Abstract
The molecular changes in the parenchyma that reflect disturbances in the function of kidney transplants are unknown. We studied the relationships among histopathology, gene expression, and renal function in 146 human kidney transplant biopsies performed for clinical indications. Impaired function (estimated GFR) correlated with tubular atrophy and fibrosis but not with inflammation or rejection. Functional deterioration before biopsy correlated with inflammation and tubulitis and was greater in cases of rejection. Microarray analysis revealed a correlation between impaired renal function and altered expression of sets of transcripts consistent with tissue injury but not with those consistent with cytotoxic T cell infiltration or IFN-γ effects. Multivariate analysis of clinical variables, histologic lesions, and transcript sets confirmed that expression of injury-related transcript sets independently correlated with renal function. Analysis of individual genes confirmed that the transcripts with the greatest positive or negative correlations with renal function were those suggestive of response to injury and parenchymal dedifferentiation not inflammation. We defined new sets of genes based on individual transcripts that correlated with renal function, and these highly correlated with the previously developed injury sets and with atrophy and fibrosis. Thus, in biopsies performed for clinical reasons, functional disturbances are reflected in transcriptome changes representing tissue injury and dedifferentiation but not the inflammatory burden.
The molecular changes in the parenchyma that reflect disturbances in renal function in kidney transplants or native kidneys are unknown. Kidney transplant function is determined by a combination of donor and recipient factors, peritransplant stress, and factors in the posttransplantation course, such as nephrotoxic drugs, infections, and recurrent renal disease.1,2 In normal kidney, we expect no inherent correlation between the GFR and the molecules expressed: Normal functioning kidneys vary widely in GFR, yet all should have a similar transcriptome, just as they have normal histology. In diseased kidneys, some structure–function relationships are known: Particularly, the histopathology features fibrosis and atrophy correlate with GFR.3–6 Renal function at the time of a biopsy reflects baseline function and recent functional changes caused by stress. It is thus conceivable that in stressed kidneys the functional disturbance is reflected in molecular changes.
Relatively few relationships between molecules and function have been established in renal disease. In native kidney disease, fibronectin and TGFβ mRNA expression were associated with function at time of biopsy and functional recovery after biopsy.7 In kidney transplants, granzyme-B-positive interstitial cells and intragraft surfactant protein-C mRNA expression were associated with more late allograft dysfunction and graft loss.8,9 In protocol transplant biopsies, higher CD3g mRNA expression correlated with worse renal function after 2 yr.10 To date, no comprehensive analysis of the relationship between transcriptome changes and renal allograft function has been performed.
The emergence of microarrays opens a new window on exploring the biologic processes in deteriorating renal allografts. To facilitate understanding of the relationship of the transcriptome to basic mechanisms and histopathology, we annotated transcripts in experimental models according to their contribution to specific biologic processes during renal allograft rejection: effector T cell infiltration, IFN-γ effects, macrophage infiltration, tissue injury and repair (IRITs), and kidney parenchymal transcripts (KTs).11–14 IRITs and KTs reflect different aspects of reversible parenchymal injury: IRITs are increased, while KTs are decreased during injury. These transcript sets provide a system for relating transcriptome changes to changes in structure, function, and outcomes. We have previously shown that these transcript sets can be successfully applied to human clinical states to provide diagnostic insight.15
Our hypothesis is that, in the abnormal kidney, disturbances in the transcriptome will reflect functional changes and will correlate with disturbances in GFR. The present study analyzed the relationship between gene expression and renal allograft function in human kidney transplant biopsies for clinical indications. We analyzed both single genes and previously identified transcript sets11–16 to relate our findings to biologic processes of known relevance for the pathogenesis of rejection and other forms of renal injury.
RESULTS
Patient Demographics and Characteristics of Estimated GFR in the Biopsy-for-Cause Population
We analyzed gene expression in relationship to renal function in biopsies for cause from 153 renal transplant patients taken between 1 wk and 32 yr posttransplant. Fifty-two biopsies were diagnosed as rejection or borderline rejection by histology (borderline T cell-mediated rejection (TCMR), n = 18 (11% of all biopsies); TCMR, n = 21 (13%); antibody-mediated rejection (ABMR), n = 10 (7%); mixed ABMR and TCMR, n = 3 (2%)). The nonrejecting biopsies displayed a wide spectrum of disease states, including ATN (n = 12 (8%)), BK virus nephropathy (n = 4 (3%)), transplant glomerulopathy (n = 11 (7%)), tubular atrophy and interstitial fibrosis otherwise not specified (n = 10 (7%)), recurrent or de novo glomerulonephritis (n = 26 (17%)), calcineurin inhibitor toxicity (n = 21 (13%)), and diabetic nephropathy (n = 4 (3%)). Demographics and clinical data are shown in Supplementary Table 1. Of 153 biopsies available for analysis, we excluded seven biopsies from patients with delayed graft function who were dialysis dependent at the time of biopsy, due to inability to calculate estimated GFR (eGFR).
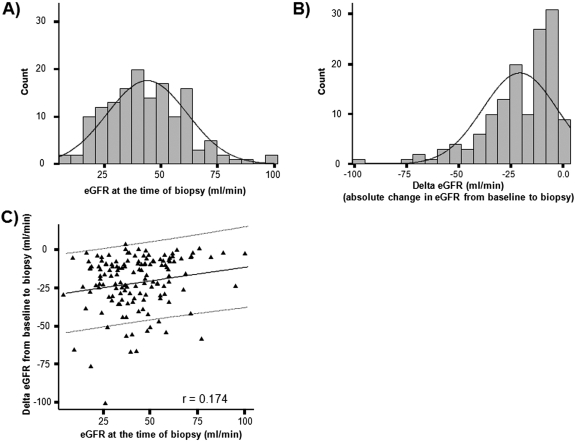
The eGFR values at biopsy ranged from 5.0 to 96.6 ml/min (mean eGFR, 43.7 ± 17.7) and were normally distributed in the patient population (Figure 1A). The degree of functional change from baseline (delta eGFR) within 4 mo before the biopsy ranged from 3.4 to −101 ml/min (mean, −21.1 ± 17.9 ml/min) (Figure 1B). Of 141 patients with baseline eGFR measurements available, 96 patients (68%) had loss of eGFR from baseline greater than 10 ml/min (delta eGFR < −10 ml/min), whereas 45 patients (32%) had relatively stable function (delta eGFR > −10 ml/min). There was a weak but significant relationship (r = 0.174, P < 0.05) between eGFR at biopsy and delta GFR from baseline to biopsy (Figure 1C).
