional regulation. H2B monoubiquitylation also has been implicated in transcription by recent genetic and cell-based assays, although the precise mechanisms involved in H2B ubiquitylation and the role(s) of ubiquitylated H2B are not fully understood (Weake and Workman, 2008). Genetic analyses of the highly conserved RAD6 and BRE1 homologues have suggested that they may function universally as the relevant ubiquitin conjugating (E2) and ubiquitin ligase (E3) enzymes, respectively. However, there are recent reports that UbcH6 serves as the E2 for H2B ubiquitylation in human cells (Zhu et al., 2005; Pavri et al., 2006).
In contrast to other histone modifications that are concentrated in the unstructured N-terminal tails, H2B ubiquitylation occurs at a residue (lysine 120) that is embedded within the C-terminal α-helix. This may account for the generally low level of H2B ubiquitylation in cells and suggests an involvement of additional mechanisms for making the site accessible to H2B ubiquitylation factors and for target gene specificity. Indeed, early studies in yeast implicated several transcription factors in H2B ubiquitylation and indicated a tight linkage between H2B ubiquitylation and transcription. Thus, RAD6 was shown to be recruited to the promoter in an activator- and BRE1-dependent manner and to co-localize with RNA polymerase II (Pol II) dependent upon BRE1 and the PAF complex (Wood et al., 2003; Xiao et al., 2005), although direct interactions of RAD6 or BRE1 with Pol II or the PAF complex were not established. Importantly, requirements of the PAF complex and the Pol II CTD serine 5 kinase Kin28 for H2B ubiquitylation have implicated H2B ubiquitylation in transcription elongation (Ng et al., 2003a; Wood et al., 2003; Xiao et al., 2005). In further support of this view, defects in H2B ubiquitylation result in sensitivity to 6-azauridine (Xiao et al., 2005) and an altered distribution of Pol II in gene coding regions (Tanny et al., 2007).
From a mechanistic viewpoint, H2B ubiquitylation is essential for specific H3K4 and H3K79 methylation events by Set1 and Dot1, respectively (Dover et al., 2002; Shilatifard, 2006), but also has H3 methylation-independent functions (Tanny et al., 2007). In this regard, Pavri et al. (2006) reported that H2B ubiquitylation directly facilitates FACT-dependent chromatin transcription in vitro. Fleming et al. (2008) also reported a connection between H2B ubiquitylation and FACT during transcription elongation in vivo but argued for a selective role for H2B ubiquitylation in chromatin reassembly by FACT following Pol II passage through a nucleosome. These results leave open the interesting question whether H2B ubiquitylation directly stimulates transcription or, conversely, whether transcription facilitates H2B ubiquitylation for subsequent functions.
Here we have employed biochemically defined systems, along with RNAi analyses, to show that RAD6 is the cognate E2 of the BRE1 complex and solely responsible for H2B ubiquitylation at lysine 120 in human cells. We further show that ongoing PAF complex-dependent transcription is required for efficient H2B ubiquitylation and that H2B ubiquitylation per se has no discernable effect on the level of p300-, PAF complex- and SII-dependent chromatin transcription. Along with further demonstrations of specific intermolecular interactions between RAD6 and components of the BRE1 and PAF complexes, these findings establish the H2B ubiquitylation factors in human cells and provide a detailed biochemical description of the mechanism of H2B ubiquitylation and function during transcription.
RESULTS
Human BRE1A Form a Complex with Human BRE1B through Its N-terminal Region
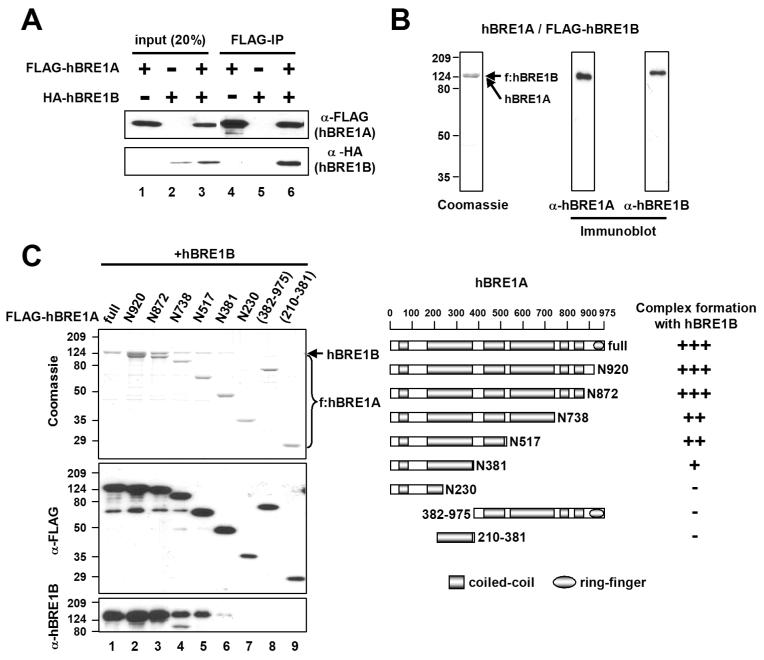
Human BRE1A/RNF20 was previously shown to serve as an E3 for H2B ubiquitylation (Kim et al., 2005) and to form a complex with the hBRE1B/RNF40 paralogue (Zhu et al., 2005). In confirmation of an intracellular interaction, ectopically expressed HA-hBRE1B was coimmunoprecipitated by anti-FLAG antibody (M2 agarose) only in the presence of FLAG-hBRE1A (Figure 1A). For more systematic studies, the heteromeric hBRE1 complex was reconstituted in insect cells by baculovirus-mediated expression of hBRE1A and FLAG-hBRE1B. The purified complex contained two polypeptides (doublet of equal densities) that were individually recognized by anti-hBRE1A and anti-hBRE1B antibodies, thus implying stoichiometric hBRE1A and hBRE1B levels in the complex (Figure 1B). A further analysis revealed that the association of hBRE1A with hBRE1B is critically dependent upon an N-terminal hBRE1A domain (residues 1-381) and further stabilized by two central regions (residues 381-517 and 738-872) that alone show no interaction with hBRE1B (Figure 1C). Overall, these observations indicate that hBRE1A, through its N-terminal region, forms a stable stoichiometric complex with hBRE1B.
Figure 1. The N-terminus of hBRE1A Interacts with hBRE1B to Form the hBRE1 Complex.
(A) Intracellular interaction of hBRE1A and hBRE1B. 293T cells were transfected with vectors expressing FLAG-hBRE1A and HA-hBRE1B proteins as indicated, derived cell extracts were incubated with M2 agarose, and bound proteins were visualized by immunoblots with indicated antibodies. (B) Coomassie blue staining and immunoblot analyses of purified hBRE1 complex with indicated antibodies. (C) hBRE1A domains responsible for the hBRE1B interaction. Insect cells were infected with baculoviruses expressing untagged-hBRE1B and the indicated FLAG-hBRE1A fragments, and M2 agarose-purified proteins were visualized by Coomassie blue staining and immunoblots with indicated antibodies (left). A schematic diagram of hBRE1A and derived fragments, with predicted coiled-coil and RING finger domains, and a summary of the hBRE1 interactions are shown (right).
RAD6 Protein Levels Correlate with H2B Ubiquitylation and H3K4-H3K79 Methylation Levels in Human Cells
Consistent with the phylogenetic sequence conservation and the ability of mouse RAD6A and RAD6B to complement yeast Rad6 (yRad6) mutants for H3K4 di-methylation (Sun and Allis, 2002), we found that both human RAD6 E2s (hRAD6A and hRAD6B), but not the hUbcH6 E2, fully complement the function of yRad6 for H2B ubiquitylation and H3K4-H3K79 di- and tri-methylation in yeast (Figure 2A). These results led us to propose a role for hRAD6, through an intrinsic E2 activity and in conjunction with hBRE1, in H2B ubiquitylation in human cells.
Figure 2. RAD6 Mediates Endogenous H2B Ubiquitylation and H3K4-H3K79 Methylation in Human Cells.
