Abstract
Vasopressin binding to the V2 receptor in renal principal cells leads to activation of protein kinase A, phosphorylation of aquaporin 2 (AQP2) at Ser256, and the translocation of AQP2 to the apical membrane, resulting in concentration of the urine. In contrast, phorbol ester–induced activation of protein kinase C pathway leads to ubiquitination of AQP2 at Lys270 and its internalization to multivesicular bodies, where it is targeted for lysosomal degradation or stored for recycling. Because little is known about the regulation of AQP2 trafficking, we used the carboxy-terminal tail of constitutively nonphosphorylated AQP2 (S256A) as a bait for interacting proteins in a yeast two-hybrid assay. We isolated lysosomal trafficking regulator–interacting protein 5 (LIP5) and found that LIP5 interacted with the proximal carboxy-terminal tail (L230-D243) of AQP2 in vitro but not with AQP3 or AQP4, which are also expressed in principal cells. Immunohistochemistry revealed that LIP5 co-localized with AQP2 in principal cells. LIP5 binding occurred independent of the state of Ser256 phosphorylation or Lys270 ubiquitination. LIP5 has been shown to facilitate degradation of the EGF receptor; here, LIP5 seemed to bind this receptor. Knockdown of LIP5 in mouse renal cells (mpkCCD) reduced the phorbol ester–induced degradation of AQP2 approximately two-fold. In summary, LIP5 binds cargo proteins and, considering the role of LIP5 in protein sorting to multivesicular bodies, plays a role in the degradation of AQP2, possibly by reducing the formation of late endosomes.
Tight regulation of the translocation of aquaporin 2 (AQP2) water channels to and from the apical membrane of renal collecting duct cells by the antidiuretic hormone arginine vasopressin (AVP) is fundamental for water homeostasis. Upon hypernatremia or hypovolemia, binding of AVP to its type 2 receptor (V2R) increases intracellular calcium and cAMP concentrations, which activate and tether protein kinase A (PKA) to AQP2-containing vesicles, resulting in phosphorylation of AQP2 and other proteins.1–7 Consequently, these vesicles dock and fuse with the apical membrane, rendering principal cells water permeable.
Regulated translocation of AQP2 to and from the apical membrane suggests the existence of proteins interacting with cytosolic segments of AQP2. Only the amino-terminal and carboxyl-terminal tails (N- and C-tails, respectively) of AQP2 extend well into the cytosol, and the C-tail of AQP2 has been shown to have an important role in its apical sorting: First, Ser256 in the AQP2 C-tail is phosphorylated in vivo by AVP stimulation, and studies in both cell and animal models revealed that this phosphorylation event is essential for AQP2 translocation to the plasma membrane.8–13 Second, all AQP2 mutants encoded in families with a dominant inheritance of nephrogenic diabetes insipidus (NDI), a disease in which the kidney is unable to concentrate urine in response to AVP, are missorted as a result of mutations in the C-tail.12,14–19 In addition, the AQP2 C-tail is mono-ubiquitinated at Lys270 upon AVP removal or PKC activation, which enhances endocytosis and degradation of AQP2.20
So far, the Rap1 GTPase-activating protein Spa1, heat-shock protein 70, and Myelin and Lymphocyte Associated Protein (MAL) are the only proteins known to bind the AQP2 C-tail, potentially playing physiologic roles in AQP2 trafficking21–23; therefore, to gain more insight in the proteins and mechanisms involved in the regulation of AQP2, we used yeast two-hybrid assays to screen a mouse kidney cDNA library for proteins interacting with the C-tail of AQP2. We found the lysosomal trafficking regulator (LYST) interacting protein 5 (LIP5; Swiss-Prot entry Q9CR26; corresponding gene name DRG-1) to interact specifically with AQP2. Interestingly, LIP5 is reported to function in multivesicular body (MVB) formation,24 although a direct interaction of LIP5 with MVB cargo proteins has never been reported. Subsequent analyses revealed the LIP5-binding region within AQP2 and the role of LIP5 in AQP2 regulation.
RESULTS
Screening for AQP2-Interacting Proteins
To isolate proteins involved in AQP2 regulation, we transfected yeast cells expressing (for clarity, expression refers to protein expression unless indicated otherwise) a fusion protein of LexA and the C-tail of AQP2-S256A with a mouse kidney cDNA library, grown under selective conditions and screened for β-galactosidase activity. Approximately 3.85 × 106 colonies were screened. Of the 22 initial positive clones, six remained positive after rescreening in combination with the AQP2-S256A bait construct but not with the empty bait plasmid. Sequence analysis of the positive clones revealed that all six clones encoded the same protein showing 100% identity to a mouse RIKEN cDNA library clone annotated Mus musculus Vps20-associated 1 homolog (Saccharomyces cerevisiae; Entrez GeneID 66201), encoding the mouse orthologue of human LIP5. A schematic representation and primary sequence of LIP5 are depicted in Supplemental Figure 1. Among the six positive clones, four different positions for LexA-fusion were observed, all of them within the first eight N-terminal LIP5 residues, indicating that the product of several independent clones interacted with the AQP2 C-tail.
Site and Specificity of the AQP2–LIP5 Interaction
To allow further characterization of the role of LIP5 in AQP2 binding and regulation, we generated antibodies directed against full-length LIP5. Immunohistochemical/cytochemical and immunoblot data (Supplemental Figure 2) and the immunoblot data from the LIP5 shRNA experiment reveal the specificity of our LIP5 antibodies.
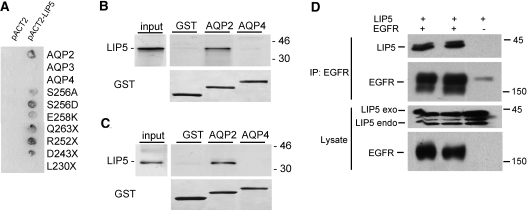
To identify the LIP5-binding region in AQP2, we conducted yeast two-hybrid assays with truncated AQP2 C-tails. β-Galactosidase assays revealed that AQP2 C-tails truncated at Q263, R252, and D243 were still able to interact with LIP5, whereas interaction was lost with AQP2 truncated at L230 (Figure 1A), indicating that region 230 to 243 of AQP2 is essential for LIP5 binding.
Figure 1.
Interaction of LIP5 with AQP2, AQP3, AQP4, and EGFR. (A) Yeast cells expressing LexA fused to the C-tail of wt-AQP2, AQP2-S256A, AQP2-S256D, AQP2-E258K, AQP3, or AQP4 proteins, all in combination with GAL4-LIP5 fusion proteins, were analyzed for interaction using β-galactosidase activity assays. Yeast cells expressing the C-tails of wt-AQP2, AQP2-S256A, AQP2-S256D, and AQP2-E258K but not those of AQP3 or AQP4 show blue staining of the colonies and thus interaction with LIP5. Sequential stop mutants of the C-tail of AQP2 revealed that only upon deletion of region L230-D243, binding of the AQP2 C-tail with LIP5 is lost. Control yeast cells transfected with bait and empty prey constructs did not show any β-galactosidase activity. (B and C) GST or GST fused to the C-tails of AQP2 (GST-AQP2) or AQP4 (GST-AQP4) were incubated with in vitro translated LIP5 (B) or dog kidney cytosol (C) and subjected to GST pulldown assays. LIP5 was visualized by autoradiography (B) or immunoblotting (C). (D) For determination of whether LIP5 also interacts with the EGFR, Xpress-tagged LIP5 was expressed alone or together with the EGFR (in duplicate) in HEK293 cells, lysed, and subjected to EGFR immunoprecipitation. Subsequent immunoblotting of the precipitates using mouse anti–Xpress-tag antibodies revealed that Xpress-tagged LIP5 co-precipitated with the EGFR but was not precipitated when expressed alone (top). EGFR immunoblotting of the lysates revealed specific signals for the EGFR in lanes of cells transfected with EGFR constructs (bottom). LIP5 immunoblotting showed Xpress-tagged (exo) and endogenous (endo) LIP5 in cells transfected with LIP5 constructs (third panel).
In the kidney, AQP2, AQP3, and AQP4 are expressed in the same cells.25,26 For investigation of the specificity of the LIP5–AQP2 interaction, LexA-AQP C-tail fusion proteins of AQP2, AQP3, and AQP4 were expressed in yeast and tested for interaction with LIP5. β-Galactosidase assays revealed that LIP5 interacted with the C-tail of wt-AQP2 but not with the C-tails of AQP3 (M264-I292) or AQP4 (V251-V323; Figure 1A). Yeast cells transfected with bait and empty pACT2 constructs revealed no staining (Figure 1A). Immunoblot analysis of the yeast cells confirmed expression of the bait and prey proteins (data not shown). For confirmation and further testing of whether LIP5 directly interacts with the AQP2 tail or requires additional proteins, 35S-methionine/cysteine-labeled LIP5 was synthesized in vitro and incubated with glutathione S-transferase (GST), GST-AQP2, or GST-AQP4 coupled to glutathione Sepharose beads. Autoradiography of the eluted proteins showed that LIP5, running at approximately 42 kD, directly interacts with GST-AQP2 but, again, not with GST-AQP4 or GST (Figure 1B).