Figure 1.
Distribution of renal function and degree of functional deterioration from baseline in the population. To assess if the range of measurements in our study population is suitable for analysis of histologic and molecular correlates of renal function, we assessed (A) the distribution of renal function (GFR) at the time of biopsy and (B) the distribution of functional deterioration from baseline (delta eGFR) in our patient population. Delta eGFR was calculated by subtracting baseline eGFR (within 4 mo before the biopsy) from eGFR at the time of biopsy. (C) We analyzed the relationship between eGFR at the time of biopsy and delta eGFR to assess how much eGFR at the time of biopsy is influenced by recent changes in eGFR.
Associations of Histologic Banff Lesions and Demographic Variables with Renal Function
In univariate analyses of clinical variables and histologic lesions, lower eGFR at biopsy was associated with interstitial fibrosis (ci-score) and tubular atrophy (ct-score) (P < 0.05) but not with inflammation, whereas functional deterioration from baseline was associated with inflammation (P < 0.05), tubulitis (P < 0.01), and peritubular capillaritis (P < 0.01) (Table 1). Donor or recipient race; donor gender, type, and age; previous transplants; HLA mismatch; panel reactive antibodies (PRA) class I or class II at time of biopsy; and cold ischemia time did not correlate with eGFR measurements. Recipient gender and age were not included in the analysis because they are used in the Cockroft–Gault equation to calculate eGFR. However, when we analyzed the correlation between these factors and reciprocal serum creatinine, no correlation was found.
Table 1.
Relationship between renal function at the time of biopsy or functional deterioration and histologic lesions
| Histologic Lesionsa | Spearman Correlation Coefficient
|
|
|---|---|---|
| Function at the Time of Biopsyb | Functional Change from Baseline to Biopsyb | |
| g | 0.010 | −0.130 |
| cg | 0.095 | 0.088 |
| i | −0.080 | −0.171* |
| ci | −0.189* | 0.093 |
| t | 0.029 | −0.251** |
| ct | −0.162* | 0.055 |
| v | 0.149 | −0.082 |
| cv | 0.047 | 0.158 |
| ah | 0.084 | 0.241** |
| mm | −0.006 | 0.058 |
| ptc | 0.049 | −0.190* |
| ptcml | −0.159 | 0.162 |
g-score, glomerulitis; cg-score, glomerulopathy; i-score, inflammation; ci-score, interstitial fibrosis; t-score, tubulitis; ct-score, tubular atrophy; v-score, intimal arteritis, cv-score, vascular fibrous intimal thickening; ah-score, arteriolar hyaline thickening; mm-score, mesangial matrix increase; ptc-score, peritubular capillaritis; ptcml, peritubular basement membrane multilayering.
delta GFR = eGFR at biopsy − eGFR at baseline; thus negative delta GFR values indicate loss of function.
P < 0.05;
P < 0.001.
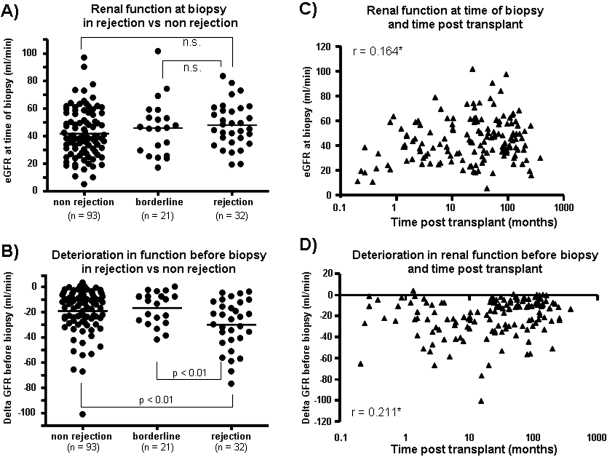
To establish the characteristics of eGFR measurements in our patient population, we assessed whether rejection or time posttransplant had an impact on GFR measurements. The eGFR at biopsy was similar between biopsies with or without rejection (Figure 2A), but the change in eGFR before the biopsy was greater in rejection (−29.9 ± 19.5 ml/min) than nonrejection biopsies (−19.1 ± 17.5 ml/min; P < 0.01) (Figure 2B). Time of the biopsy posttransplant showed a significant but weak correlation with eGFR at the time of biopsy (r = 0.164, P < 0.05) (Figure 2C). The difference in eGFR values between early and late biopsies was mainly driven by very early biopsies taken within 1 mo posttransplant that had low eGFR values. After these biopsies were eliminated, there was no relationship between eGFR and time.
Figure 2.
Relationship between renal function at biopsy and functional deterioration before biopsy to diagnosis of rejection or time posttransplant. We assessed whether biopsies with rejection (A) had different levels of eGFR at the time of biopsy compared with biopsies without rejection or borderline rejection or (B) differed in the degree of functional deterioration from baseline. Lines represent the mean in each group. The relationships between time posttransplant and (C) eGFR at biopsy and (D) functional deterioration from baseline are also shown. * P < 0.05.