(A) Human RAD6 can fully complement the function of yeast Rad6 for H2B ubiquitylation and H3K4-H3K79 methylation. Yeast whole cell extracts from a wild-type strain (containing a chromosomal FLAG-H2B gene), its isogenic Δrad6 strain, and Δrad6 strains that harbor the indicated E2 expression plasmids (driven by the natural yeast Rad6 promoter) were subjected to immunoblotting with the indicated antibodies. Two different amounts of cell extracts were loaded. (B) Effects of siRNA-mediated knockdown of H2B ubiquitylation factors on histone modifications. 293T cells were treated with control, hBRE1A, hBRE1B, hRAD6A, hRAD6B and hUbcH6 siRNAs indicated. Total cell lysates were subjected to immunoblot analyses with indicated antibodies. A nonspecific band in the hUbcH6 immunoblot is indicated by an asterisk. (C) Total RNAs from siRNA-treated cells were subjected to RT-PCR analyses for mRNA levels of indicated genes.
In further functional analyses, an hBRE1A siRNA, but not a control siRNA, resulted in the loss of both hBRE1A (Figure 2B, first panel, lane 2) and hBRE1B (second panel, lane 2). In contrast, an hBRE1B siRNA resulted in a large reduction of hBRE1B (second panel, lane 3) with no effect on hBRE1A (first panel, lane 3). These data are consistent with the existence of an hBRE1A·hBRE1B complex (hBRE1 complex) and an associated stabilization of hBRE1B by hBRE1A. Similarly, hRAD6A and combined hRAD6A plus hRAD6B siRNAs decreased the level of hRAD6 (third panel, lanes 5 and 7), whereas an hRAD6B siRNA did not result in any obvious change in the overall level of hRAD6 (third panel, lane 6; note that the anti-hRAD6 antibody does not discriminate hRAD6A and hRAD6B due to their near-identical amino acid sequences). The latter result is not due to an inefficiency of hRAD6B siRNA since the hRAD6A and hRAD6B siRNAs decreased corresponding mRNA levels to nearly the same extent (Figure 2C). Therefore, consistent with a previous report (Koken et al., 1996), we conclude that hRAD6A comprises the majority of the endogenous hRAD6 protein pool. Finally, an hUbcH6 siRNA effected an almost complete loss of hUbcH6 protein without causing any changes in the levels of the other proteins tested (Figure 2B, fourth panel, lane 8).
Reductions of endogenous hBRE1A and hBRE1B, independently or together, also resulted in significant concomitant decreases of H2B ubiquitylation (probed both by an anti-H2B antibody and by an anti-ubH2B antibody that specifically recognizes ubiquitylated H2B at lysine 120 (Minsky et al., 2008)), H3K4 di- and tri-methylation, and H3K79 di-methylation, but little or no changes in H3K4 and H3K79 mono-methylation (Figure 2B, lanes 2-4 versus lane 1). Most importantly, hRAD6A siRNA or combined hRAD6A plus hRAD6B siRNAs resulted in significant decreases in H2B ubiquitylation and H3K4 tri-methylation and a moderate decrease in H3K79 di-methylation (lanes 5 and 7 versus lane 1). In contrast, but consistent with its negligible effect on the total hRAD6 level, an hRAD6B siRNA caused no significant changes in any of these histone modifications (lane 6 versus lane 1). Notably, an almost complete reduction of endogenous hUbcH6 by an hUbcH6 siRNA failed to effect any detectable changes in these histone modifications (lane 8). Beyond global histone modifications, we also observed a clear correlation between hRAD6 and H2B ubiquitylation levels, as well as H3K4 methylation levels, on the p21 gene (Figure S1). Taken together, these observations show strong correlations between the level of hRAD6, but not hUbcH6, and the levels of H2B ubiquitylation and H3K4 and H3K79 methylation.
The Human BRE1 Complex Specifically Interacts with Human RAD6
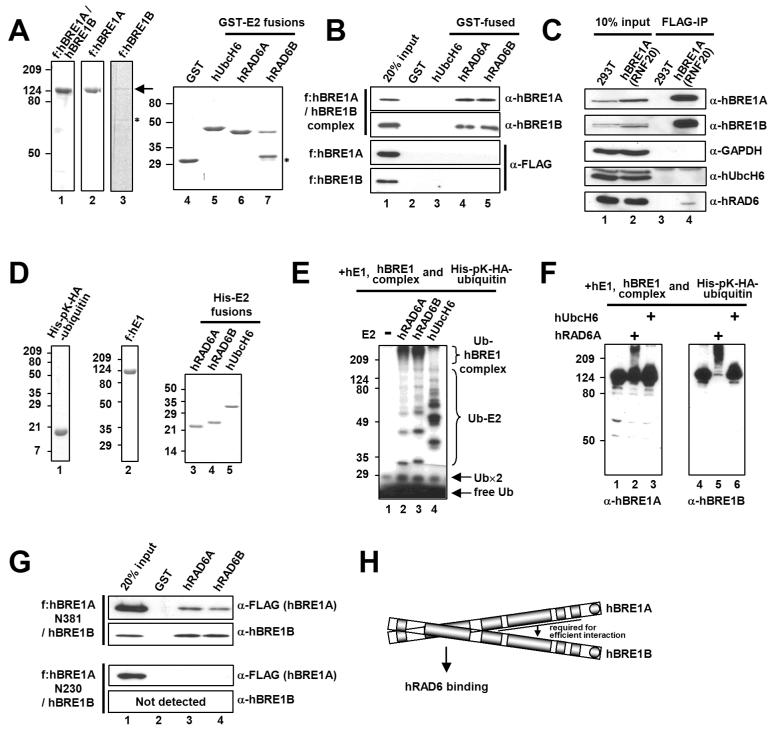
Each E3 specifically and directly binds to its cognate E2 (Hershko and Ciechanover 1998). To identify an E2 that directly binds to the hBRE1 complex, we examined interactions of the purified hBRE1 complex with several human E2s (Figure 3A; Figure S2A). Among the nine tested E2s, including hUbcH6, only hRAD6A and hRAD6B directly bound to the hBRE1 complex (Figure 3B; Figure S2B). These data contrast with those of an earlier report that hUbcH6 specifically interacts with the hBRE1 complex (Zhu et al., 2005). However, the specific physical interaction described here was also confirmed by other approaches (Figure S3; Figure S4). In addition, and importantly, hRAD6 was found to interact with the hBRE1 complex but not with individual hBRE1A or hBRE1B polypeptides (Figure 3B; Figure S3B; Kim et al., 2005), indicating that formation of the heteromeric hBRE1 complex is critical for direct interaction with hRAD6. In a further demonstration of intracellular interactions (Figure 3C), endogenous hRAD6 protein, but not hUbcH6, was coimmunoprecipitated (by anti-FLAG antibody) with hBRE1A and hBRE1B from extracts derived from human cells that express FLAG-hBRE1A/RNF20. Similar interactions were observed following baculovirus-mediated expression of hBRE1, hRAD6 and hUbcH6 proteins in insect cells (Figure S4).
Figure 3. The hBRE1 Complex Directly and Specifically Interacts with hRAD6A and hRAD6B.
(A) Analyses of purified hBRE1 complex, FLAG-hBRE1A, FLAG-hBRE1B, GST and GST-E2 proteins by Coomassie blue staining. Asterisk, degradation product. (B) Selective binding of the hBRE1 complex to hRAD6A and hRAD6B. GST pull down assays employed the purified proteins shown in (A) and bound proteins were scored by immunoblotting with indicated antibodies. (C) Intracellular binding of the hBRE1 complex to hRAD6. Total cell lysates from a FLAG-hBRE1A (RNF20) 293T cell line were incubated with M2 agarose, and bound proteins were visualized by immunoblotting with indicated antibodies. (D) Analysis of purified His-pK-HA-ubiquitin, FLAG-hE1, and His-E2 proteins by Coomassie blue staining. (E and F) Ubiquitylation of the hBRE1 complex by hRAD6. The purified hBRE1 complex was analyzed in an E3 ubiquitylation assay with indicated E2 enzymes in the presence of either 32P-lablled (E) or unlabelled (F) ubiquitin, respectively, and ubiquitylation of the hBRE1 complex was monitored by autoradiography (E) or immunoblot (F), respectively. Ub×2 indicates a ubiquitin dimer. (G) Binding of the purified hBRE1 complex containing either FLAG-hBRE1A N381 or N230 fragments (Figure 1C) to hRAD6A and hRAD6B. (H) Schematic of direct binding of the hBRE1 complex to hRAD6.