To investigate whether renal LIP5 would specifically interact with the AQP2 tail, we incubated dog kidney cytosol with GST, GST-AQP2, or GST-AQP4 coupled to glutathione Sepharose beads. LIP5 immunoblotting of the isolated interacting proteins showed a renal protein of approximately 42 kD specifically interacting with GST-AQP2 and not with GST or GST-AQP4 (Figure 1C). These results thus showed that LIP5 binding to the C-tail of AQP2 was specific in relation to the C-tails of AQP3 and AQP4.
LIP5 Interaction with the EGF Receptor
Our data reveal AQP2 as the first cargo protein identified to interact with LIP5; however, LIP5 has been reported to facilitate EGF receptor (EGFR) degradation and might therefore also interact with EGFR.24 For testing this, EGFR and Xpress-tagged LIP5 were coexpressed in HEK293 cells. After EGFR immunoprecipitation, immunoblotting for the Xpress-tag indeed demonstrated that tagged LIP5 co-precipitated with EGFR (in duplicate; Figure 1D, lanes 1 and 2), indicating that LIP5 interacts with EGFR. Note that immunoblotting for LIP5 of the cell lysates shows endogenous LIP5 of approximately 42 kD, besides a strong and weak band of Xpress-tagged LIP5. Our data thus reveal that, besides AQP2, the cargo protein EGFR interacts with LIP5.
LIP5 Expression in the Kidney
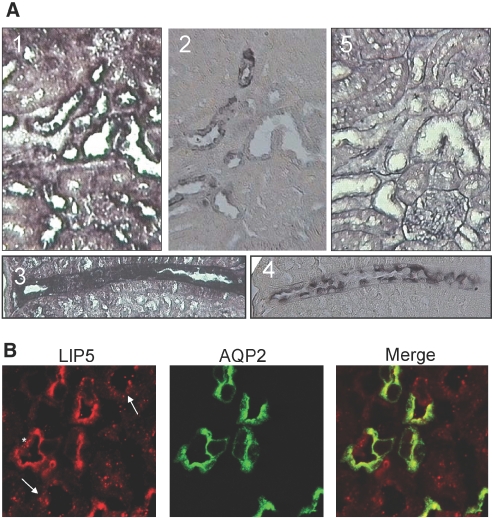
For LIP5 to have a role in the regulation of AQP2 in vivo, it needs to be present in renal principal cells. We performed LIP5 in situ hybridization and AQP2 immunohistochemistry on alternating mouse kidney sections. Microscopic analysis revealed that renal principal cells (positive for AQP2; Figure 2A, 2 and 4) indeed also contain LIP5 mRNA (Figure 2A, 1 and 3). Besides these cells, LIP5 mRNA was detected in epithelial cells of other renal tubules and collecting duct cells. A sense probe of LIP5 cRNA, which was taken along as a negative control, revealed no staining (Figure 2A, 5).
Figure 2.
Localization of LIP5 in mouse kidney. (A) Alternating mouse kidney sections were used to visualize the co-localization of LIP5 mRNA (by in situ hybridization; 1 and 3) and AQP2 (by immunohistochemistry; 2 and 4). LIP5 mRNA is detected in most epithelial cells. In situ hybridization using a sense probe did not reveal any specific staining (5). (B) Renal sections of mice receiving water ad libitum were subjected to immunohistochemistry for LIP5 (red) and AQP2 (green). In renal principal cells, which express AQP2, LIP5 shows similar localization to that of AQP2 (middle). In intercalating cells (*) and epithelial cells of other tubules (arrows), LIP5 staining is more punctuate.
Next, AQP2 and LIP5 co-localization was determined. Immunohistochemistry for AQP2 and LIP5 on mouse kidney sections and confocal laser scanning microscopy revealed that AQP2 co-localizes with LIP5 in the apical region of renal principal cells (Figure 2B). Interestingly, in AQP2-negative tubules (arrows in Figure 2B) and intercalating cells (asterisks in Figure 2B), which are the AQP2-negative cells of collecting ducts, LIP5 showed a punctuate staining, suggesting a vesicular localization of LIP5 in these cells.
Effect of Physiologic Modification in the AQP2 C-Tail on Its Interaction with LIP5
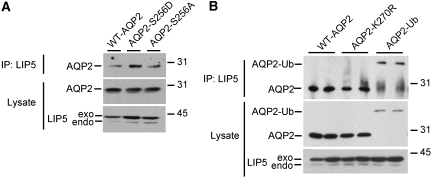
In vivo, redistribution of AQP2 from intracellular vesicles to the apical membrane coincides with phosphorylation of AQP2 at Ser256.10,11 Subsequent studies in MDCK cells showed that phosphorylation at Ser256 is essential and sufficient for apical membrane localization of AQP2, because AQP2-S256A, which mimics nonphosphorylated AQP2, is localized in intracellular vesicles, whereas AQP2-S256D, mimicking phosphorylated AQP2, is localized in the apical membrane.8 Also, AQP2 mutants in dominant NDI have missense or frame-shift mutations in the C-tail, one of which is AQP2-E258K.15,16 For determination of whether LIP5 binding to the AQP2 C-tail depends on the AQP2 phosphorylation state and whether binding is lost with AQP2 mutants in dominant NDI, the C-tails of AQP2-S256A, AQP2-S256D, and AQP2-E258K were expressed with LIP5 in yeast and tested for interaction. β-Galactosidase assays revealed a positive signal for the S256A, S256D, and E258K C-tails (Figure 1A), suggesting that LIP5 binding is independent of the AQP2 phosphorylation state and is maintained in the AQP2-E258K mutant. Using HEK293 cells, transiently-expressing AQP2-S256A or AQP2-S256D, co-immunoprecipitation assays confirmed that interaction of LIP5 with AQP2 is independent of AQP2 phosphorylation state (Figure 3A).
Figure 3.
LIP5 interaction with physiologically modified AQP2. (A) For determination of whether LIP5 binding to AQP2 depends on the phosphorylation state of AQP2, LIP5 was expressed together with wt-AQP2, AQP2-S256D, and AQP2-S256A in HEK293 cells; lysed; and subjected to LIP5 immunoprecipitation. Subsequent immunoblotting of the precipitates demonstrated that wt-AQP2, AQP2-S256D, and AQP2-S256A co-precipitated with LIP5 (top). Immunoblotting of the lysates revealed similar signals for AQP2 in lanes of cells transfected with AQP2 protein constructs (middle) and for endogenous (endo) and exogenous (exo) LIP5 in cells transfected with LIP5 constructs (bottom). (B) For determination of whether LIP5 binding to AQP2 depends on the ubiquitination state of AQP2, LIP5 was expressed together with wt-AQP2, AQP2-K270R, and AQP2-Ub in HEK293 cells. Subsequent cell lysis and LIP5 immunoprecipitation followed by immunoblotting showed that wt-AQP2, AQP2-K270R, and AQP2-Ub co-precipitated with LIP5 (top). Immunoblotting of the lysates revealed equal signals for AQP2 in lanes of cells transfected with wt-AQP2, AQP2-K270R, and AQP2-Ub constructs (middle) and for endogenous (endo) and exogenous (exo) LIP5 in cells transfected with LIP5 constructs LIP5 (bottom).
Besides phosphorylation, AQP2 is subject to mono-ubiquitination at Lys270, which enhances endocytosis and degradation of AQP2.20 To determine whether LIP5 interaction with AQP2 depends on the ubiquitination state of AQP2, wt-AQP2, AQP2 that cannot be ubiquitinated (AQP2-K270R), or constitutively ubiquitinated AQP2 (AQP2-Ub) were expressed in HEK293 cells and tested for interaction. Co-immunoprecipitation assays and subsequent immunoblotting revealed that LIP5 binds both AQP2-K270R and AQP2-Ub, indicating that LIP5 binding to AQP2 is independent of AQP2 ubiquitination (Figure 3B).
Role of LIP5 in AQP2 Abundance
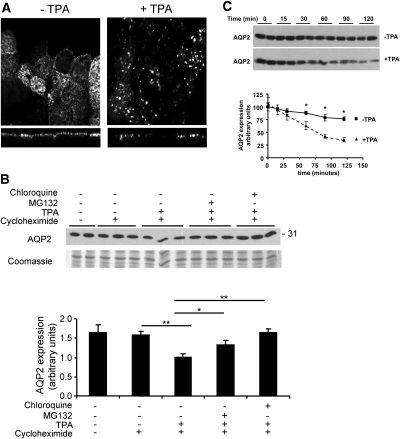
LIP5 is implicated in sorting of proteins to the internal vesicles of MVBs after endocytosis, from where proteins are targeted for lysosomal degradation,24 and LIP5 knockdown decreases lysosomal degradation of EGFR.24 In polarized MDCK cells, AQP2 is also degraded through the lysosomal pathway. In addition, activation of the protein kinase C (PKC) pathway by the phorbol-ester 12-tetradecanoylphorbol-13-acetate (TPA) counteracts the AVP-induced translocation of AQP2 to the apical membrane by inducing AQP2 internalization and degradation.8,20 To test whether LIP5 has a similar function with regard to AQP2, we tested whether mpkCCD cells (murine collecting duct cells that yield expression of endogenous AQP2 protein upon treatment with dDAVP27,28) show internalization of AQP2 upon TPA treatment. Immunocytochemistry and confocal laser scanning microscopy analysis showed presence of endogenous AQP2 in the apical membrane with dDAVP, which was redistributed to intracellular vesicles upon 45 min of TPA treatment (Figure 4A). Furthermore, immunoblot analysis showed that 2 h of TPA treatment in the presence of the protein synthesis inhibitor cycloheximide significantly increases the degradation of existing AQP2, which was partially blocked by the proteasomal inhibitor MG132 and completely blocked by chloroquine, which inhibits lysosomal degradation (Figure 4B).