The delta eGFR before biopsy correlated weakly with time posttransplant (r = 0.211, P < 0.05) (Figure 2D). Biopsies taken early (<12 mo posttransplant) had more change in eGFR (−25.6 ± 17.2 ml/min) before the biopsy than those taken late (−18.8 ± 17.9; P = 0.03), but both groups showed similar ranges of delta eGFR. More kidneys biopsied late had relatively stable function (delta eGFR > −10 ml/min) than those biopsied early: 37% of late biopsies versus 21% of early biopsies.
Relationship between Pathogenesis-Based Transcript Expression and Renal Allograft Function at Biopsy
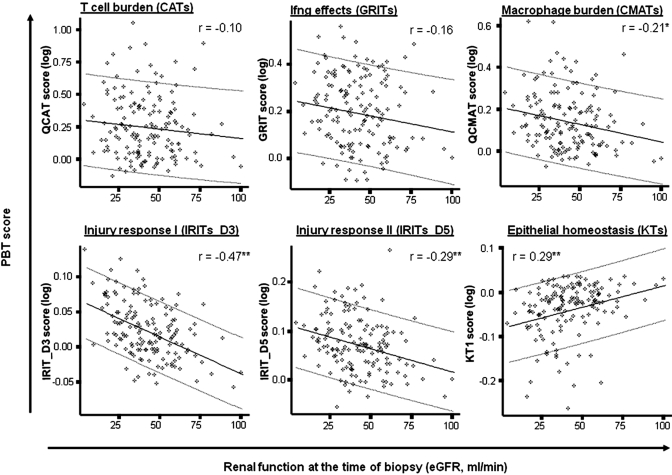
We compared eGFR at the time of biopsy and expression of transcript sets associated with T cell infiltration (CATs), IFN-γ effects (GRITs), macrophage infiltration/activation (MATs), and tissue injury (IRITs or KTs) (Figure 3). The strongest correlation was observed with injury-related transcript sets: Low eGFR was associated with increased expression of IRITs and loss of KTs (IRIT_D3, r = −0.47; KTs, r = 0.29). Low eGFR was also associated with high expression of macrophage transcript sets, albeit weakly (r values between −0.18 and −0.27). The inflammatory T cell burden, represented by CATs and GRITs, showed no significant relationship to eGFR at the time of biopsy. The delta eGFR had weaker relationships with injury transcript sets (IRITs_D3: r = −0.26, IRITs_D5: r = −0.22, KTs: r = 0.23, P < 0.01) but significant correlations with inflammation, including macrophage-associated transcript sets (CMATs, r = −0.29; IMATs, r = −0.32; P < 0.05), T cell infiltration (CATs, r = −0.23; P < 0.05), and IFN-γ effects (GRITs, r = −0.29; P < 0.01). These results were similar in biopsies with or without rejection and were not changed by excluding very early biopsies (<1 mo posttransplant).
Figure 3.
Relationship between eGFR and expression of PBTs. To assess whether there is any relationship between eGFR at the time of biopsy and molecular changes in the biopsy, we analyzed the correlation between eGFR and expression of previously defined PBTs that reflect the T cell burden (CATs), IFN-γ effects (GRITs), macrophage burden (CMATs), the tissue injury response (IRITs_D3 and IRITs_D5), and epithelial cell integrity (KTs). Expression of each transcript set within each biopsy was summarized as the geometric mean expression of all transcripts in that set to derive a gene set score. The gene set score for each biopsy is plotted against the corresponding eGFR value for that biopsy. Numbers represent Spearman rank correlation coefficients. * P < 0.05; ** P < 0.01.
To assess if transcript expression reflects renal function independent of clinical or histologic variables, we performed a multivariate analysis using clinical variables, histologic lesions, and transcript set scores as input variables (Table 2). This analysis showed that, in addition to time posttransplant and the presence of atrophy and fibrosis, expression of injury-related transcripts (IRIT_D3) independently correlated with eGFR at biopsy. This observation was also confirmed for biopsies with or without Banff rejection and for either early or late biopsies (Table 2). The KTs were not retained in this model, which is most likely due to the high correlation between IRITs and KTs (r = −0.713 (KTs and IRITs_D3) and r = −0.768 (KTs and IRITs_D5)).
Table 2.
Multivariate analysis of association between demographic variables, Banff lesions, PBT expression, and renal allograft function at biopsy
| Biopsy Groups | Variables Included in the Model | Slope | P value | R2 (%) |
|---|---|---|---|---|
| All biopsies | ||||
| IRIT_D3 expression | −180.3 | <0.01 | 28.3 | |
| Time posttransplant | 5.9 | <0.01 | ||
| Interstitial fibrosis (ci-score)a | −8.1 | 0.01 | ||
| Biopsies with rejection versus biopsies without rejection | ||||
| With rejection (n = 32) | IRIT_D5 expression | −152.1 | <0.01 | 34.6 |
| Peritubular capillaritis (PTC-score) | −11.4 | 0.02 | ||
| Without rejection (n = 93) | IRIT_D3 expression | −208.7 | <0.01 | 24.0 |
| Biopsies early versus biopsies late posttransplant | ||||
| Early (<12 mo) (n = 55) | IRIT_D3 expression | −109.3 | 0.047 | 17.3 |
| Arteriolar hyalinosis (ah-score) | −5.7 | 0.024 | ||
| Late (≥12 mo) (n = 98) | IRIT_D3 expression | −230.5 | <0.001 | 33.5 |
| Tubular atrophy (ct-score) | −6.7 | 0.040 |
Could be replaced by tubular atrophy (ct-score) and slope. P value and R2 value remain unchanged.