Direct interactions between RING finger-containing E3 and cognate E2 proteins result in both substrate and E3 ubiquitylation (Lorick et al., 1999). In this regard, and further confirming the functional relevance of the physical interaction between hRAD6 and the hBRE1 complex, incubation of the hBRE1 complex with purified ubiquitin, E1 and E2 proteins (Figure 3D) revealed a hRAD6-dependent poly-ubiquitylation of the hBRE1 complex that was visualized by autoradiography (Figure 3E, lanes 2 and 3) and immunoblot (Figure 3F, lanes 2 and 5) and shown to be dependent upon the catalytic activity of hRAD6 (Figure S5). Although purified hUbcH6 exhibited a strong ubiquitin conjugating activity, it failed to mediate any detectable poly-ubiquitylation of the hBRE1 complex (Figure 3E, lane 4; Figure 3F, lanes 3 and 6; Figure S5B).
Several reports showed that the signature RING finger domains of E3s serve as platforms for the E2 binding that is essential for protein ubiquitylation (Lorick et al., 1999). Surprisingly, we found that the C-terminal RING fingers in hBRE1A and hBRE1B are both dispensable for ubiquitylation of the hBRE1 complex (Figure S6), suggesting that hRAD6 interacts with the hBRE1 complex through a region(s) other than the RING finger domain. As expected from the results of Figure 3B, an hBRE1 complex containing intact hBRE1B and the minimal hBRE1A fragment (N381) capable of complex formation with hBRE1B (Figure 1C) was shown to interact with hRAD6 whereas hBRE1B and an hBRE1A fragment (N230) that is unable to form a complex with hBRE1B could not (Figure 3G). These results demonstrate unequivocally that hRAD6, but not hUbcH6, interacts specifically and functionally with the hBRE1 complex, both in vitro and in vivo, through a region other than the RING finger domain in the hBRE1 complex (Figure 3H; see Discussion).
Human RAD6 Ubiquitylates Chromatin-associated H2B at Lysine 120 Only in the Presence of the Human BRE1 Complex
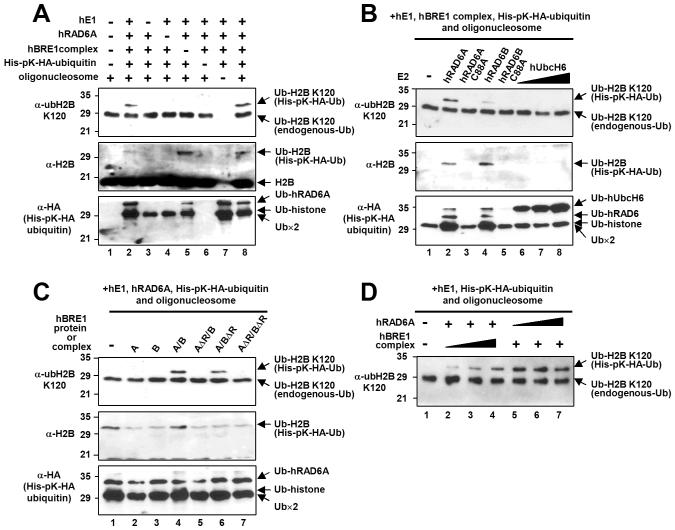
To clearly demonstrate a direct hRAD6 E2 function in H2B ubiquitylation, we employed an in vitro chromatin ubiquitylation assay. Complete reactions containing purified E1, hRAD6A, the hBRE1 complex, ubiquitin (His- and HA-tagged) and oligonucleosomes derived from HeLa cells generated a lysine 120-ubiquitylated H2B (ubH2B) (Figure 4A, top panel, lanes 2 and 8), whereas reactions with omission of any of these components did not (top panel, lanes 3-7). Although a significant level of H2B ubiquitylation was detected in the absence of the hBRE1 complex, this clearly represents H2B ubiquitylation at a site(s) other than lysine 120 (compare top panel, lane 5 versus middle panel, lane 5). This is consistent with the ability of RAD6 to ubiquitylate free core histones (Jentsch et al., 1987; Figure S7), as well as both histone octamer and nucleosomal substrates (Figure S7), in the absence of an E3. This appears to reflect a non-specific RAD6 ubiquitin conjugating activity that is manifested in a purified assay system lacking constraints that normally control accessibility of RAD6 to histones. These results also strengthen the validity of the anti-ubH2B antibody for monitoring precise in vitro H2B ubiquitylation events.
Figure 4. RAD6 Is the E2 Ubiquitin Conjugating Enzyme for H2B Lysine 120 Ubiquitylation in Human Cells.
(A-D) In vitro chromatin ubiquitylation assays. Reactions containing 5 μg oligonucleosome, 100 ng hE1, 200 ng E2, 600 ng hBRE1 complex and 2.8 μg His-pK-HA-tagged ubiquitin, unless otherwise indicated, were subjected to immunoblot with antibodies indicated on the left of each panel. (A) Collective requirement of factors for H2B ubiquitylation. Note that endogenous ubiquitylated H2B (in the oligonucleosome substrate) is not detectable by anti-H2B antibody (middle panel). (B) Comparison of hRAD6 and hUbcH6 E2 activities for H2B ubiquitylation. Reactions contained 200 ng wild-type or mutant hRAD6 (lanes 2-5) and 100 ng (lane 6), 200 ng (lane 7) or 600 ng (lane 8) hUbcH6. (C) hBRE1 complex RING finger requirement for H2B ubiquitylation. FLAG-hBRE1A, FLAG-hBRE1B and hBRE1 complexes (isolated via FLAG-hBRE1A) with intact or deleted (either or both) RING fingers are indicated. (D) The hBRE1 complex is limiting for H2B ubiquitylation. Reactions contained 50 ng (lane 2), 150 ng (lane 3) or 450 ng (lane 4) hBRE1 complex and 50 ng (lane 5), 100 ng (lane 6) or 200 ng (lane 7) hRAD6A.
In relation to specificity and possible redundancy of the ubiquitylation enzymes, hRAD6A (more efficiently) and hRAD6B (less efficiently) were both found to ubiquitylate H2B at lysine 120 in a catalytic activity-dependent manner (Figure 4B, lanes 1-5). Notably, hUbcH6 failed to generate any detectable ubH2B (lanes 6-8), even at a three-fold higher dose than hRAD6 (lane 8). Unlike individual RAD6 proteins, individual hBRE1A and hBRE1B polypeptides failed to mediate ubiquitylation of H2B (Figure 4C, lanes 1-4), thus demonstrating that formation of the heteromeric BRE1 complex and the resulting interaction with RAD6 are critical for the E3 function of the hBRE1s. In a further analysis of purified hBRE1 complexes with combinations of RING finger deletions (Figure S6), H2B ubiquitylation was abrogated by deletion either of both RING fingers (Figure 4C, lane 7 versus lane 4) or of the hBRE1A RING finger (lane 5 versus lane 4), but was relatively unaffected by deletion of the hBRE1B RING finger (lane 6 versus lane 4). Overall these results indicate redundant functions for RAD6A and RAD6B, but distinct functions for the essential hBRE1A and hBRE1B proteins (and their individual RING fingers), in H2B ubiquitylation (see Discussion).
In a further analysis of the efficiency of the in vitro H2B ubiquitylation reactions, we observed a dose-dependent increase in H2B ubiquitylation in response to varying amounts of the hBRE1 complex (Figure 4D, lanes 2-4), but not of hRAD6A (lanes 5-7). These data suggest that the hBRE1 complex is more limiting for H2B ubiquitylation, at least in vitro, and may explain enhanced H2B ubiquitylation by ectopic hBRE1A, but not by ectopic hRAD6, in a cell-based assay (Kim et al., 2005).