Figure 4.
TPA induces internalization and lysosomal degradation of AQP2 in mpkCCD cells. (A) TPA induced internalization of AQP2. For testing whether phorbol esters also induce internalization of AQP2 in mpkCCD cells, a monolayer of polarized mpkCCD cells stably expressing exogenous AQP2 (mpkCCD-AQP2) was stimulated with forskolin for 45 min followed by stimulation with forskolin only (−TPA) or together with TPA (+TPA) for 45 min. Subsequent immunocytochemistry and confocal laser scanning microscopy demonstrated internalization of AQP2 upon TPA treatment. (B) TPA induces lysosome-mediated degradation of AQP2. MpkCCD-AQP2 cells were stimulated with forskolin for 45 min, followed by incubation with forskolin alone or together with cycloheximide. Moreover, cells treated with forskolin and cycloheximide were additionally treated with or without TPA alone or TPA together with the proteasome blocker MG132 or the lysosome blocker chloroquine. All treatments after stimulation with forskolin were for 2 h, after which the cells were lysed. Immunoblotting for AQP2 revealed that co-incubation with chloroquine but not MG132 counteracted the TPA-induced increase in AQP2 degradation. Immunoblotting was performed in at least two independent experiments performed in triplicate. Quantification of AQP2 levels showed significant effects (two-tailed t test; *P < 0.05, **P < 0.01). (C) TPA reduces the half-life of existing AQP2. MpkCCD-AQP2 cells were grown and stimulated to induce AQP2 expression as described in A. Cells were then treated with cycloheximide alone or together with TPA for the indicated periods. Cells were then lysed and analyzed using Western blotting using AQP2 antibodies (top), followed by semiquantification using densitometry. The data are plotted as a percentage of the control (mpkCCD cells grown 4 d in the presence of dDAVP). Quantification of AQP2 levels showed significant effects (two-tailed t test; *P < 0.05).
To investigate the effect of TPA on the half-life of AQP2, we incubated vasopressin-stimulated mpkCCD cells for various periods with or without TPA in the continuous presence of cycloheximide and immunoblotted for AQP2 (Figure 4C). After 60 min of TPA treatment, AQP2 levels were significantly (P < 0.05) decreased for TPA-treated versus control cells. Calculations revealed that TPA reduced the half-life of AQP2 from 234.5 ± 32.7 to 66.2 ± 14.3 min.
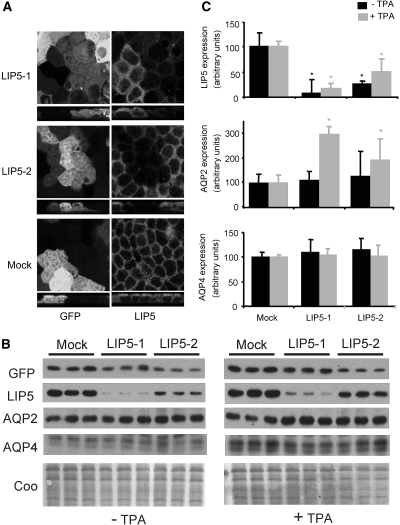
To investigate whether LIP5 plays a role in AQP2 degradation, we made lentiviruses driving the expression of two different murine LIP5 (LIP5–1/2) shRNAs or a random sequence (mock). For recognition of infected cells, the viral DNA also encoded cytomegalovirus (CMV) promoter–driven GFP. Testing the viruses in untransfected, dDAVP-stimulated mpkCCD cells revealed that most cells had lost the shRNA constructs at the time of appropriate AQP2 abundance (at 4 d of dDAVP treatment after 4 d of cell polarization); therefore, mpkCCD cell lines stably expressing AQP2 were generated. To perform experiments with physiologically relevant AQP2 protein levels, we selected clones that express AQP2 at a similar or lower level than endogenous AQP2 after stimulation with 1 nM dDAVP by immunoblotting (data not shown). After infection of mpkCCD-AQP2 cells with shRNA viruses, immunocytochemistry showed a strongly decreased LIP5 abundance in cells infected with the LIP5-specific shRNAs (LIP5-1, LIP5-2; Figure 5A), whereas cells infected with a virus encoding the mock shRNA (Figure 5A) showed no difference in LIP5 protein abundance compared with uninfected cells. This indicated that LIP5-specific shRNAs but not viral infection itself affects LIP5 protein abundance. Unfortunately, we did not succeed in co-staining for LIP5 and AQP2 in the GFP background.
Figure 5.
LIP5 silencing results in reduced lysosomal degradation of AQP2. MpkCCD cells stably transfected with AQP2 were infected with viruses expressing two different LIP5 shRNAs (LIP5-1, LIP5-2) at a multiplicity of infection of 20. Four days later and after stimulation with forskolin for 45 min, the cells were incubated with cycloheximide in the absence or presence of TPA for 2 h. (A) Cells were subjected to immunocytochemistry for GFP (left) and LIP5 (right). In mock shRNA-expressing cells, LIP5 abundance was not affected. LIP5 shRNA expressing cells showed decreased LIP5 staining. (B) Cell lysates were immunoblotted for GFP, LIP5, and AQP2. Immunoblotting for GFP serves as a measure for infection efficiency, whereas Coomassie staining demonstrates equal protein loading. A representative immunoblot of two experiments is shown. (C) Quantification of the immunoblot signals of two independent experiments performed in triplicate. The signals were scanned, and the amounts of LIP5, AQP2, AQP4, and total protein were quantified in arbitrary units ± SEM. *Significant difference from mock-treated cells (two-tailed t test; P < 0.05).
To determine the effect of LIP5 knockdown on AQP2 degradation, we treated cells with cycloheximide in the absence or presence of TPA for 2 h and lysed them. Consistent with immunocytochemistry, subsequent immunoblot analysis for LIP5 revealed that LIP5 shRNAs but not mock shRNA yielded significantly reduced LIP5 protein levels (90 and 70% for LIP5-1 and LIP5-2, respectively; Figure 5, B and C). Upon TPA treatment, AQP2 levels were significantly higher with both shRNA virus–infected cells compared with mock virus–infected cells (Figure 5, B and C, +TPA). Assuming a similar half-life for AQP2 in non–TPA-treated cells as found for non–TPA-treated mpkCCD cells (Figure 4C), the reduced decrease of AQP2 with TPA in LIP5 shRNA-expressing cells indicated that knockdown of LIP5 increased the TPA-induced half-life approximately 1.5- to 2.5-fold. Because Coomassie staining confirmed equal protein loading (Figure 5, B and C) and there was no effect on LIP5 or AQP2 abundance levels in mpkCCD cells infected with mock shRNA virus (Figure 5A, Mock), these data indicate that LIP5 indeed facilitates degradation of AQP2.
Interestingly, mpkCCD cells endogenously express AQP4, which is not bound by LIP5 (Figure 1). Analysis of AQP4 under all of these conditions revealed that AQP4 abundance is not affected by LIP5 knockdown (Figure 5, B and C), indicating that the effect of LIP5 is specific for AQP2.
DISCUSSION
LIP5 Specifically Interacts with the Proximal Region of the AQP2 C-Tail Independent of AQP2 Phosphorylation or Mono-ubiquitination
We identified LIP5 to interact with the C-tail of AQP2-S256A in yeast two-hybrid assays. Whereas AQP2 is coexpressed with AQP3 and AQP4 in renal principal cells and all have a similar gross architecture, yeast two-hybrid and GST pulldown analyses revealed that LIP5 interacts directly and specifically with the AQP2 C-tail only, not with the C-tails of AQP3 or AQP4 (Figure 1, A through C). Further analysis of the AQP2 C-tail revealed that LIP5 binds region L230 to D243 of AQP2 (Figure 1A). Consistent with binding to this region of AQP2, the interaction of LIP5 to AQP2 is independent of AQP2 phosphorylation (at Ser256) and mono-ubiquitination (at K270) and is also not affected by an AQP2 mutation causing NDI, because the sites of these modifications are outside the LIP5 binding region (Figure 3). The precise mode of interaction and amino acids involved remains to be established.
LIP5 Interacts with Cargo Proteins
The MVB machinery is composed of three complexes—ESCRT-I, -II, and -III—which are sequentially recruited to sites of MVB sorting and vesicle formation. Subsequent release of ESCRT-III from the membrane requires Vps4 ATPase activity and allows the ESCRT machinery to recycle through multiple rounds of luminal vesicle formation. So far, LIP5 has been shown to interact with Vps4 and several ESCRT-III components, including CHMP5, CHMP1B, CHMP2A, and CHMP3.24,29–33 Our data reveal for the first time that LIP5 also interacts with cargo proteins, because AQP2 as well as the EGFR specifically co-precipitated with LIP5 (Figure 1). It will be interesting to unravel to which LIP5 segment these two proteins bind and whether these two proteins compete with CHMPs for binding to Vps4.