Relationship between Individual Transcript Expression and Function at Biopsy
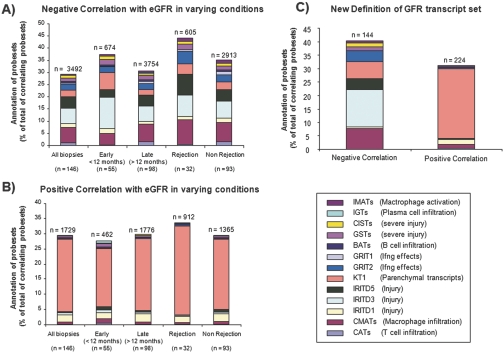
We analyzed the relationship between individual transcripts and eGFR at biopsy, using all transcripts passing a variance-based filter, independent of their membership in previously annotated transcript sets. This analysis identified many transcripts correlating positively (n = 1798) or negatively (n = 3602) with eGFR (uncorrected P < 0.05). Of the transcripts with negative correlations (i.e., increased expression with low eGFR), 29.4% had been annotated by our previous algorithms; the majority of these were injury-related (IRITs or KTs) or macrophage-associated transcripts (Figure 4A). Of the transcripts positively correlated with eGFR (i.e., decreased expression with low eGFR), 29.5% had been annotated; these were mainly decreased parenchymal transcripts (KTs) (Figure 4B). The top 30 transcripts with positive or negative correlations are shown in Table 3. The associations between transcripts and eGFR were similar in biopsies with or without rejection and in early and late biopsies (Figure 4, A and B). Thus, renal function at biopsy is reflected in the transcriptome and is represented by the molecular response of the tissue to injury but not by the inflammatory burden.
Figure 4.
Single gene analysis of molecular correlates of eGFR. We analyzed the associations between eGFR at the time of biopsy and transcript changes independent of previous annotations. We identified all transcripts with a significant correlation (uncorrected P value <0.05) between their expression level and eGFR values. To consider transcripts with increased or decreased expression, we analyzed positive (i.e., decreased with low eGFR) and negative (i.e., increased with low eGFR) correlations separately. To account for possible differences in biopsies with or without rejection or between early and late biopsies, the analysis was performed separately for these subgroups in addition to an analysis across all biopsies, resulting in a total of ten transcript sets (five with positive and five with negative correlations with eGFR). We searched these transcript sets for transcripts that had previously been identified as part of a PBT. The distribution of these annotations among those transcripts correlating with eGFR is shown as a percent of total correlating transcripts (A) for those with negative correlations (increased expression with low eGFR) and (B) for those transcripts with positive correlations (decreased expression with eGFR). Numbers represent the total number of correlating transcripts in each group. The distribution of these annotations was very similar between the biopsy subgroups, indicating that the presence or absence of rejection and the time of biopsy do not have a significant impact on the relationship between transcript expression and eGFR. (C) To create gene sets with robust correlations between transcript expression and eGFR across all biopsy subgroups, we identified those transcripts that were identified in the analyses shown in Panels A and B in all subgroup analyses. The resulting gene lists then comprise a new eGFR transcript set that can be used to assess its performance in the biopsies in relationship to histologic lesions and clinical parameters.
Table 3.
Top 30 transcripts that showed strongest correlation with eGFR at biopsy (all P values <0.001)
| Positively Correlated
|
Negatively Correlated
|
||||||
|---|---|---|---|---|---|---|---|
| AffyID | Gene Symbol | PBTsa | Correlation | AffyID | Gene Symbol | PBTsa | Correlation |
| 227560_at | SFXN2 | 0.47 | 209529_at | PPAP2C | −0.59 | ||
| 226325_at | ADSSL1 | 0.47 | 226535_at | ITGB6 | KT1 | −0.57 | |
| 206878_at | DAO | KT1 | 0.46 | 203892_at | WFDC2 | IRIT_D3 | −0.57 |
| 209368_at | EPHX2 | KT1 | 0.46 | 241031_at | NLF1 | −0.55 | |
| 214433_s_at | SELENBP1 | KT1 | 0.46 | 227372_s_at | BAIAP2L1 | −0.53 | |
| 221675_s_at | CHPT1 | KT1 | 0.46 | 226621_at | FGG | −0.53 | |
| 218025_s_at | PECI | 0.46 | 229435_at | GLIS3 | −0.52 | ||
| 209757_s_at | MYCN | 0.46 | 201596_x_at | KRT18 | IRIT_D3 | −0.50 | |
| 227417_at | MOSC2 | 0.46 | 225105_at | OCC-1 | −0.50 | ||
| 204430_s_at | SLC2A5 | KT1 | 0.46 | 210986_s_at | TPM1 | −0.50 | |
| 217973_at | DCXR | KT1 | 0.45 | 203021_at | SLPI | −0.50 | |
| 219732_at | RP11–35N6.1 | 0.45 | 202430_s_at | PLSCR1 | −0.50 | ||
| 206457_s_at | DIO1 | KT1 | 0.45 | 227530_at | AKAP12 | IRIT_D3 | −0.50 |
| 204294_at | AMT | 0.45 | 201510_at | ELF3 | IRIT_D3 | −0.50 | |
| 213967_at | RALYL | 0.45 | 227875_at | KLHL13 | −0.50 | ||
| 241949_at | ACOT6 | 0.45 | 209008_x_at | KRT8 | IRIT_D3 | −0.50 | |
| 205364_at | ACOX2 | KT1 | 0.45 | 213506_at | F2RL1 | IRIT_D3 | −0.49 |
| 206840_at | AFM | KT1 | 0.45 | 225520_at | MTHFD1L | −0.49 | |
| 221163_s_at | MLXIPL | KT1 | 0.45 | 232850_at | DCDC2 | −0.49 | |
| 230949_at | SLC23A3 | 0.45 | 225872_at | SLC35F5 | −0.49 | ||
| 205216_s_at | APOH | 0.44 | 204259_at | MMP7 | IRIT_D5 | −0.49 | |
| 242956_at | IDH1 | 0.44 | 201428_at | CLDN4 | IRIT_D3 | −0.49 | |
| 235512_at | CDKL1 | KT1 | 0.44 | 225478_at | MFHAS1 | −0.49 | |
| 206325_at | SERPINA6 | 0.44 | 205466_s_at | HS3ST1 | −0.48 | ||
| 205075_at | SERPINF2 | KT1 | 0.44 | 206600_s_at | SLC16A5 | −0.48 | |
| 236774_at | DDC | KT1 | 0.44 | 209154_at | TAX1BP3 | IRIT_D3 | −0.48 |
| 215726_s_at | CYB5A | 0.44 | 228284_at | TLE1 | −0.48 | ||
| 244627_at | DAK | KT1 | 0.44 | 203476_at | TPBG | −0.48 | |
| 205983_at | DPEP1 | KT1 | 0.44 | 221261_x_at | MAGED4B | −0.48 | |
| 211303_x_at | PSMAL | 0.44 | 208700_s_at | TKT | −0.48 | ||
IRIT_D3 and IRIT_D5, injury and repair induced transcripts that peak at day 3 and day 5, respectively; KT1, renal transcripts with decreased expression in injury.