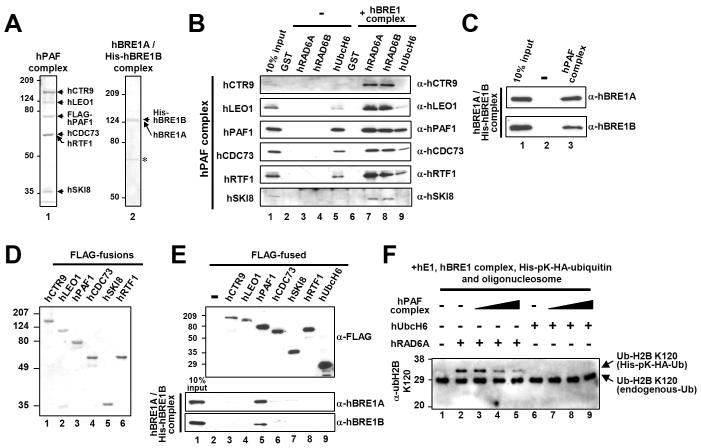
RAD6 Association with the PAF Complex Is Dependent Upon the BRE1 Complex, Which Interacts Directly with the PAF Complex Through the PAF1 Subunit
The PAF elongation factor complex is required for both H2B ubiquitylation and H3K4-K79 methylation in yeast (Krogan et al., 2003; Ng et al., 2003a; Wood et al., 2003; Xiao et al., 2005). To test how the human PAF (hPAF) complex links H2B ubiquitylation and transcription processes, we first examined the interaction between highly purified E2 proteins (Figure 3A) and hBRE1 and hPAF complexes (Figure 5A). Interestingly, hRAD6A and hRAD6B were shown to associate with the purified hPAF complex in the presence, but not in the absence, of the hBRE1 complex (Figure 5B, lanes 7 and 8 versus lanes 3 and 4). Given the earlier demonstration of direct hRAD6-hBRE1 complex interactions (above), these results suggest that the hBRE1 complex mediates the association of hRAD6 with the hPAF complex. In confirmation, the purified hBRE1 complex was found to bind directly to the purified hPAF complex (Figure 5C). A further analysis with individual subunits of the hPAF complex (Figure 5D) showed a strong selective binding of the purified hBRE1 complex to the hPAF1 subunit (Figure 5E), thus implicating hPAF1 as the major subunit responsible for interaction of the hPAF complex with the hBRE1 complex. Although hUbcH6 was shown to interact weakly (potentially non-specifically; below), with the hPAF complex, inclusion of the hBRE1 complex in the reaction did not alter binding efficiency (Figure 5B, lane 5 versus lane 9).
Figure 5. The hPAF Complex Directly Interacts with the hBRE1 Complex.
(A and D) Analyses of purified proteins by Coomassie blue staining. Reconstituted hPAF (isolated via FLAG-hPAF1) and hBRE1 (isolated via His-hBRE1B) complexes (A) and individual FLAG-tagged subunits of the hPAF complex (D) were expressed from baculovirus vectors. Asterisk, non-specific bands. (B, C and E) Binding of the purified hPAF complex or isolated subunits to H2B ubiquitylation factors. The purified hPAF complex was tested for binding to GST-fused E2 proteins in the presence and in the absence of the hBRE1 complex (B). The purified hBRE1 complex was tested for binding to the M2 agarose-immobilized hPAF complex (C) or to individual hPAF complex subunits (E). Bound proteins were scored by immunoblot with indicated antibodies. (F) Effect of the hPAF complex on H2B ubiquitylation. The chromatin ubiquitylation assay was performed as in Figure 4. Reactions contained 150 ng (lanes 3 and 7), 300 ng (lanes 4 and 8) or 600 ng (lanes 5 and 9) hPAF complex and hRAD6A or hUbcH6 additions as indicated.
In view of the direct association of the hRAD6-hBRE1 complex with the hPAF complex, we tested whether the hPAF complex directly stimulates H2B ubiquitylation. Somewhat surprisingly, the hPAF complex was found to decrease hRAD6-hBRE1 complex-mediated H2B ubiquitylation within an oligonucleosome substrate (Figure 5F, lanes 3-5 versus lane 2), presumably because a strong competitive binding of the hPAF complex in this context prevents the hBRE1 complex from binding effectively to histones. This suggests that the PAF complex plays a role in H2B ubiquitylation other than stimulating catalysis by RAD6-BRE1. Consistent with our inability to see any role for hUbcH6 in H2B ubiquitylation, hUbcH6 still did not effect any detectable H2B ubiquitylation in our assays even in the presence of the hPAF complex (Figure 5F, lanes 6-9).
The PAF Complex Plays a Role in H2B Ubiquitylation Within Chromatin Templates Through an Effect on Transcription
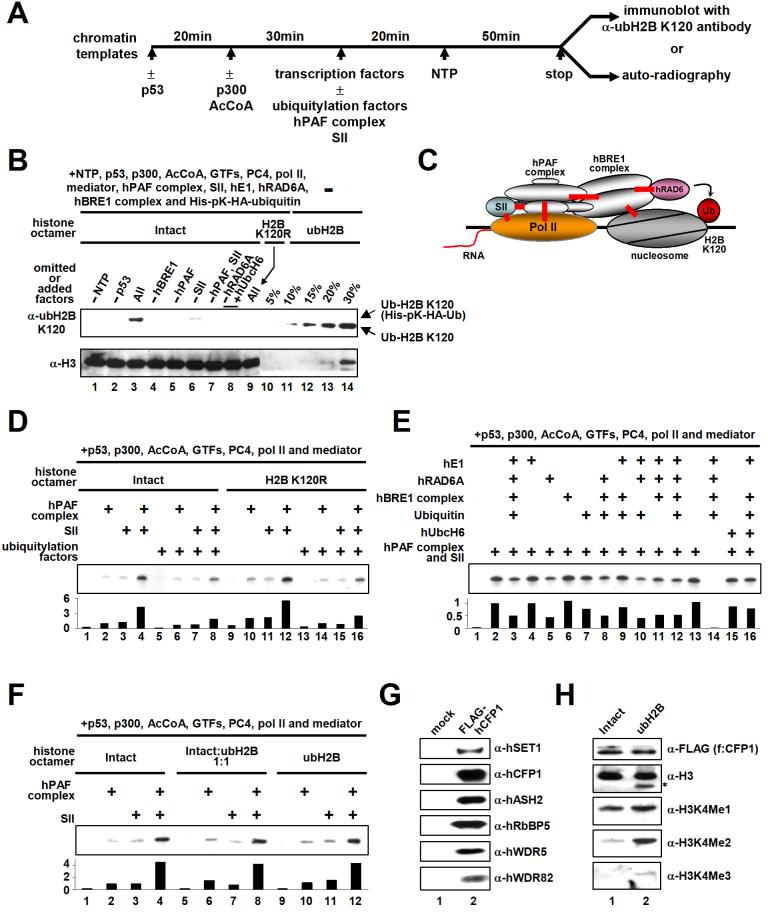
In order to test the role of the PAF complex in H2B ubiquitylation during transcription, we employed an in vitro assay (schematized in Figure 6A) containing purified factors and a chromatinized pML array template that contains p53 binding sites (Figure S8). An assay under transcription conditions with all components present generated a very significant level of H2B ubiquitylation at lysine 120 (Figure 6B, compare lanes 3 and 9). This corresponds to ubiquitylation of about 15 % of the total H2B used for transcription, as judged by comparison with histone octamer standards (compare lane 3 versus lanes 10-14) containing fully ubiquitylated H2B (McGinty et al., 2008). We also confirmed a requirement for the hBRE1 complex (Figure 6B; lane 4) and an inability of hUbcH6 to substitute for the E2 function of hRAD6 (lane 8). Importantly, this efficient H2B ubiquitylation is coupled to activator-dependent transcription because omission of either p53 or nucleoside triphosphates (but with ATP present) reduced ubH2B to an undetectable level (lanes 1 and 2). Similarly, omission of the hPAF complex resulted in an undetectable level of ubH2B (lanes 5 and 7) whereas SII omission significantly decreased the efficiency of H2B ubiquitylation (lane 6). As shown in Figure 6D, and as will be detailed elsewhere (J.K., M.G. and R.G.R., unpublished observations), SII and the hPAF complex act independently to effect equivalent low levels of transcription, and synergistically to effect high levels of transcription, in this assay (lanes 1-4). Hence, the efficiency of H2B ubiquitylation generally correlates with the transcription level. These results thus suggest that the PAF complex facilitates H2B ubiquitylation through an effect on transcription and thereby directly links transcription to H2B ubiquitylation factors (Figure 6C). However, the fact that omission of the hPAF complex completely eliminates H2B ubiquitylation, while omission of SII does not, indicates that H2B ubiquitylation is also dependent upon hPAF complex functions other than those directly connected to transcription.