LIP5 Is Involved in the Lysosomal Degradation Pathway of AQP2
Membrane proteins targeted for lysosomal degradation are sorted into vesicles that bud into MVBs. These MVBs can serve as long-term storage compartments, fuse with lysosomes to deliver the internal vesicles and their contents for degradation, or fuse with the plasma membrane to release the vesicles as extracellular exosomes. MVB vesicle formation and protein sorting require a set of class-E vacuolar protein sorting (VPS) proteins.34–36 Most class-E proteins function as components of one of the three ESCRT complexes, which are sequentially recruited to sites of MVB sorting and vesicle formation. The ESCRT-III proteins are the last to assemble, forming a membrane-associated lattice that functions in the final stages of this process. Via direct protein–protein interactions, Vps4 is recruited to MVBs, which induces the release of the ESCRT-III complex from the membrane.34,36
Emerging evidence points toward a role for LIP5 dimers in the disassembly of the ESCRT-III complex by stimulating Vps4 ATPase activity, which occurs directly through interaction between the conserved C-terminal VSL domain of LIP5 and Vps4.31–33 Consistently, deletion of Vta1 (the yeast LIP5 orthologue) in yeast results in altered vacuolar morphology,29 and knockdown of LIP5 in mammalian cells facilitates downregulation of EGFR.24
Several data suggest a similar role for LIP5 in AQP2 regulation in principal cells: First, LIP5 is coexpressed with AQP2 in renal principal cells (Figure 2) and LIP5 has been identified in AQP2-containing exosomes isolated from urine (My012 protein).37 Second, renal LIP5 interacts specifically with the C-terminal tail of AQP2 (Figure 1). Third, LIP5 facilitates the lysosomal degradation of AQP2: Whereas induced LIP5 overexpression in MDCK-AQP2 cells did not affect AQP2 abundance (Supplemental Figure 3) or translocation to the apical membrane, knockdown of LIP5 abundance resulted in a 1.5- to 2.5-fold increase in the AQP2 half-life after TPA treatment (Figure 5, B and C), a condition known to induce AQP2 internalization and degradation8 (Figure 4). Considering our finding that AQP2 levels seemed somewhat increased in LIP5 shRNA versus mock cells without TPA treatment (Figure 5, B and C), the half-life of AQP2 in LIP5 knockdown cells without TPA treatment may be higher than that of AQP2 in dDAVP-stimulated mpkCCD cells (Figure 4C); therefore, the estimated increase in AQP2 half-life with LIP5 knockdown after TPA treatment should be taken as a rough estimate.
Under the tested conditions, the effect of LIP5 knockdown is clear upon treatment with TPA but not without TPA treatment, which can be explained as follows: At any moment in time, AQP2 resides at several locations in the cell (endoplasmic reticulum, Golgi, storage vesicles, plasma membrane, recycling vesicles, MVBs, and lysosomes), and the steady-state localization of AQP2 is a balance regulated by extracellular and intracellular signals, such as the presence of AVP, and cAMP-dependent phosphorylation or activation of the PKC pathway. TPA-induced degradation of AQP2 in mpkCCD cells mainly occurs via lysosomes (Figure 4B) and, as illustrated by the large half-life of AQP2 of nearly 4 h (Figure 4C), only a small fraction of AQP2 is targeted for lysosomal degradation in the tested 2-h period under non-TPA conditions. With the TPA-induced internalization and lysosomal degradation of AQP220 (Figure 4), which decreases the AQP2 half-life to approximately 70 min, more AQP2 will thus pass LIP5 on its way to MVBs/lysosomes for the 2 h measured, and, therefore, the effect of (the absence of) LIP5 on AQP2 is larger and better detectable under these conditions. The partial inhibition of AQP2 degradation in our LIP5 shRNA experiments may suggest that, like its yeast orthologue Vta1,33 LIP5 has only a modulatory role in the lysosomal degradation of AQP2; however, this cannot be deduced from our experiments, because as a result of incomplete infection and/or shRNA knockdown, our shRNA-expressing cells were not devoid of LIP5 (Figure 5B).
On the basis of these data, we propose the following model for the role for LIP5 in the regulation of AQP2 in principal cells: After stimulation with AVP and translocation of AQP2 to the apical membrane, renal water reabsorption will occur, which may be increased by an extended presence of AQP2 in the plasma membrane through an interaction with MAL.22 With removal of AVP or activation of the PKC pathway, which is thought to follow binding of hormones such as endothelin, PGE2, and ATP, AQP2 is ubiquitinated at Lys270, which signals its endocytosis.20 After recruitment to clathrin-coated vesicles, AQP2 is then endocytosed from the apical membrane involving direct interaction with heat-shock protein 70.23 Via early endosomes, AQP2 is then sorted via ESCRT proteins and interaction with LIP5 to inner vesicles of MVBs, from where it can be targeted to lysosomes for degradation or released as exosomes from the cells into urine (Figure 6).37 Unfortunately, we were not able to co-immunoprecipitate AQP2 with LIP5 from mouse kidney homogenates, which may be due to a reduced abundance of LIP5 or low level of AQP2–LIP5 interaction in the kidney.
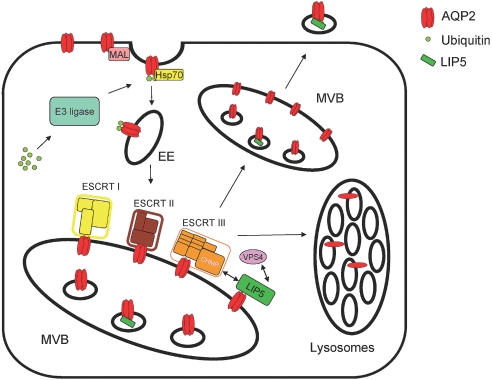
Figure 6.
Model for LIP5 function in the regulation of AQP2. On the basis of our findings, we propose the following model for LIP5 function in AQP2 regulation: After AVP-induced translocation of AQP2 to the apical membrane, water will be reabsorbed from the pro-urine. Interaction with MAL may increase water reabsorption further by extending the presence of AQP2 in the plasma membrane. Upon removal of AVP or activation of the PKC pathway, AQP2 is mono-ubiquitinated by a presently unknown ubiquitin E3 ligase, and, subsequently, endocytosis will occur. AQP2 is recruited to clathrin-coated vesicles and endocytosed from the apical membrane involving direct interaction with heat-shock protein 70 (HSP70). Via early endosomes (EE), AQP2 is then sorted via ESCRT-I, -II and, -III protein complexes to the limiting membrane of MVBs. There, interaction with LIP5 and LIP5-facilitated activity of VPS4 AATPase mediates translocation of AQP2 from the limiting membrane to inner vesicles of MVBs, from where it can be targeted to lysosomes for degradation or released as exosomes from the cells into urine.
The binding of LIP5 to the proximal region of the AQP2 C-tail and its independence of the ubiquitination and phosphorylation status of AQP2 (Figure 3) is in line with the finding that TPA-induced internalization of AQP2 constitutively phosphorylated at S256 still leads to its degradation8,38 and that AQP2 degradation is well detectable only at 2 h after TPA treatment, at which point AQP2 is not detectably ubiquitinated anymore.20 Recently, AQP2 was found to be phosphorylated at other sites in its C-terminus.39 It remains to be established whether phosphorylation of these sites influences LIP5 binding to AQP2.
Interestingly, LIP5 has also been reported to interact with the lysosomal trafficking regulator LYST,40 which is a cytosolic protein of 425 kD with a putative function in lysosome-related organelles.41 Yeast two-hybrid screens with LYST identified several putative partners, some of which have been shown to co-localize (HRS)42 or be involved (calmodulin)1 in the regulation of AQP2. If LYST is expressed in collecting duct cells, then LYST may be a scaffolding protein involved in the degradation of AQP2.
Moreover, calmodulin has been shown to interact with proximal C-terminal tail of AQP0,43 the region bound by LIP5 in AQP2. Although these regions are similar between AQP0 and AQP2, we could not detect binding between heterologously expressed calmodulin and the AQP2 C-tail in GST pulldown experiments (data not shown).
In summary, we have identified that LIP5 is coexpressed with AQP2 in renal principal cells, interacts with the proximal C-tail of AQP2, and facilitates its lysosomal degradation. Our data thus indicate that LIP5 plays an important role in the MVB/lysosomal targeting of AQP2, as induced by AVP-counteracting hormones, after which it will be degraded or expelled from the cells into urine. It will be interesting to see how LIP5 structurally couples to AQP2, whether the degradation of other cargo proteins is facilitated by direct interaction with LIP5, and whether LIP5 also affects AQP2 stability in vivo.