Defining a New eGFR-Associated Transcript Set
We used the associations between transcript disturbances and eGFR disturbances to create two new eGFR-associated transcript sets (eGFRTs). These were defined as those transcripts correlating either positively or negatively with eGFR in all biopsy subgroups (all biopsies, early and late biopsies, and biopsies with or without rejection). Those transcripts positively correlated with eGFR (i.e., decreased when eGFR was lower) were summarized in one transcript set (n = 224). Those transcripts correlated negatively with eGFR (i.e., increased in kidneys with lower eGFR) were summarized in a second transcript set (n = 144). The transcripts included in these sets and their correlation with eGFR are shown in Supplementary Tables 2 and 3.
Similar to the transcripts correlating with eGFR in the subgroups (described above), the majority of annotated transcripts in these sets were parenchymal transcripts (IRITs and KTs) (Figure 4C). We confirmed the statistical significance of the enrichment of KTs and IRITs_D3 in the GFR transcript sets by a hypergeometric test (Supplementary Table 4).
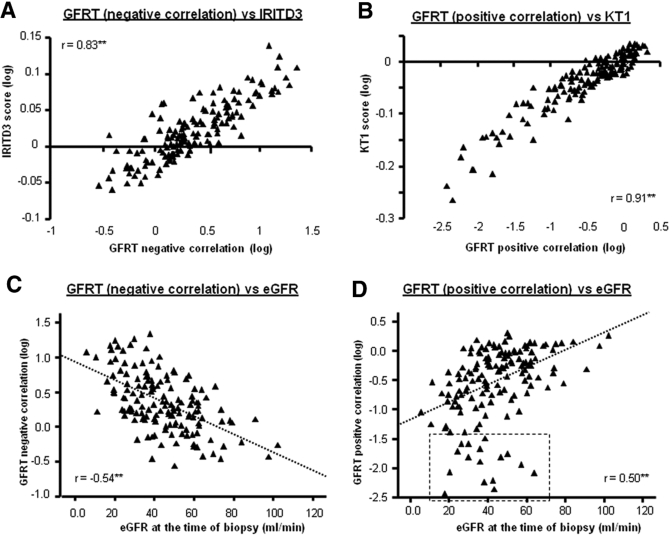
Analogous to the previously defined pathogenesis-based transcript sets (PBTs), we calculated a score for eGFRTs for each biopsy and compared these to observed eGFRs, other transcript sets, and histologic lesions. Although correlations were observed for all PBTs, the most striking correlations were seen with IRITs_D3 (r = 0.83; P < 0.01)) (Figure 5A) and KTs (r = 0.91) (Figure 5B). The high correlations were surprising, considering that only 30% of transcripts within each eGFRT had previously been annotated as members of transcripts sets. The correlations with other PBTs are shown in Supplementary Figures S1 and S2.
Figure 5.
Performance of eGFR transcript sets in relationship to previously defined transcript sets and eGFR at the time of biopsy. We assessed the relationship of the eGFR transcript sets in relation to our previously identified PBTs. The most striking relationships were observed with transcript sets reflecting tissue injury (IRITs_D3) and epithelial integrity (KTs). The results are shown in Panels A and B). We assess the performance of the eGFR transcript sets in relation to the actual eGFR of the biopsy (Panels C and D). An assessment of the outliers is presented in the text.
The eGFRTs correlated with the same lesions as eGFR (ci-score and ct-score), but in addition they also correlated with some lesions associated with functional deterioration (i-score, t-score, and ah-score) (Table 4). The correlations of the eGFRTs with the actual eGFR in each biopsy were moderate but significant (r = −0.54 and r = 0.50) (Figure 5, C and D), similar to the IRIT_D3 set. Some biopsies were outliers, deviating from the regression line due to more disturbed eGFRT expression than expected for their eGFR (box in Figure 5D). The biopsies with a higher loss of eGFRT transcripts than reflected in the loss of eGFR displayed more inflammation (i-score) (P < 0.01) and tubulitis (t-score) (P < 0.05), less arteriolar hyalinosis (ah-score) (P < 0.05), and greater functional deterioration from baseline (delta eGFR) (P < 0.05).
Table 4.