Figure 6. Transcription-dependent H2B Ubiquitylation.
(A) Schematic representation of the standard in vitro transcription assay. Transcription factors included TFIIA, TFIIB, TFIIE, TFIID, TFIIF, TFIIH, PC4, Mediator and Pol II. Ubiquitylation factors included E1, E2 (hRAD6A or hUbcH6), the hBRE1 complex and ubiquitin. Chromatin-based assays also contained the components (ACF1, ISWI, NAP1 and TOPO1) employed for chromatin assembly. Reactions contained 10 ng p53, 20 ng p300, 240 ng hPAF complex, 10 ng SII, 25 ng E1, 25 ng E2, 75 ng hBRE1 complex, and 350 ng ubiquitin where indicated. (B) Transcription-coupled H2B ubiquitylation assays with deletions and additions as indicated (lanes 1-9). In each case, nine standard transcription reactions were combined, concentrated and subjected to immunoblots with indicated antibodies. Note that all four unmodified nucleoside triphosphates were used in this assay. Histone octamers containing fully ubiquitylated H2B (indicated as % of histone octamers present in the transcription assay) were loaded in lanes 10-14. (C) Schematic model of transcription coupled-H2B ubiquitylation. Verified direct interactions are depicted by red lines. (D, E and F) Effects of H2B ubiquitylation factors (D and E) or fully ubiquitylated (semi-synthetic) H2B (F) on chromatin transcription. Relative transcription levels were quantitated by phosphoimager and normalized to that of SII alone (D and F) or to that of the hPAF complex and SII (E). (G and H) H2B ubiquitylation directly stimulates hSET1 complex-mediated H3K4 di- and tri-methylation. (G) Analyses of purified hSET1 complex (isolated via FLAG-hCFP1 by M2 agarose from a stable FLAG-hCFP1 cell line) by immunoblot. (H) Chromatin templates were assembled with unmodified H2B (lane 1) or ubH2B (lane 2) octamers and subjected to in vitro methylation assay with purified hSET1 complex. H3 methylation status was probed by indicated antibodies. The fast migrating band (asterisk) detected by an anti-H3 antibody in lane 2 is thought be N-terminus-deleted H3 because this band is not detected by H3K4 modification-specific antibodies.
Effect of H2B Ubiquitylation on Chromatin Transcription and H3K4 Methylation
We next tested the effect of H2B ubiquitylation on hPAF- and SII-dependent transcription of chromatin by p53. Surprisingly, we found that addition of ubiquitylation factors (ubiquitin, E1, hRAD6A and the hBRE1 complex) actually effected a moderate decrease in transcription (Figure 6D, lanes 2, 3 and 4 versus lanes 6, 7 and 8, respectively). However, this inhibition is not due to H2B ubiquitylation at lysine 120 since these inhibitory effects were also observed in transcription of chromatin reconstituted with the H2B K120R mutant (lanes 10, 11 and 12 versus lanes 14, 15 and 16, respectively). In a test for contributions of individual factors to the inhibitory effect (Figure 6E), we found transcription inhibition mainly in reactions containing hRAD6A (lanes 2 and 13 versus lanes 3, 5, 8, 10, 11 and 12). We suspect that this modest non-specific hRAD6-dependent transcription inhibition is due both to a lack of normal constraints to interfering interactions (such as non-specific binding of hRAD6 to histones; Figure S7) and to a lack of positively acting effectors that recognize ubH2B in the purified assay system. In addition, hUbcH6 alone (lane 15) or with other ubiquitylation factors (lane 16) did not increase, but instead slightly decreased, transcription.
To test more directly for an effect of H2B ubiquitylation on chromatin transcription in the absence of the chromatin ubiquitylation machinery, we assembled chromatin with histone octamers containing a semi-synthetic H2B fully ubiquitylated at lysine 120 (McGinty et al., 2008; Figure S9). Interestingly, chromatin templates assembled with intact H2B versus ubH2B octamers showed similar micrococcal nuclease (MNase) digestion patterns (Figure S9), indicating that the bulky ubiquitin adduct does not alter overall recombinant chromatin structure. More interestingly, in a test for effects of ubH2B on transcription of chromatin (Figure 6F), the level of basal (coactivator-independent) transcription and the levels of hPAF complex- and SII-dependent transcription from completely H2B-ubiquitylated chromatin were indistinguishable from those observed with unmodified chromatin (lane 2 versus lanes 6 and 10; lane 3 versus lanes 7 and 11; lane 4 versus lanes 8 and 12). Thus, ubH2B itself does not result in any structural changes that noticeably affect either the repressed state of the reconstituted chromatin or its activation by the factors in our defined assay system (see Discussion).
Our observation that H2B ubiquitylation is both transcription-dependent and unable alone to directly affect transcription through chromatin suggests that ubH2B must affect a downstream factor(s) that in turn influences transcription. To test a potential role for ubH2B in directly facilitating H3K4 methylation by SET1, the hSET1 complex was purified from HeLa cells that stably express a FLAG-tagged subunit (hCFP1) unique to the hSET1 methyltransferase complex. The composition of the isolated complex was similar to that reported by Lee and Skalnik (2005), using the same approach, and the immunoblot in Figure 6G confirms the presence of established subunits (including hSET1) of this complex. The hSET1 complex exhibits efficient H3K4 mono-methylation activity on both unmodified and H2B ubiquitylated chromatins (Figure 6H). Remarkably, however, the presence of ubH2B on chromatin significantly stimulates hSET1 complex-dependent H3K4 di- and tri-methylation (lane 2 versus lane 1). Taken together, these data strongly suggest that transcription-coupled H2B ubiquitylation directly stimulates H3K4 di- and tri-methylation by the hSET1 complex and thus explain why H2B ubiquitylation is concentrated on actively-transcribing genes (Minsky et al., 2008) and why H2B ubiquitylation-dependent H3K4 tri-methylation marks recent transcription events (Ng et al., 2003b).
Distribution of H2B Ubiquitylation Factors and Ubiquitylated H2B on Endogenous Genes during Transcription Activation
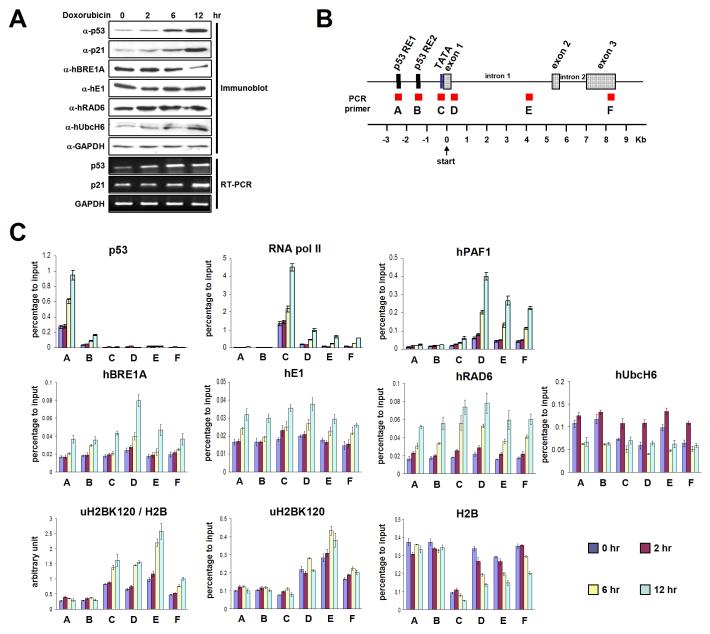
Given our previous demonstration of physical and functional interactions of p53 and hBRE1 (Kim et al., 2005), the p53-dependent doxorubicin-inducible p21 gene was chosen for analysis by chromatin immunoprecipitation (ChIP) of the region-specific accumulation of ubH2B and related factors during gene activation. Following verification of the expected doxorubicin-dependent increases in levels of p53 and p21 proteins and transcripts (Figure 7A), six regions of the p21 locus were probed with corresponding amplicons (summarized in Figure 7B). Consistent with previous studies by others, we observed (i) a basal level of p53 at RE1 (A) and RE2 (B) regions, followed by continuous doxorubicin-induced increases in p53 occupancy only at these sites over a 12 hour period, (ii) a basal level of paused Pol II at the promoter region (C) followed by time-dependent increases both at the promoter (C) and (at reduced levels) along the coding regions (D, E and F) in response to doxorubicin, the latter representing movement of Pol II toward 3' region during the transcription process.