CONCISE METHODS
Yeast Two-Hybrid
cDNAs encoding wild-type and mutant AQP2 C-termini starting at amino acid Phe224 were generated by standard PCR reactions using AQP2-C-EcoRI (5′-GATCGGAATTCCCGCCAGCCAAGAGCCT-3′) as a forward primer and AQP2-C-XhoI as a reverse primer. As templates, pCB6-wtAQP2, pCB6-AQP2-S256A, and pCB6-AQP2-S256D8 were used. cDNAs encoding truncated AQP2 C-tail proteins were generated similarly using AQP-C-EcoRI as a forward primer and AQP2-L230X (5′-GATCCTCGAGCTAGCTCTTGGCTGGCGGT-3′), AQP2-D243X (5′-GATCCTCGAGCTACGGCTCCAGGCCCTTCAG-3′), AQP2-R252X (5′-GATCCTCGAGTCACACCTCGCGCTCCTCCC-3′), or AQP2-Q263X (GATCCTCGAGCTACGGCGAGTGCAGCTCCAC-3′) as a reverse primer (in all mutagenesis primers introduced, mutations are underlined and restriction sites are in italic). The obtained fragments were digested with EcoRI and XhoI and cloned into the EcoRI and SalI sites of pBTM116.44 The C-terminal tails of AQP3 (amino acids M264-I292) and AQP4 (amino acids V251 through V323) were cut from pGBT9-AQP3 and pGBT9-AQP445 using EcoRI and PstI and cloned into the corresponding sites of pBTM116. For all constructs, introduction of only the desired mutation was confirmed by DNA sequence analysis.
To identify AQP2 interacting proteins, we screened a GAL4 cDNA library of adult mouse whole kidneys (MATCHMAKER, Clontech, Palo Alto, CA) against a bait of the AQP2-S256A C-terminal tail. For this, L40 yeast cells (MATa, trp1, leu2, his3, LYS2::lexA-HIS3, URA3::lexA-lacZ) were transfected with the pBTM116-S256A construct. After 3 d, colonies were picked, grown in medium, pelleted, and analyzed for their LexA-C-tail expression using AQP2 and LexA immunoblot analysis. For library screening, four independent colonies expressing LexA-AQP2-S256Awere grown to A546 >2 at 30°C, 260 rpm in 100 ml of SD-Trp medium and used to inoculate 1 L of YPDA to 0.2 A600. At 0.8 A600, the cells were collected by centrifugation at 4200 × g at 4°C for 15 min, washed twice with sterile distilled H2O, pelleted at 1300 × g (10 min, 4°C), and transfected according to Gietz et al.46 using 50 μg of Library DNA. After incubation for 30 min at 30°C and a 30 min heat shock at 42°C, cells were harvested by centrifugation at 1900 × g for 3 min, washed with distilled H2O, resuspended in 20 ml of distilled H2O, and plated on 25 selective SD-Trp-His-Leu dishes of 245 × 245 mm. After 5 d, colonies were tested for β-galactosidase expression by a colony lift assay.47 For this, colonies were transferred onto 3-mm Whatmann filters, permeabilized in liquid N2, and overlaid with 0.2 mg/ml X-gal in TBSY (150 mM NaCl, 50 mM Tris-HCl [pH 7.4], and 0.8% agarose). Colonies that turned blue were transferred to selective plates and rescreened for LacZ activity as already described. From colonies that remained positive after rescreening, DNA was isolated,48 electroporated to electrocompetent KC8 cells (2.3 kV, 25 μF, and 200 Ω), and plated on M9-Leu plates. After checking insert sizes of DNA from three independent colonies by BglII digestion, L40 cells were retransformed with prey DNA in combination with either pBTM116 or pBTM116-AQP2-S256A to verify specificity of the isolated prey plasmid for binding to the AQP2-S256A C-tail by a colony lift assay. Prey plasmids giving rise to blue colonies when transformed with pBTM116-AQP2-S256A/D but not with pBTM116 were subjected to DNA sequence analysis to identify the prey cDNA. Transformation of one or two plasmids to L40 yeast cells at a time was done by using 1 μg of plasmid equivalents in a downscaled library transformation protocol and a selection for colonies on appropriate SD plates for 3 to 4 d.
Pulldown Assays
For the expression of wt-AQP2 C-tail as a GST fusion protein, a standard PCR was done using the forward primer AQP2-CtermFWD (5′-GGAATTCCCAGCCAAGAGCCTGTCG-3′) and the reverse primer AQP2-C-XhoI on a pBS-AQP249 template. The PCR fragment was digested with EcoRI-XhoI and cloned into corresponding sites of pGEX4T-1 (Amersham Biosciences, Freiburg, Germany). For GST-AQP4 expression, we used a pGEX1 vector containing the C-tail of AQP4.50 To make a GST-LIP5 expression construct, pACT2 clone 14b (identified from library screening) was cut with EcoRI and XhoI, and the cDNA fragment encoding full-length LIP5 was isolated and cloned into the corresponding sites of pGEX6.1 (Amersham Biosciences, Freiberg, Germany).
GST, GST-AQP2, and GST-AQP4 production in BL21-DE3 bacteria was induced by IPTG and isolated with prewashed Glutathione-Sepharose 4B beads. LIP5 was produced by in vitro transcription/translation using the Riboprobe-T7 system (Promega, Leiden, Netherlands) in the presence of 35S-methionine/cysteine (Redivue Promix; Amersham Biosciences, Uppsala, Sweden) and incubated with the GST fusion protein beads, rotating overnight at 4°C. Bound LIP5 was eluted by boiling the samples in Laemmli sample buffer and analyzed by SDS-PAGE. Pulled-down LIP5 was visualized using a STORM phosphor imager (Amersham Biosciences, Uppsala, Sweden).
Cytosol of dog kidneys was prepared by crushing the tissue in a blender and subsequent homogenization in 20 mM HEPES, 100 mM NaCl, 5 mM MgCl2, 1 mM dithiothreitol, 1 mM PMSF, 5 μg/ml leupeptin, and 5 μg/ml pepstatin using a potter (PotterS, Braun Biotech, Melsungen, Germany). Cytosol was cleared by centrifugation at 33,000 rpm in a Ti45 rotor in a Beckman ultracentrifuge for 1 h at 4°C. The supernatant was preincubated for 1 h with glutathione-Sepharose 4B beads containing GST and subsequently incubated with GST, GST-AQP2, or GST-AQP4 glutathione-Sepharose 4B beads overnight at 4°C. After binding, the beads were washed with 25 mM Tris (pH 7.5), 0.5 mM EDTA, and 1% Triton-X100, and bound proteins were eluted by shaking in 1.5 M NaCl, 25 mM Tris (pH 7.5), and 0.5 mM EDTA at room temperature for 1 h. Samples were desalted using Biospin columns (Biorad, Hercules, CA). Isolated proteins were analyzed by SDS-PAGE and immunoblotting.
Empty or transfected COS cells were lysed in 300 μl of lysis buffer with 1% NP40. For LIP5 depletion, lysates were incubated for 2 h with 4 μl of LIP5 antiserum and 40 μl of protein A-agarose beads (Kem-En-Tec A/S, Copenhagen, Denmark). Lysates were then incubated with GST-AQP2 glutathione-Sepharose 4B beads and further processed as described already.
Expression in Eukaryotic Cells
For expression of LIP5 in eukaryotic cells, an EcoRI-XhoI fragment encoding full-length LIP5 was isolated from the pACT2 clone 14b, identified from library screening, and ligated into the corresponding sites of pCDNA3. Expression constructs encoding wild-type AQP2 or the S256A, S256D, K270R, and AQP2-Ub mutants were as described previously.7,20 pCDNA3-EGFR was provided by Prof. Y. Yarden (Department of Biologic Regulation, Weizmann Institute of Science, Rehovot, Israel).51 HEK293 cells were maintained in DMEM (Biowittaker, Verviers, Belgium) supplemented with 10% FCS. For transient transfections, HEK293 cells were seeded at 1.2 × 105 cells/cm2 in six-well plates. Four hours after seeding, cells were transfected using polyethylenimine (Polysciences, Eppelheim, Germany). A total of 3.5 μg of circular DNA and 14 μl of polyethylenimine (1 μg/μl) were added to 80 μl of OptiMEM medium (Life Technologies Europe, Breda, Netherlands), vortexed, and incubated at room temperature for 20 min. Subsequently, the mixture was added to the cells and incubated overnight. Three days after transfection, cells were lysed in IP lysis buffer (1% Triton X-100, 150 mM NaCl, and 25 mM HEPES [pH 7.4]) and incubated with rabbit anti-LIP5 antibodies, mouse anti-Xpress, or mouse anti-EGFR 528 (Santa Cruz Biotechnology, Santa Cruz, CA) immobilized on protein A-agarose beads (Kem-En-Tec A/S) overnight at 4°C. Immunoprecipitated proteins were analyzed by immunoblotting.