Performance of the GFRTs in relationship to eGFR and lesions
| Correlation Coefficients | GFRT Negative | GFRT Positive |
|---|---|---|
| Histologic lesions (Banff scores)a | ||
| g | 0.09 | −0.10 |
| cg | 0.03 | 0.03 |
| i | 0.33** | −0.32** |
| ci | 0.29** | −0.20** |
| t | 0.16 | −0.19* |
| ct | 0.29** | −0.20* |
| v | −0.02 | −0.02 |
| cv | −0.09 | 0.17* |
| ah | −0.17* | 0.21* |
| mm | 0.17 | 0.05 |
| ptc | 0.07 | −0.08 |
| Renal function | ||
| eGFR at biopsy | −0.54** | 0.50** |
| Functional deterioration from baseline (delta eGFR) | −0.28** | −0.30** |
g-score, glomerulitis; cg-score, glomerulopathy; i-score, inflammation; ci-score, interstitial fibrosis; t-score, tubulitis; ct-score, tubular atrophy; v-score, intimal arteritis; cv-score, vascular fibrous intimal thickening; ah-score, arteriolar hyaline thickening; mm-score, mesangial matrix increase; ptc-score, peritubular capillaritis.
P < 0.05;
P < 0.01.
Validation of Results
We validated the results in an independent validation set. The demographics of the patient population in the validation set (n = 132) were comparable to those in the original data set, with a similar distribution of eGFR values.
We repeated the gene set and single gene analyses in the validation set. This analysis confirmed the significant correlation of injury-related gene sets (IRITs and KTs) with eGFR (IRITs_D3, r = −0.32; P < 0.01; KTs, r = 0.26; P < 0.01), whereas inflammatory gene sets (QCATs, GRITs, and QMATs) did not correlate significantly with eGFR. Single gene analysis confirmed that the majority of annotated transcripts that correlated with eGFR were annotated as injury-related transcripts (IRITs and KTs).
In a second validation step, we used the transcripts identified in the original data set and assessed their performance in the validation set. This analysis confirmed a significant correlation with eGFR for 28 of the 30 top transcripts that positively correlated with eGFR and for 28 of the top 30 transcripts that negatively correlated with eGFR in the original data set. When we applied the GFR-related gene sets (identified in the original data set) to the validation set, we confirmed that the GFRTs recapitulate expression of the injury-related genes sets (IRITs (r = 0.65; P < 0.01) and KTs (r = 0.90; P < 0.01)) and correlate significantly with eGFR in the validation set (r = 0.35 and r = −0.29 for the eGFRT with negative or positive correlation, respectively (P < 0.01)). In contrast, none of the histologic lesions correlated with eGFR in the validation set.
DISCUSSION
The present study assessed the relationship between changes in gene expression and disturbances in renal allograft function. Using previously annotated transcript sets that reflect biologic events, we found that impaired function correlated with increased expression of tissue injury gene sets and decreased expression of kidney transcripts, indicating dedifferentiation of the epithelium. These results were confirmed in multivariate analysis and were consistent in biopsies with or without rejection and in those taken early or late posttransplant. Surprisingly, lower renal function did not correlate with gene sets reflecting inflammation. Analysis of the correlations of eGFR with individual transcripts (independent of their membership in transcript sets) yielded similar conclusions: Many transcripts identified in this unbiased analysis had already been annotated as transcripts increased with injury (IRITs) or decreased after injury (KT1s), whereas transcripts reflecting inflammation were not detected. Thus, the examination of all transcripts in the biopsy independently confirmed the important relationship of the IRIT and KT changes with functional disturbances.
Transcriptome abnormalities that correlate with eGFR provide new information beyond the information obtained from histology and demographics, as shown in multivariate analysis. The transcript changes correlating with eGFR disturbances in transplants reflect the injury response of the parenchyma, with expression of developmental genes and loss of transcripts associated with differentiated epithelium. The correlations are impressive considering that renal function in a normal kidney population depends on variables such as kidney size, age, and gender, which are not strongly represented as transcriptome differences. Thus normal kidneys—with normal histology, normal function, and normal transcriptome—should display no correlation between transcripts and eGFR. The transcript changes correlating with GFR disturbances in biopsies for cause measure perturbations in the parenchyma and are indicators of an ongoing and potentially reversible epithelial injury process13,14 and potentially the determinants of atrophy–scarring versus normal repair, which in turn will be associated with loss of function.
The GFR-related transcript sets correlated strongly with the behavior of the previously annotated IRIT and KT injury transcript sets, but the fact that only approximately one-third of the GFR transcripts overlapped with KTs and IRITs points to heterogeneity in the injury-induced transcriptome changes. We anticipated heterogeneity in the IRITs from our previous studies in mouse models of immune and nonimmune injury.13 We had not analyzed differential expression of the KTs in mouse models, but the results of the single gene analysis in the present study shows that a subgroup of KTs has considerably stronger correlation with renal function than others, indicating extensive heterogeneity in the KTs in relationship to renal function disturbances. The varying degrees of correlations of individual IRITs and KTs with renal function open the possibility of understanding the biologic processes that contribute to epithelial deterioration and warrant further investigation.
The consistent relationship between injury transcripts and renal function across all biopsy subgroups indicates that the tissue injury response is a common pathway linking mechanisms of renal injury (inflammation and noninflammatory disease states) to renal dysfunction. The changes in parenchymal transcript expression are a stereotyped response of the tissue to injury, representing a selective structured program that leads to loss of function. This is in keeping with the fact that these transcripts are readily induced in allografts as well as in isografts in a mouse model13 and are observed even in injured kidneys lacking histologic changes such as necrosis or inflammation that are destined for full recovery. Some of these transcripts, such as SELENBP1,17 matrix metallopeptidase 7,18 and ITGB619 have previously been reported to be associated with renal disease. Thus, the transcript changes reflect a perturbation in kidney biology that correlates with disturbed function but is not related to a particular mechanism of injury. Mechanisms directly altering the epithelium could include soluble effector T cell or macrophage products. In addition, the epithelium may change in response to changes in the extracellular matrix or the microcirculation. The lack of correlation between renal function and inflammation-related transcripts implies that the determinant of the functional disturbance in inflammation is the parenchymal response.