Figure 7. Distribution of the H2B Ubiquitylation Factors and Ubiquitylated H2B on the p21 Locus during p53-Dependent Transcription.
(A) Doxorubicin induction of p53 and the p21 target gene. HCT116 cells were treated with 0.5 μM doxorubicin for the indicated times, and proteins and mRNA levels were analyzed by immunoblot and RT-PCR, respectively. (B) Schematic representation of the p21 locus indicating six amplicons used for real-time PCR. (C) ChIP analyses on the p21 locus. Cells were treated as in (A) and ChIP analyses were performed with indicated antibodies. Error bars denote standard deviations from three independent PCR reactions from a single ChIP that is representative of several that were performed. [Note: these data are part of a comprehensive ChIP analysis and limited parts (e.g., p53 and p21 expression data and p53 and Pol II ChIP data) are expected to be published, for cross reference, with more extensive data on SII and individual hPAF complex subunit analyses.]
As will also be reported elsewhere in conjunction with an analysis of SII and all hPAF complex subunits (J.K., M.G. and R.G.R., unpublished observations), hPAF1 occupancy at the promoter region (C) was not significant at the uninduced (0 hr) stage, but was increased at the promoter (C) and, especially, the adjacent (D) and downstream (E and F) transcribed regions in response to doxorubicin. This suggests that Pol II-mediated recruitment (or subsequent stabilization) of the hPAF complex at the promoter and downstream regions is tightly regulated by on-going transcription. In addition, the higher levels of hPAF1 accumulation at transcribed regions (D, E and F), relative to the promoter region (C), is consistent with the proposed function of the hPAF complex as a transcription elongation factor. The hBRE1A pattern was somewhat similar to that observed for hPAF1, consistent with hPAF complex-dependent recruitment of the hBRE1 complex to the chromatin, but with the exception of an obvious accumulation at RE1, RE2 and promoter regions. The latter result may reflect the hBRE1 potential for direct interactions with p53 (Kim et al., 2005; Figure S10). Interestingly, hE1 showed an increased accumulation, beyond a high basal level, at all tested regions upon gene activation. This might reflect the universal requirement of E1 for all other ubiquitylation reactions (beyond H2B ubiquitylation), as shown for a proteasome complex that is recruited to the genes after activation (Gonzalez et al., 2002). hRAD6 recruitment showed a broad increase over a 12 hour period, with highest level at the 5' transcribed region (D) and a pattern somewhat similar to that observed for hBRE1A. This may reflect hBRE1- and hPAF complex-dependent recruitment of hRAD6. However, RAD6 accumulation may also reflect other pathways since RAD6 serves as the E2 for ubiquitylation of proteins other than H2B and, related, since DNA damage induces localization of RAD6 and its cognate E3 RAD18 to chromatin (Lyakhovich and Shekhar, 2004). Importantly, although hUbcH6 showed a broad distribution across the p21 gene in the uninduced state, as well as a weak interaction with the hPAF complex (Figure 5B), the level of associated hUbcH6 actually decreased upon doxorubicin-induced transcription of the p21 gene.
Analyses of p21-associated ubH2B (normalized to total H2B levels) revealed significant levels of ubH2B at both promoter (C) and transcribed (D, E and F) regions, which were increased following p21 induction. Importantly, the level of ubH2B was highest at the middle coding region (E) and considerably lower at the 3' coding region, consistent with a recent report (Minsky et al., 2008). The lower level of ubH2B at the 5' transcribed region (D) relative to the middle coding region (E), despite higher levels of H2B ubiquitylation factors at the D region than at the E region, may be related to the preferential recruitment and action of an H2B deubiquitylation factor at the early transcribed region (Zhang et al., 2008). It also is noteworthy that whereas significant amounts of H2B ubiquitylation factors are present in untranscribed (A and B) regions, ubH2B is concentrated in the transcribed regions. This supports the notion that efficient H2B ubiquitylation (or stabilization) is coupled to transcription events.
DISCUSSION
The increasingly appreciated roles of H2B ubiquitylation in diverse transcription events must ultimately be understood at a mechanistic level, necessitating both biochemical and genetic analyses with properly defined (physiological) components of the ubiquitylation machinery. Here, our elucidation of the factors essential for robust H2B K120 ubiquitylation in human cells and their deployment in biochemically defined chromatin-templated assays have revealed significant mechanistic aspects of H2B ubiquitylation during transcription. Our results show (i) that RAD6 is the cognate E2 for the hBRE1 E3 complex and solely responsible for H2B ubiquitylation in human cells, (ii) that the hPAF complex is required for efficient H2B ubiquitylation and acts by recruiting the H2B ubiquitylation machinery to the transcription machinery, through direct interactions with the hBRE1 complex and with RNA polymerase II, and not by directly stimulating RAD6-BRE1 catalytic activity, (iii) and importantly, that on-going transcription is required for efficient H2B ubiquitylation, (iv) that, despite its bulk, ubH2B itself does not affect hPAF complex- and SII-mediated chromatin transcription directly, consistent with a downstream function and, related, (v) that ubH2B can directly enhance H3K4 di- and tri- methylation by the hSET1 complex. By identifying the essential factors for robust H2B ubiquitylation in human cells and by elaborating the mechanistic basis for H2B ubiquitylation, these results provide new insights into the complex mechanism of H2B ubiquitylation during transcription.
The BRE1 Complex as a Functional E3 Ubiquitin Ligase for H2B Ubiquitylation
In an extension of earlier work (Kim et al., 2005; Zhu et al., 2005), functional studies have shown that the human BRE1A and BRE1B paralogues are both required for H2B ubiquitylation in vivo and in vitro. Additional studies indicated that the RING finger-type hBRE1A and hBRE1B E3s interact through an N-terminal region of hBRE1A and that complex formation is important for stability of individual components (notably hBRE1B). Given recent demonstrations of a heteromeric complex between the two essential BRE1 paralogues in fission yeast (Tanny et al., 2007) and a multimeric complex of the single Bre1 in budding yeast (Figure S11), it is likely that the formation of a multimeric BRE1 complex is conserved from yeast to human and is essential for its E3 function.
Although hBRE1A and hBRE1B are both required for H2B ubiquitylation, the hBRE1A RING finger is critical whereas the hBRE1B RING finger is dispensable. Thus, despite similar structures, the two hBRE1 homologues play distinct roles in H2B ubiquitylation. In addition, the fact that hRAD6 binds to the hBRE1 complex but not to hBRE1A or hBRE1B alone, may explain in part why hBRE1B, but not its RING finger, is required for H2B ubiquitylation.
Identification of RAD6 as the E2 Ubiquitin Conjugating Enzyme for H2B Ubiquitylation in Human Cells
Consistent with expectations from the phylogenetic conservation of the RAD6 protein and from genetic studies in several organisms (reviewed in Weake and Workman, 2008), but in contrast to reports of a role for the human UbcH6 E2 (Zhu et al., 2005; Pavri et al., 2006), our current studies provide unequivocal evidence that RAD6 functions as the physiological E2 for H2B ubiquitylation in human cells. First, hRAD6 protein levels correlate with both global and gene-specific H2B ubiquitylation in RNAi analyses. Second, hRAD6 directly interacts with the hBRE1 complex via a specific domain. Third, hRAD6 exhibits an E2 activity for robust H2B ubiquitylation at lysine 120 both in pure assays with natural oligonucleosome substrates and in transcription-coupled assays with recombinant chromatin templates. Fourth, ChIP analyses show that hRAD6 is actively recruited to a transcribed gene upon gene induction. Since none of these parameters were evident for UbcH6, its role in the cell may be restricted to a minor H2B, or more likely a distinct, ubiquitylation pathway.
The strong homology (over 95% similarity) between hRAD6A and hRAD6B is suggestive of redundant functions. Although a concomitant decrease in H2B ubiquitylation and total RAD6 protein was only observed with hRAD6A siRNA, and not with hRAD6B siRNA, this reflects the fact that hRAD6A is much more abundant than hRAD6B in cells (Koken et al., 1996). In addition, our in vitro assays with chromatin substrates clearly showed that hRAD6A and hRAD6B both have the ability to ubiquitylate H2B. Therefore, our results suggest that the normal H2B ubiquitylation observed in RAD6B knockout mice (Baarends et al., 2007) is due to the redundant function of intact RAD6A.