In Situ Hybridization
LIP5 cDNA templates with a T7 RNA polymerase promoter sequence (underlined in primer sequences) at the 5′ or 3′ end were used to generate LIP5 cRNA probes. These templates were made by a standard PCR reaction, using 5′-TAATACACTCACT-ATAGGGAGAGGCCCCTCTGCCGCCGCTGC-3′ or 5′-CGTCCAGCTCGATCTTCATTGTCTGCATACA-3′ (sense) together with 5′-GGCCCCTCTGCCGCCGCTGC-3′ or 5′-TAATACGACTCACTATAGGGAGACGTCCAGCTCGATCTTCATTGTCTGCATACA-3′ (antisense) primers, respectively. 11-UTP dioxigenin (DIG)-labeled probes of these templates were made in vitro using T7 RNA polymerase (Roche Diagnostics, Almere, Netherlands). For preparation of mouse kidney sections, mice were transcardially perfused with ice-cold 0.6% NaCl solution for 5 min, followed by Bouin's fixative for 15 min. After dissection, kidneys were postfixed in Bouin's fixative for 24 h. Then, kidneys were dehydrated in a graded series of ethanol and xylene and embedded in paraffin. Seven-micrometer sections were mounted on poly-l-lysine–coated slides and dried for 16 h at 37°C before they were rehydrated in a graded series of ethanol. Tissue penetration was enhanced by incubation in 0.1% pepsin in 0.2 M HCl for 15 min at 37°C, followed by fixation in 4% paraformaldehyde in PBS for 5 min and incubation in 1% hydroxylammoniumchloride for 15 min. After this, sections were dehydrated in ethanol and air dried. Hybridization took place for 1 h at 55°C in hybridization buffer (10% sodium dextran sulfate, 50% formamide, 4× salt and sodium citrate (SSC), 1× Denhardt's and 200 μg/ml yeast tRNA; 1× SSC = 0.15 M NaCl and 0.015 M sodium citrate) with 500 ng/ml DIG-labeled probe. After stringency washes in 2× SSC, 1× SSC, 0.5× SSC, and 0.1× SSC for 30 min at 37°C, sections were rinsed for 10 min in Tris-buffered saline (TBS), blocked in blocking solution (1% BSA and 2% normal goat serum in TBS) for 30 min, and incubated with alkaline phosphatase (AP)-conjugated sheep anti-DIG Fab fragments (1:500; Roche Diagnostics) in blocking solution for 16 h at 4°C. After three washes of 10 min in TBS and one wash of 5 min in AP buffer (100 mM Tris and 100 mM NaCl [pH 9.5]), sections were stained in 350 μg/ml 4-nitro blue tetrazolium chloride and 175 μg/ml 5-bromo-4-chloro-3-indolyl-phosphate (Roche Diagnostics) in AP buffer until color development was sufficient. Then, sections were washed twice with distilled H2O and mounted in Mowiol. Alternating sections were used for AQP2 immunohistochemistry. After mounting of fixed sections on poly-l-lysine–coated slides and rehydration in graded series of ethanol, sections were washed with TBS, incubated with 1% H2O2 in TBS for 15 min to quench endogenous peroxidase activity, rinsed with TBS, incubated with 1:100 diluted rabbit anti-AQP2 antibodies in blocking buffer, washed three times with TBS, and incubated for 1 h in 1:100 diluted donkey anti-rabbit antibodies coupled to horseradish peroxidase (Jackson ImmunoResearch Laboratories, West Grove, PA). After washing three times in TBS, sections were incubated in diaminobenzidene until color development was sufficient. Then, sections were dehydrated in graded series of alcohol and ethylene and mounted in Entellan (Merck, Darmstadt, Germany). A sense LIP5 cRNA probe was taken along as a negative control for the specificity of the LIP5 mRNA hybridization.
mpkCCD Cells and shRNA Assays
MpkCCD cells (clone 1427) were grown in modified defined medium (DMEM:Ham's F12 1:1 vol/vol; 60 nM sodium selenate, 5 μg/ml transferrin, 2 mM glutamine, 50 nM dexamethasone, 1 nM triiodothyronine, 10 ng/ml EGF, 5 μg/ml insulin, 20 mM d-glucose, 2% FCS, and 20 mM HEPES [pH 7.4]) at 37°C in an air atmosphere of 5% CO2. The medium was replaced every 2 d. Exponentially growing cells (at approximately 70% confluence) were trypsinized and seeded at a density of 1.5 × 105 cells/cm2 on semipermeable filters (Transwell, 0.4-μm pore size; Corning Costar, Cambridge, MA) of 1.13 cm2. The cells remained in culture for a total period of 8 d before being analyzed. The cells were treated for the last 96 h with 1 nM dDAVP to only the basolateral side to induce AQP2 expression maximally. TPA (0.1 μM), cycloheximide (50 μM), MG132 (20 μM), and chloroquine (100 μM) were administered to both the apical and the basolateral compartments for the indicated time periods.
To make pLV-CMV-GFP-shRNA-LIP5 constructs for the LIP5 shRNA assays, we first created the pTER-LIP5 construct. Phosphorylated LIP5-specific oligonucleotides (10 μg) were annealed in 100 μl of 25 mM KCl by incubating the mixture at 95°C for 2 min and slowly cooling to room temperature. A total of 1 μl of the mixture was ligated into the BglII- and HindIII-digested pTER vector.52 The following oligonucleotides were used: 5′-GATCCCCGACAATGACTCCTGTGTGGTTCAAGAGACC-3′, 5′-CTTGAACCACACAGGAGTCATTGTCGGG-3′, 5′-ACACAGGAGTCATTGTC TTTTTGGAAA-3′, and 5′-AGCTTTTCCAAAAAGACAATGACTCCTGTGTGGTCT-3′ for pTER-LIP5–1; 5′GATCCCCTGAAGATCGAGCTGGACGATTCAAGAGATC-3′, 5′-CTTGAATCGTCCAGCTCGATCTTCAGGG-3′, 5′-GTCCAGCTCGATCTTCATTTTTGGAAA-3′, and 5′-AGCTTTTCCAAAAATGAAGATCGAGCTGGACGATCT-3′ for pTER-LIP5–2. To generate the lentiviral LIP5 shRNA constructs, we inserted the PstI fragments from the pTER-LIP5 constructs containing the H1 promoter and LIP5 shRNA sequences into the corresponding site of pLV-CMV-GFP.53
MpkCCD cells were stably transfected with pCB6-AQP2 using the calcium-phosphate precipitation technique as described previously.54 Third-generation lentiviruses were produced by co-transfection of the packaging vectors pRSV-Rev, pMDL g/p RRE, and pMD2G (Tronolab, Lausanne, Switzerland) and the transfer vector pLV-CMV-GFP-shRNA-LIP5 into human embryonic kidney 293T cells as described previously.55 The titer was determined by a p24 HIV ELISA (Murex Diagnostics, Dartford, United Kingdom). MpkCCD cells stably expressing AQP2 were infected with lentivirus immediately before being plated in the presence of Polybrene (8 μg/ml) using a multiplicity of infection of 20. The next day, medium was replaced. Immunoblotting and immunocytochemistry were performed 4 d after infection. The half-life of AQP2 was determined on the basis of the best fitting model of regression.
LIP5 Antibodies
To obtain rabbit anti-LIP5 antibodies, we induced expression of soluble GST-LIP5 with IPTG in DH5α bacteria transfected with pGEX6.1-LIP5 (made as described in the Pulldown Assays section) and isolated using glutathione-Sepharose 4B beads (Amersham Pharmacia Biotech, Uppsala, Sweden). After complete bleeding of the rabbits, anti-GST antibodies were removed by passing the serum over a GST-coupled Affi-gel 15 column (Amersham Pharmacia Biotech). Then, the flow-through was passed over a GST-LIP5–coupled Affi-gel 15 column, and the antibodies were eluted with 0.1 M glycine (pH 2.8), after which they were directly neutralized in 5× PBS (pH 7.4).
Immunohistochemistry
Mice were perfused with 1% (wt/vol) paraformaldehyde-lysine-periodate.56 The kidneys were removed, cut into 2- to 3-mm sections, and incubated in paraformaldehyde-lysine-periodate for 2 h. After fixation, the kidneys were dehydrated and embedded in paraffin. Five-micrometer sections were cut, stretched in 37°C water, and dried on gelatin-coated object glass (Menzel Gläser, Braunschweig, Germany) for at least 1 h at 37°C. Then, the sections were deparaffinized with xylol; rehydrated subsequently with 100, 96, 90, 80, 70, and 50% ethanol and water; and mounted in mowiol. Immunocytochemistry was done as described for MDCK cells.57 For detection of LIP5 and AQP2, the sections were incubated with affinity-purified rabbit anti-LIP5 antibodies (1:25), goat anti-rabbit antibodies coupled to Alexa 594 (Molecular Probes, Eugene, OR; 1:100), and, subsequently, with affinity-purified guinea pig anti-AQP258 and goat anti-guinea pig antibodies coupled to Alexa 488 (Molecular Probes; 1:100).
Immunoblotting
Immunoblotting was done as described previously.59 As antibodies, affinity-purified rabbit anti-AQP2 (1:300058), rabbit anti-LIP5 (1:1000), rabbit anti-AQP4 (1:150058), rabbit anti-GFP (1:5000; provided by Dr. B. Wieringa, UMC Nijmegen, Nijmegen, Netherlands), guinea pig anti-AQP2 (1:400058), mouse anti-Xpress (1:5000), or rabbit anti-EGFR 1005 (1:1000; Santa Cruz Biotechnology) was used. As secondary antibodies, goat anti-rabbit, goat anti-guinea pig, or sheep anti-mouse antibodies coupled to horseradish peroxidase (Sigma, St. Louis, MO; 1:5000) were used.