The GFRT transcript sets showed some correlation with inflammation and inflammatory transcript sets in addition to their correlation with eGFR and with GFR-related histologic lesions—fibrosis and atrophy—indicating that some of these transcripts can be changed without immediately impacting function. This interpretation is also supported by the fact that some biopsies diverge from the regression line between GFR transcript sets and eGFR, thus showing more disturbance in the GFR transcript set than reflected in the GFR. Changes in GFR probably require relatively severe damage to the epithelium, and not every molecular disturbance of the epithelium immediately results in GFR changes. It is possible that the parenchymal transcriptome changes are a more sensitive indicator of parenchymal disturbances than the GFR, and the molecular changes provide a more direct indication of ongoing parenchymal disturbance than eGFR measurements. Such changes are reversible if the insult is removed13,14 but probably progress to more severe epithelial changes with shutdown and loss of nephrons if the insult is sustained.
The results of this study are based on observations in transplant biopsies taken for clinical indications. However, it seems likely that these relationships between transcript changes and function are applicable to native kidney disease and will find more general utility. We anticipate that some protocol biopsies will show similar changes in GFR transcript expression that will not be reflected in GFR changes in those biopsies, which by definition have stable function. By reflecting the intensity of the parenchymal response to injury and functional perturbation, the injury- and eGFR-related transcripts represent a window on the pathogenesis of epithelial changes in human disease states.
CONCISE METHODS
Patient Population Specimens and Data Collection
The study was approved by the institutional review board of the University of Alberta (issue no. 5299). Written informed consent was obtained from all study patients. All renal transplant biopsies for cause from each patient (deterioration in function, stable impaired function, or proteinuria) as standard of care between January 2004 and October 2007 were included. In the case of multiple biopsies from one patient, only the first biopsy was included in the analysis. To exclude effects of antirejection treatment on the transcriptome, we excluded biopsies that received treatment before the biopsy (intravenous steroid within 7 d, depleting or nondepleting antibody, intravenous Ig, or plasmapheresis within 1 mo before biopsy).
We validated out results in an independent validation set, which consists of n = 132 kidney transplant biopsies from n = 132 patients. The biopsies in the validation set were taken at the University of Alberta between March 2007 and May 2008 (n = 21), at the University of Illinois at Chicago between January 2007 and August 2007 (n = 13), at the University of Minnesota Medical Center at Fairview between October 2006 and September 2007 (n = 82), and at the Hennepin County Medical Center, Minneapolis, Minnesota, between September 2006 and August 2007 (n = 16). All biopsies were taken for clinical indication. None or these biopsies and none of these patients were included in the data set used for our original analysis because microarray results on these biopsies were not available at the time of our original analyses.
Biopsies were obtained under ultrasound guidance by spring-loaded needles (ASAP Automatic Biopsy, Microvasive, Watertown, MA). In addition to the cores obtained for pathology, we collected one core for gene expression. Clinical data were collected for each patient and entered into a Laboratory Information Management System (LIMS) developed in our laboratory. Histologically normal renal cortical tissue from eight native nephrectomies performed for renal carcinoma served as controls. The biopsy sample processing was performed as described in detail in our previous study.20
Histopathologic Diagnosis
All biopsies were assessed using the Banff 97 criteria21 by a pathologist (B.S.) who was blinded to the results of molecular studies. All biopsies had adequate cortical tissue for analysis by Banff criteria with the exception of six biopsies (two biopsies had one artery and four biopsies had no arteries).
Assessment of Renal Function
Renal function was defined by eGFR (Cockcroft–Gault) as the lowest eGFR within 1 wk before or after the time of biopsy (eGFR at biopsy). Baseline function was defined as the highest eGFR within 4 mo before the biopsy. Deterioration in renal allograft function before the biopsy (delta eGFR) was defined as the difference between eGFR at biopsy and baseline eGFR. We repeated the analysis using either reciprocal serum creatinine or the Modification of Diet in Renal Disease (MDRD) equation as estimates of renal function; these analyses yielded similar results in terms of the associations of molecular changes with eGFR.
Pathogenesis-Based Transcript Sets
We previously identified PBTs that reflect major biologic processes during renal allograft rejection: effector T cell infiltration (QCATs, n = 25),22 IFN-γ effects (GRIT1s, n = 68),12 macrophage infiltration (QCMATs, n = 68), different stages of tissue injury and repair (IRITs_D3, n = 820; IRITs_D5, n = 578),13 and loss of epithelial integrity reflected by loss of kidney parenchymal transcripts (KT1s, n = 1481).14 Thus, IRITs and KTs reflect different aspects of ongoing but probably reversible parenchymal injury. The IRITs were defined by being increased in isograft kidneys (i.e., subjected to the stresses of surgery, anesthesia, and ischemia but ultimately all destined for full recovery and lacking histologic changes such as necrosis or inflammation). Subsets (IRITs_D3 and IRITs_D5) were defined by the day of peak expression in mice (day 3 or day 5 posttransplant, respectively). The KTs were defined by high expression in normal mouse kidney but not in inflammatory cells. We confirmed in previous experiments that the loss of parenchymal transcripts during rejection was not attributable to dilution by other cell types but probably reflects a selective and structured program of the nephron.14
For annotation of individual transcripts identified by correlation with eGFR, we used additional transcript sets from previous studies reflecting severe injury (CISTs and GSTs),23 B cell infiltration (BATs), and plasma cell infiltration (IGTs).24 The transcript sets were originally derived from mouse kidneys and cell cultures and were translated into the corresponding human transcripts (http://www.affymetrix.com/). The algorithms and probe sets for each transcript are available at (http://transplants.med.ualberta.ca/).
Data Analysis
Data files were preprocessed using robust multichip averaging in Bioconductor and subjected to variance-based filtering25 as described previously.12 Expression of individual genes was analyzed as fold change compared with control kidneys.