For some E3s, target protein ubiquitylation involves direct interaction of a resident RING finger with the corresponding E2 (Lorick et al., 1999). However, the Ubr1 N-end rule E3 (Xie and Varshavsky, 1999) and the DNA repair-related RAD18-RAD5 E3 (Ulrich and Jentsch, 2000) interact with their cognate E2, RAD6, in a RING finger-independent manner. We also found that the BRE1 complex interacts with RAD6 in a RING finger-independent manner both in human (Figure 3) and in yeast (J.K. and R.G.R., unpublished observations). The fact that RAD6 is the cognate E2 for these E3s indicates that RAD6 is a non-canonical E2 that binds to its cognate E3 via regions other than the RING finger and further suggests that the role of the RING finger in E3 is not merely to recruit E2 to the vicinity of the target proteins.
Mechanistic View of the Role of the PAF Complex in H2B Ubiquitylation
The PAF complex requirement for both H2B ubiquitylation and downstream H3K4 and H3K79 methylation has been well-documented by yeast genetics (Krogan et al., 2003; Ng et al., 2003a; Wood et al., 2003; Xiao et al., 2005). However, there has been no clear biochemical evidence that explains the direct role of the PAF complex in H2B ubiquitylation. In the present study, our chromatin-templated ubiquitylation and transcription assays with defined factors have revealed distinct PAF complex functions in H2B ubiquitylation. First, the PAF complex can recruit H2B ubiquitylation factors to chromatin through direct interactions with both the BRE1 complex and Pol II. Second, the PAF complex couples transcription and efficient H2B ubiquitylation through its intrinsic chromatin transcription enabling activity rather than through a direct stimulation of RAD6-BRE1 activity. It is plausible that the PAF complex-enhanced passage of Pol II through nucleosomes allows H2B ubiquitylation factors (recruited by the PAF complex) easier access to the H2B ubiquitylation site. The fact that H2B ubiquitylation is further enhanced by SII, which synergistically increases transcription with the hPAF complex, strengthens our claim that efficient H2B ubiquitylation is coupled to transcription. This relationship (hRAD6→hBRE1 complex→hPAF complex→Pol II) nicely fits the yeast genetic data wherein yBre1 deletion completely abrogates yRad6 association with the entire body of Pol II-dependent genes (Wood et al., 2003; Xiao et al., 2005) and where deletion of the PAF complex subunit yRtf1 leads to dissociation of yRad6 from coding regions (Xiao et al., 2005). Along with these observations, our data contrast with a previous report (Zhu et al., 2005) that hUbcH6, rather than the hBRE1 complex, interacts with the hPAF complex to physically link the H2B ubiquitylation machinery to Pol II and that the hPAF complex enhances hUbcH6-mediated H2B ubiquitylation in the absence of on-going transcription.
Role of Histone H2B Ubiquitylation in Transcription and H3 Methylation
Cell-based assays have shown that H2B ubiquitylation is required for proper activation of several inducible genes and, conversely, that a number of factors implicated in transcription initiation and elongation are required for H2B ubiquitylation (Weake and Workman, 2008). An intriguing question raised by these results is whether H2B ubiquitylation itself stimulates transcription or whether transcription facilitates H2B ubiquitylation for purpose of a subsequent ubiquitylated H2B function. Relevant to this issue, our transcription-coupled chromatin ubiquitylation assays with biochemically defined factors clearly show that on-going transcription is required for efficient H2B ubiquitylation (Figure 6B). Of note, the overall level of H2B ubiquitylation in this assay (about 15%) is markedly higher than the level (<1%) observed with oligonucleosome substrates in the absence of transcription. Moreover, since only a small portion of the chromatin template is transcribed in vitro, the level of H2B ubiquitylation may be much greater than 15% in the transcribed region. In addition, our demonstration of low-level transcription-independent H2B ubiquitylation with natural HeLa cell-derived oligonucleosomes, but not with recombinant chromatin (data not shown), raises the possibility that natural transcriptionally active chromatins (with associated histone modifications) may serve as preferential substrates for H2B ubiquitylation in vitro. Our demonstration of a transcription requirement for H2B ubiquitylation also provides a plausible explanation for why H2B ubiquitylation-dependent H3K4 methylation marks recent transcription (Ng et al., 2003b).
Strikingly, our transcription assays with a recombinant, fully H2B-ubiquitylated chromatin template demonstrate that H2B ubiquitylation per se has no demonstrable effect on the level of hPAF complex- and SII-enhanced transcription mediated by p53 in conjunction with p300. This contrasts with a previously reported observation that hUbcH6-mediated H2B ubiquitylation directly stimulates FACT-dependent histone displacement and transcription elongation in vitro (Pavri et al., 2006). Although we failed to observe any effect of FACT, in the presence or absence of RAD6-dependent H2B ubiquitylation, on the overall level of transcription (data not shown), this may reflect in part the utilization of NAP1, rather than FACT, as a histone chaperone in our assays. Our results are consistent with those of Fleming et al. (2008), who showed an in vivo role for H2B ubiquitylation in the efficient reassembly of nucleosomes after Pol II passage, thereby repressing cryptic transcription initiation, but found no role in nucleosome disassembly during Pol II passage. Hence, H2B ubiquitylation was proposed to function in the wake of elongating Pol II rather than by directly stimulating Pol II elongation. Nontheless, and of note, our inability to see an effect of robust H2B ubiquitylation on the overall level of transcription or on transcription-related events may relate to our use of a defined transcription system that lacks downstream factors that act in conjunction with ubH2B (see below). Thus, our results overall indicate that H2B ubiquitylation does not directly affect the function of the transcriptional machinery, but that it is a consequence of transcription that is important for events following passage of Pol II.
Current observations suggest several possibilities for a positive role for H2B ubiquitylation in transcription-related events. First, ubH2B may provide a binding platform for a factor(s) that is responsible for downstream events such as histone modification and chromatin remodeling during transcription elongation. Second, and potentially related to the first possibility, H2B ubiquitylation-dependent H3 methylation may affect transcription. In this regard, H2B ubiquitylation was shown to directly stimulate both hDOT1L-mediated H3K79 methylation (McGinty et al., 2008) and hSET1 complex-mediated H3K4 di- and tri-methylation (Figure 6H). In relation to transcription, H3K4 tri-methyl marks are known to be recognized, for example, by PHD fingers in factors affecting chromatin remodeling or histone modifications (Ruthenburg et al., 2007). Third, H2B ubiquitylation, followed by deubiquitylation, may also be required for promoter proximal transcription events on certain genes (Zhang et al., 2008), and these requirements could be imposed by other unknown factors. The defined transcription-H2B ubiquitylation system described here should prove critical for a further characterization of transcription-related factors that are dependent upon H2B ubiquitylation.
EXPERIMENTAL PROCEDURES
RNA Interference
293T or HCT116 cells were treated with siRNA duplex (Dharmacon) using oligofectamine according to the manufacturer's instruction (Gibco-Invitrogen).
Protein Interaction Assays
For GST-pull down assays, 4 μg of GST-fused proteins and 200 ng of purified factors were mixed with glutathione-Sepharose 4B beads in binding buffer (20 mM Tris-Cl (pH 7.9), 150 mM KCl, 0.2 mM EDTA, 20 % glycerol, 0.1 % NP-40, 0.5 mg/ml BSA and 0.5 mM PMSF). For the interaction studies in Figure 5C and Figure 5E, M2 agarose preparations previously coupled with either 5 μg purified FLAG-hPAF complex or 2 μg purified FLAG-proteins were incubated with 200 ng purified hBRE1 complex in the same binding buffer (except for the inclusion of 300 mM KCl) as above. After 3 hr incubation at 4°C, the beads were washed and bound proteins were analyzed by immunoblots. Coimmunoprecipitation assays were performed as described (Kim et al., 2005).
In Vitro E3 Ubiquitylation Assays
Reactions containing 100 ng E1, 200 ng E2, 150 ng hBRE1 complex, 1.3 μg His-pK-HA-ubiquitin that can be radio-labeled by protein kinase (Kim et al., 2002; pK indicates protein kinase recognition site), 50 mM Tris-Cl (pH 7.9), 5 mM MgCl2, 2 mM NaF, 0.4 mM DTT and 4 mM ATP in 20 μl were incubated at 37 °C for 1 h, resolved by SDS-PAGE and subjected to auto-radiography or immunoblots.