DISCLOSURES
None.
Supplementary Material
Acknowledgments
P.M.T.D. is a recipient of a VICI grant (865.07.002) of the Netherlands Organization for Scientific Research (NWO). This research was supported by grants from the Dutch Organization of Scientific Research (NWO-MW 902-18-292) to P.M.T.D. and P.v.d.S., and from the European Union (RTN aquaglyceroporins; 035995-2), kidney foundation (C03.2060), UMC St Radboud (2004-55) and NWO (865.07.002) to P.M.T.D.
We thank Ronnie Wismans and Tony Coenen for superb technical support and Dr. S. Gisler (Department Physiology, University of Zurich, Zurich, Switzerland) for sending us material and sharing his technical expertise on our yeast two-hybrid analyses.
Published online ahead of print. Publication date available at www.jasn.org.
B.W.M.v.B., M.B., and G.H contributed equally to this work.
B.W.M.v.B.'s current affiliations is Department of Vascular Medicine, University Medical Center Utrecht, Utrecht, Netherlands. G.H.'s current affiliation is Department of Toxicogenetics, Leiden University Medical Center, Leiden, Netherlands. E.-J.K.'s current affiliation is Department of Human Genetics, Radboud University Medical Center Nijmegen, Nijmegen, Netherlands. H.C.S.'s current affiliation is Department of Otorhinolaryngology, University Medical Center Utrecht, Utrecht, Netherlands.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1.Chou CL, Yip KP, Michea L, Kador K, Ferraris JD, Wade JB, Knepper MA: Regulation of aquaporin-2 trafficking by vasopressin in renal collecting duct: Roles of ryandoine-sensitive Ca2+ stores and calmodulin. J Biol Chem 275: 36839–36846, 2000 [DOI] [PubMed] [Google Scholar]
- 2.Agre P, King LS, Yasui M, Guggino WB, Ottersen OP, Fujiyoshi Y, Engel A, Nielsen S: Aquaporin water channels: From atomic structure to clinical medicine. J Physiol 542: 3–16, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deen PMT, Van Balkom BWM, Kamsteeg EJ: Routing of the aquaporin-2 water channel in health and disease. Eur J Cell Biol 79: 523–530, 2000 [DOI] [PubMed] [Google Scholar]
- 4.Chou CL, Christensen BM, Frische S, Vorum H, Desai RA, Hoffert JD, de Lanerolle P, Nielsen S, Knepper MA: Non-muscle myosin II and myosin light chain kinase are downstream targets for vasopressin signaling in the renal collecting duct. J Biol Chem 279: 49026–49035, 2004 [DOI] [PubMed] [Google Scholar]
- 5.Stefan E, Wiesner B, Baillie GS, Mollajew R, Henn V, Lorenz D, Furkert J, Santamaria K, Nedvetsky P, Hundsrucker C, Beyermann M, Krause E, Pohl P, Gall I, MacIntyre AN, Bachmann S, Houslay MD, Rosenthal W, Klussmann E: Compartmentalization of cAMP-dependent signaling by phosphodiesterase-4D is involved in the regulation of vasopressin-mediated water reabsorption in renal principal cells. J Am Soc Nephrol 18: 199–212, 2007 [DOI] [PubMed] [Google Scholar]
- 6.McSorley T, Stefan E, Henn V, Wiesner B, Baillie GS, Houslay MD, Rosenthal W, Klussmann E: Spatial organisation of AKAP18 and PDE4 isoforms in renal collecting duct principal cells. Eur J Cell Biol 85: 673–678, 2006 [DOI] [PubMed] [Google Scholar]
- 7.Kamsteeg EJ, Heijnen I, van Os CH, Deen PMT: The subcellular localization of an aquaporin-2 tetramer depends on the stoichiometry of phosphorylated and nonphosphorylated monomers. J Cell Biol 151: 919–930, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Balkom BWM, Savelkoul PJ, markovich D, Hofman E, Nielsen S, van der Sluijs P, Deen PMT: The role of putative phosphorylation sites in the targeting and shuttling of the aquaporin-2 water channel. J Biol Chem 277: 41473–41479, 2002 [DOI] [PubMed] [Google Scholar]
- 9.Katsura T, Gustafson CE, Ausiello DA, Brown D: Protein kinase A phosphorylation is involved in regulated exocytosis of aquaporin-2 in transfected LLC-PK1 cells. Am J Physiol 41: F816–F822, 1997 [PubMed] [Google Scholar]
- 10.Nishimoto G, Zelenina M, Li D, Yasui M, Aperia A, Nielsen S, Nairn AC: Arginine vasopressin stimulates phosphorylation of aquaporin-2 in rat renal tissue. Am J Physiol 276: F254–F259, 1999 [DOI] [PubMed] [Google Scholar]
- 11.Christensen BM, Zelenina M, Aperia A, Nielsen S: Localization and regulation of PKA-phosphorylated AQP2 in response to V(2)-receptor agonist/antagonist treatment. Am J Physiol Renal Physiol 278: F29–F42, 2000 [DOI] [PubMed] [Google Scholar]
- 12.De Mattia F, Savelkoul PJ, Kamsteeg EJ, Konings IB, van der SP, Mallmann R, Oksche A, Deen PM: Lack of arginine vasopressin-induced phosphorylation of aquaporin-2 mutant AQP2–R254L explains dominant nephrogenic diabetes insipidus. J Am Soc Nephrol 16: 2872–2880, 2005 [DOI] [PubMed] [Google Scholar]
- 13.McDill BW, Li SZ, Kovach PA, Ding L, Chen F: Congenital progressive hydronephrosis (cph) is caused by an S256L mutation in aquaporin-2 that affects its phosphorylation and apical membrane accumulation. Proc Natl Acad Sci U S A 103: 6952–6957, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuwahara M, Iwai K, Ooeda T, Igarashi T, Ogawa E, Katsushima Y, Shinbo I, Uchida S, Terada Y, Arthus MF, Lonergan M, Fujiwara TM, Bichet DG, Marumo F, Sasaki S: Three families with autosomal dominant nephrogenic diabetes insipidus caused by aquaporin-2 mutations in the C-terminus. Am J Hum Genet 69: 738–748, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mulders SM, Bichet DG, Rijss JPL, Kamsteeg EJ, Arthus MF, Lonergan M, Fujiwara M, Morgan K, Leijendekker R, van der Sluijs P, van Os CH, Deen PMT: An aquaporin-2 water channel mutant which causes autosomal dominant nephrogenic diabetes insipidus is retained in the Golgi complex. J Clin Invest 102: 57–66, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kamsteeg EJ, Wormhoudt TA, Rijss JPL, van Os CH, Deen PMT: An impaired routing of wild-type aquaporin-2 after tetramerization with an aquaporin-2 mutant explains dominant nephrogenic diabetes insipidus. EMBO J 18: 2394–2400, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SW, Jeon YS, Lee JU, Kang DG, Kook H, Ahn KY, Kim SZ, Cho KW, Kim NH, Han JS, Choi KC: Diminished adenylate cyclase activity and aquaporin 2 expression in acute renal failure rats. Kidney Int 57: 1643–1650, 2000 [DOI] [PubMed] [Google Scholar]
- 18.Kuwahara M, Iwai K, Uchida S, Gu Y, Terada Y, Sato K, Asai T, Bichet D, Sasaki S, Marumo F: A novel mutation in the aquaporin-2 (AQP2) gene causing autosomal dominant nephrogenic diabetes insipidus [Abstract]. J Am Soc Nephrol 9: 390A, 1998 [Google Scholar]
- 19.Marr N, Bichet DG, Lonergan M, Arthus MF, Jeck N, Seyberth HW, Rosenthal W, van Os CH, Oksche A, Deen PMT: Heteroligomerization of an aquaporin-2 mutant with wild-type aquaporin- 2 and their misrouting to late endosomes/lysosomes explains dominant nephrogenic diabetes insipidus. Hum Mol Genet 11: 779–789, 2002 [DOI] [PubMed] [Google Scholar]
- 20.Kamsteeg EJ, Hendriks G, Boone M, Konings IB, Oorschot V, van der SP, Klumperman J, Deen PM: Short-chain ubiquitination mediates the regulated endocytosis of the aquaporin-2 water channel. Proc Natl Acad Sci U S A 103: 18344–18349, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Noda Y, Horikawa S, Furukawa T, Hirai K, Katayama Y, Asai T, Kuwahara M, Katagiri K, Kinashi T, Hattori M, Minato N, Sasaki S: Aquaporin-2 trafficking is regulated by PDZ-domain containing protein SPA-1. FEBS Lett 568: 139–145, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Kamsteeg EJ, Duffield AS, Konings IB, Spencer J, Pagel P, Deen PM, Caplan MJ: MAL decreases the internalization of the aquaporin-2 water channel. Proc Natl Acad Sci U S A 104: 16696–16701, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu HA, Sun TX, Matsuzaki T, Yi XH, Eswara J, Bouley R, McKee M, Brown D: Heat shock protein 70 interacts with aquaporin-2 and regulates its trafficking. J Biol Chem 282: 28721–28732, 2007 [DOI] [PubMed] [Google Scholar]
- 24.Ward DM, Vaughn MB, Shiflett SL, White PL, Pollock AL, Hill J, Schnegelberger R, Sundquist WI, Kaplan J: The role of LIP5 and CHMP5 in multivesicular body formation and HIV-1 budding in mammalian cells. J Biol Chem 280: 10548–10555, 2005 [DOI] [PubMed] [Google Scholar]
- 25.Deen PMT, van Os CH: Epithelial aquaporins. Curr Opin Cell Biol 10: 435–442, 1998 [DOI] [PubMed] [Google Scholar]
- 26.Nielsen S, Frokiaer J, Marples D, Kwon TH, Agre P, Knepper MA: Aquaporins in the kidney: From molecules to medicine. Physiol Rev 82: 205–244, 2002 [DOI] [PubMed] [Google Scholar]
- 27.Hasler U, Mordasini D, Bens M, Bianchi M, Cluzeaud F, Rousselot M, Vandewalle A, Feraille E, Martin PY: Long-term regulation of aquaporin-2 expression in vasopressin-responsive renal collecting duct principal cells. J Biol Chem 277: 10379–10386, 2002 [DOI] [PubMed] [Google Scholar]
- 28.Li Y, Shaw S, Kamsteeg EJ, Vandewalle A, Deen PM: Development of lithium-induced nephrogenic diabetes insipidus is dissociated from adenylyl cyclase activity. J Am Soc Nephrol 17: 1063–1072, 2006 [DOI] [PubMed] [Google Scholar]
- 29.Shiflett SL, Ward DM, Huynh D, Vaughn MB, Simmons JC, Kaplan J: Characterization of Vta1p, a class E Vps protein in Saccharomyces cerevisiae. J Biol Chem 279: 10982–10990, 2004 [DOI] [PubMed] [Google Scholar]
- 30.Shim S, Merrill SA, Hanson PI: Novel interactions of ESCRT-III with LIP5 and VPS4 and their implications for ESCRT-III disassembly. Mol Biol Cell 19: 2661–2672, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Azmi IF, Davies BA, Xiao J, Babst M, Xu Z, Katzmann DJ: ESCRT-III family members stimulate Vps4 ATPase activity directly or via Vta1. Dev Cell 14: 50–61, 2008 [DOI] [PubMed] [Google Scholar]
- 32.Xiao J, Xia H, Zhou J, Azmi IF, Davies BA, Katzmann DJ, Xu Z: Structural basis of Vta1 function in the multivesicular body sorting pathway. Dev Cell 14: 37–49, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Azmi I, Davies B, Dimaano C, Payne J, Eckert D, Babst M, Katzmann DJ: Recycling of ESCRTs by the AAA-ATPase Vps4 is regulated by a conserved VSL region in Vta1. J Cell Biol 172: 705–717, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Scott A, Gaspar J, Stuchell-Brereton MD, Alam SL, Skalicky JJ, Sundquist WI: Structure and ESCRT-III protein interactions of the MIT domain of human VPS4A. Proc Natl Acad Sci U S A 102: 13813–13818, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Babst M: A protein's final ESCRT. Traffic 6: 2–9, 2005 [DOI] [PubMed] [Google Scholar]
- 36.Morita E, Sundquist WI: Retrovirus budding. Annu Rev Cell Dev Biol 20: 395–425, 2004 [DOI] [PubMed] [Google Scholar]
- 37.Pisitkun T, Shen RF, Knepper MA: Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci U S A 101: 13368–13373, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nejsum LN, Zelenina M, Aperia A, Frokiaer J, Nielsen S: Bidirectional regulation of AQP2 trafficking and recycling: Involvement of AQP2–S256 phosphorylation. Am J Physiol Renal Physiol 288: F930–F938, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA: Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci U S A 103: 7159–7164, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tchernev VT, Mansfield TA, Giot L, Kumar AM, Nandabalan K, Li Y, Mishra VS, Detter JC, Rothberg JM, Wallace MR, Southwick FS, Kingsmore SF: The Chediak-Higashi protein interacts with SNARE complex and signal transduction proteins. Mol Med 8: 56–64, 2002 [PMC free article] [PubMed] [Google Scholar]
- 41.Faigle W, Raposo G, Tenza D, Pinet V, Vogt AB, Kropshofer H, Fischer A, Saint-Basile G, Amigorena S: Deficient peptide loading and MHC class II endosomal sorting in a human genetic immunodeficiency disease: The Chediak-Higashi syndrome. J Cell Biol 141: 1121–1134, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shukla A, Hager H, Corydon TJ, Bean AJ, Dahl R, Vajda Z, Li H, Hoffmann HJ, Nielsen S: SNAP-25-associated Hrs-2 protein colocalizes with AQP2 in rat kidney collecting duct principal cells. Am J Physiol Renal Physiol 281: F546–F556, 2001 [DOI] [PubMed] [Google Scholar]
- 43.Girsch SJ, Peracchia C: Calmodulin interacts with a C-terminus peptide from the lens membrane protein MIP26. Curr Eye Res 10: 839–849, 1991 [DOI] [PubMed] [Google Scholar]
- 44.Bartel PL, Fields S: Analyzing protein-protein interactions using two-hybrid system. Methods Enzymol 254: 241–263, 1995 [DOI] [PubMed] [Google Scholar]
- 45.Madrid R, Le Maout S, Barrault MB, Janvier K, Benichou S, Merot J: Polarized trafficking and surface expression of the AQP4 water channel are coordinated by serial and regulated interactions with different clathrin-adaptor complexes. EMBO J 20: 7008–7021, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gietz RD, Schiestl RH, Willems AR, Woods RA: Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360, 1995 [DOI] [PubMed] [Google Scholar]
- 47.Dalton S, Treisman R: Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell 68: 597–612, 1992 [DOI] [PubMed] [Google Scholar]
- 48.Hoffman CS, Winston F: A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene 57: 267–272, 1987 [DOI] [PubMed] [Google Scholar]
- 49.Deen PMT, Verdijk MAJ, Knoers NVAM, Wieringa B, Monnens LAH, van Os CH, van Oost BA: Requirement of human renal water channel aquaporin-2 for vasopressin-dependent concentration of urine. Science 264: 92–95, 1994 [DOI] [PubMed] [Google Scholar]
- 50.van Balkom BW, Van Raak M, Breton S, Pastor-Soler N, Bouley R, van der SP, Brown D, Deen PM: Hypertonicity is involved in redirecting the aquaporin-2 water channel into the basolateral, instead of the apical, plasma membrane of renal epithelial cells. J Biol Chem 278: 1101–1107, 2003 [DOI] [PubMed] [Google Scholar]
- 51.Karunagaran D, Tzahar E, Liu N, Wen D, Yarden Y: Neu differentiation factor inhibits EGF binding: A model for trans-regulation within the ErbB family of receptor tyrosine kinases. J Biol Chem 270: 9982–9990, 1995 [DOI] [PubMed] [Google Scholar]
- 52.van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H: Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep 4: 609–615, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Seppen J, Rijnberg M, Cooreman MP, Oude Elferink RP: Lentiviral vectors for efficient transduction of isolated primary quiescent hepatocytes. J Hepatol 36: 459–465, 2002 [DOI] [PubMed] [Google Scholar]
- 54.Deen PMT, Nielsen S, Bindels RJM, van Os CH: Apical and basolateral expression of aquaporin-1 in transfected MDCK and LLC-PK cells and functional evaluation of their transcellular osmotic water permeabilities. Pflugers Arch 433: 780–787, 1997 [DOI] [PubMed] [Google Scholar]
- 55.Dull T, Zufferey R, Kelly M, Mandel RJ, Nguyen M, Trono D, Naldini L: A third-generation lentivirus vector with a conditional packaging system. J Virol 72: 8463–8471, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McLean IW, Nakane PK: Periodate-lysine-paraformaldehyde fixative: A new fixation for immunoelectron microscopy. J Histochem Cytochem 22: 1077–1083, 1974 [DOI] [PubMed] [Google Scholar]
- 57.Deen PMT, Van Balkom BWM, Savelkoul PJ, Kamsteeg EJ, Van Raak M, Jennings ML, Muth TR, Rajendran V, Caplan MJ: Aquaporin-2: COOH terminus is necessary but not sufficient for routing to the apical membrane. Am J Physiol Renal Physiol 282: F330–F340, 2002 [DOI] [PubMed] [Google Scholar]
- 58.Deen PMT, van Aubel RA, van Lieburg AF, van Os CH: Urinary content of aquaporin 1 and 2 in nephrogenic diabetes insipidus. J Am Soc Nephrol 7: 836–841, 1996 [DOI] [PubMed] [Google Scholar]
- 59.Deen PMT, Croes H, van Aubel RA, Ginsel LA, van Os CH: Water channels encoded by mutant aquaporin-2 genes in nephrogenic diabetes insipidus are impaired in their cellular routing. J Clin Invest 95: 2291–2296, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Deen PMT, Rijss JPL, Mulders SM, Errington RJ, van Baal J, van Os CH: Aquaporin-2 transfection of Madin-Darby canine kidney cells reconstitutes vasopressin-regulated transcellular osmotic water transport. J Am Soc Nephrol 8: 1493–1501, 1997 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.