In Vitro Chromatin Ubiquitylation Assays
Reactions containing the indicated amounts of HeLa cell-derived oligonucleosomes, hE1, E2, the hBRE1 complex, His-pK-HA-ubiquitin, 50 mM Tris-Cl (pH 7.9), 5 mM MgCl2, 2 mM NaF, 0.4 mM DTT, 4 mM ATP in 20 μl were incubated at 37 °C for 10 h, resolved by SDS-PAGE and subjected to immunoblotting.
In Vitro Chromatin Assembly, Transcription and Methyltransferase Assays
Procedures for ACF-mediated assembly of chromatin with recombinant histone octamers were as described (An et al., 2004). Transcription assays with the highly purified factors on chromatin templates were as described (Guermah et al., 2006). For chromatin methyltransferase assays, reactions containing 6 μl of purified hSET1 complex, 100 μM S-adenosylmethionine (SAM) and 700 ng of recombinant chromatin (assembled as above except for inclusion of 50 mM Tris-Cl (pH 8.5)) in 77 μl were incubated at 30 °C for 6 h with addition of 1 μl of 7.5 mM SAM every 2 h, resolved by SDS-PAGE and subjected to immunoblotting.
Chromatin Immunoprecipitation (ChIP) Assays
HCT116 cells grown in McCoy's 5A (Gibco-Invitrogen) were treated with 0.5 μM doxorubicin (Sigma) for the indicated times. ChIP analyses were performed according to the manufacturer's instruction (Upstate). Primers for quantitative PCR were adopted from Donner et al. (2007) and are summarized in Table S2.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. C.D. Allis for helpful discussion; Drs. N. Minsky and M. Oren for anti-ubH2B antibody; Drs. O. Rozenblatt-Rosen and M. Meyerson for anti-hCTR9 antibody; Dr. D. Reinberg for anti-hSKI8 antibody and the FLAG-hBRE1A/RNF20 cell line. This work was supported by NIH grants (CA129325, DK071900 and CA113872) to R.G.R. and by a Leukemia and Lymphoma Society SCOR grant.
REFERENCES
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Baarends WM, Wassenaar E, Hoogerbrugge JW, Schoenmakers S, Sun ZW, Grootegoed JA. Increased phosphorylation and dimethylation of XY body histones in the Hr6b-knockout mouse is associated with derepression of the X chromosome. J. Cell Sci. 2007;120:1841–1851. doi: 10.1242/jcs.03451. [DOI] [PubMed] [Google Scholar]
- Donner AJ, Szostek S, Hoover JM, Espinosa JM. CDK8 is a stimulus-specific positive coregulator of p53 target genes. Mol. Cell. 2007;27:121–133. doi: 10.1016/j.molcel.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dover J, Schneider J, Tawiah-Boateng MA, Wood A, Dean K, Johnston M, Shilatifard A. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J. Biol. Chem. 2002;277:28368–28371. doi: 10.1074/jbc.C200348200. [DOI] [PubMed] [Google Scholar]
- Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol. Cell. 2008;31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Gonzalez F, Delahodde A, Kodadek T, Johnston SA. Recruitment of a 19S proteasome subcomplex to an activated promoter. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- Guermah M, Palhan VB, Tackett AJ, Chait BT, Roeder RG. Synergistic functions of SII and p300 in productive activator-dependent transcription of chromatin templates. Cell. 2006;125:275–286. doi: 10.1016/j.cell.2006.01.055. [DOI] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu. Rev. Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Jentsch S, McGrath JP, Varshavsky A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Science. 1987;329:131–135. doi: 10.1038/329131a0. [DOI] [PubMed] [Google Scholar]
- Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Mol. Cell. 2005;20:759–770. doi: 10.1016/j.molcel.2005.11.012. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim JH, Lee SH, Kim DH, Kang HY, Bae SH, Pan ZQ, Seo YS. The novel human DNA helicase hFBH1 is an F-box protein. J. Biol. Chem. 2002;277:24530–24537. doi: 10.1074/jbc.M201612200. [DOI] [PubMed] [Google Scholar]
- Koken MH, Hoogerbrugge JW, Jasper-Dekker I, de Wit J, Willemsen R, Roest HP, Grootegoed JA, Hoeijmakers JH. Expression of the ubiquitin-conjugating DNA repair enzymes HHR6A and B suggests a role in spermatogenesis and chromatin modification. Dev. Biol. 1996;173:119–132. doi: 10.1006/dbio.1996.0011. [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol. Cell. 2003;11:721–729. doi: 10.1016/s1097-2765(03)00091-1. [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J. Biol. Chem. 2005;280:41725–41731. doi: 10.1074/jbc.M508312200. [DOI] [PubMed] [Google Scholar]
- Lorick KL, Jensen JP, Fang S, Ong AM, Hatakeyama S, Weissman AM. RING fingers mediate ubiquitin-conjugating enzyme (E2)-dependent ubiquitination. Proc. Natl. Acad. Sci. USA. 1999;96:11364–11369. doi: 10.1073/pnas.96.20.11364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyakhovich A, Shekhar MP. RAD6B overexpression confers chemoresistance: RAD6 expression during cell cycle and its redistribution to chromatin during DNA damage-induced response. Oncogene. 2004;23:3097–3106. doi: 10.1038/sj.onc.1207449. [DOI] [PubMed] [Google Scholar]
- McGinty RK, Kim J, Chatterjee C, Roeder RG, Muir TW. Chemically ubiquitylated histone H2B stimulates hDOT1L-mediated intranucleosomal methylation. Nature. 2008;453:812–816. doi: 10.1038/nature06906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky N, Shema E, Field Y, Schuster M, Segal E, Oren M. Monoubiquitinated H2B is associated with the transcribed region of highly expressed genes in human cells. Nat. Cell. Biol. 2008;10:483–488. doi: 10.1038/ncb1712. [DOI] [PubMed] [Google Scholar]
- Ng HH, Dole S, Struhl K. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 2003a;278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K. Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol. Cell. 2003b;11:709–719. doi: 10.1016/s1097-2765(03)00092-3. [DOI] [PubMed] [Google Scholar]
- Pavri R, Zhu B, Li G, Trojer P, Mandal S, Shilatifard A, Reinberg D. Histone H2B monoubiquitination functions cooperatively with FACT to regulate elongation by RNA polymerase II. Cell. 2006;125:703–717. doi: 10.1016/j.cell.2006.04.029. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu. Rev. Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Sun ZW, Allis CD. Ubiquitination of histone H2B regulates H3 methylation and gene silencing in yeast. Nature. 2002;418:104–108. doi: 10.1038/nature00883. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Erdjument-Bromage H, Tempst P, Allis CD. Ubiquitylation of histone H2B controls RNA polymerase II transcription elongation independently of histone H3 methylation. Genes Dev. 2007;21:835–847. doi: 10.1101/gad.1516207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrich HD, Jentsch S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 2000;19:3388–3397. doi: 10.1093/emboj/19.13.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol. Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A. The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 2003;278:34739–34742. doi: 10.1074/jbc.C300269200. [DOI] [PubMed] [Google Scholar]
- Xiao T, Kao CF, Krogan NJ, Sun ZW, Greenblatt JF, Osley MA, Strahl BD. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 2005;25:637–651. doi: 10.1128/MCB.25.2.637-651.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Varshavsky A. The E2-E3 interaction in the N-end rule pathway: the RING-H2 finger of E3 is required for the synthesis of multiubiquitin chain. EMBO J. 1999;18:6832–6844. doi: 10.1093/emboj/18.23.6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang XY, Varthi M, Sykes SM, Phillips C, Warzecha C, Zhu W, Wyce A, Thorne AW, Berger SL, McMahon SB. The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression. Mol. Cell. 2008;29:102–111. doi: 10.1016/j.molcel.2007.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Pham AD, Mandal SS, Erdjument-Bromage H, Tempst P, Reinberg D. Monoubiquitination of human histone H2B: the factors involved and their roles in HOX gene regulation. Mol. Cell. 2005;20:601–611. doi: 10.1016/j.molcel.2005.09.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.