We used two strategies to explore the association between gene expression and renal function: (1) comparison of the relationship between eGFR and expression of PBTs and (2) analysis of the relationship between eGFR and individual genes, independent of their membership in any previously identified gene sets. For the analysis of PBT expression, expression of transcripts within each PBT was summarized as the geometric mean of fold changes across all probe sets, resulting in the PBT score. The relationship between PBT scores and eGFR was then analyzed by Spearman rank correlation. The influence of multiple variables on renal function was analyzed by multiple regression. A P value <0.05 was considered statistically significant. To identify individual transcripts that correlate with eGFR, we assessed the correlation of all transcripts passing the variance-based filter with eGFR and selected those with a P value <0.05 for further analysis. To assess whether the resulting gene list was significantly enriched in transcripts reflecting any particular biologic process, we assessed the enrichment of individual gene sets within the GFR transcript sets by a hypergeometric test (implemented in Bioconductor software). This test assesses the number of transcripts from a given gene set (e.g., KTs or IRITs) that were found within the GFR transcript sets and compares it to the number that would have been expected (based on the number of genes in the transcript sets that entered the selection for the GFR transcript set), thus permitting us to test if the GFR transcript sets contain more genes from any given gene list than would have been expected by chance alone.
Relationships between eGFR and transcript changes were assessed separately in a priori defined subgroups according to time posttransplant or the presence of rejection. For this purpose, rejection was defined as a histopathologic diagnosis of ABMR, TCMR, or mixed ABMR and TCMR. Biopsy without rejection was defined by other than biopsy with rejection but excluded biopsy with suspicious ABMR or borderline TCMR.
We used log transformation to correct data skewness in those variables that were not normally distributed, including cold ischemia time, time posttransplant, and PBT scores. We used independent t test to compare the mean eGFR between groups. Pearson's or Spearman's correlation coefficient was used to correlate eGFR with variables that were normally or non-normally distributed, respectively. Statistical analyses were performed using SPSS 14.0 statistical software package (SPSS, Inc., Chicago, IL).
DISCLOSURES.
The authors have no competing financial interests.
Supplementary Material
Acknowledgments
The authors give special thanks and appreciation to Dr. Arthur Matas, Department of Surgery, University of Minnesota, and to Bertram L. Kasiske, Department of Medicine, University of Minnesota, for providing renal transplant biopsy tissue samples and corresponding clinical data for the validation set of this study. The authors also thank Dr. Zija Jacaj, Debra Lieberman, and Patricia West-Thielke for help with collection of the clinical data and Vido Ramassar, Anna Hutton, Stacey Lacoste, and Sujatha Guttikonda for technical support. This research has been supported by funding or resources from Genome Canada, Genome Alberta, the University of Alberta, the University of Alberta Hospital Foundation, Alberta Advanced Education and Technology, Roche Molecular Systems, Hoffmann-La Roche Canada Ltd., the Alberta Ministry of Advanced Education and Technology, the Roche Organ Transplant Research Foundation, the Kidney Foundation of Canada, and Astellas Canada. P.F.H. also holds a Canada Research Chair in Transplant Immunology and the Muttart Chair in Clinical Immunology.
Published online ahead of print. Publication date available at www.jasn.org.
S.B. and G.E. contributed equally to this work.
Supplemental information for this article is available online at http://www.jasn.org/.
REFERENCES
- 1.Suri D, Meyer TW: Influence of donor factors on early function of graft kidneys. J Am Soc Nephrol 10: 1317–1323, 1999 [DOI] [PubMed] [Google Scholar]
- 2.Keith DS, DeMattos A, Golconda M, Prather J, Cantarovich M, Paraskevas S, Tchervenkov J, Norman DJ: Factors associated with improvement in deceased donor renal allograft function in the 1990s. J Am Soc Nephrol 16: 1512–1521, 2005 [DOI] [PubMed] [Google Scholar]
- 3.Agodoa L, Eknoyan G, Ingelfinger J, Keane W, Mauer M, Mitch W, Striker G, Wilcox C: Assessment of structure and function in progressive renal disease. Kidney Int 52: S144–S150, 1997 [PubMed] [Google Scholar]
- 4.Bohle A, Grund KE, Mackensen S, Tolon M: Correlations between renal interstitium and level of serum creatinine. Morphometric investigations of biopsies in perimembranous glomerulonephritis. Virchows Arch A Pathol Anat Histol 373: 15–22, 1977 [DOI] [PubMed] [Google Scholar]
- 5.Bohle A, Christ H, Grund KE, Mackensen S: The role of the interstitium of the renal cortex in renal disease. Contrib Nephrol 16: 109–114, 1979 [DOI] [PubMed] [Google Scholar]
- 6.Schainuck LI, Striker GE, Cutler RE, Benditt EP: Structural-functional correlations in renal disease. II. The correlations. Hum Pathol 1: 631–641, 1970 [DOI] [PubMed] [Google Scholar]
- 7.Eikmans M, Baelde HJ, Hagen EC, Paul LC, Eilers PHC, De Heer E, Bruijn JA: Renal mRNA levels as prognostic tools in kidney diseases. J Am Soc Nephrol 14: 899–907, 2003 [DOI] [PubMed] [Google Scholar]
- 8.Mengel M, Mueller I, Behrend M, von Wasielewski R, Radermacher J, Schwarz A, Haller H, Kreipe H: Prognostic value of cytotoxic T-lymphocytes and CD40 in biopsies with early renal allograft rejection. Transpl Int 17: 293–300, 2004 [DOI] [PubMed] [Google Scholar]
- 9.Eikmans M, Roos-van Groningen MC, Sijpkens YW, Ehrchen J, Roth J, Baelde HJ, Bajema IM, de Fijter JW, De Heer E, Bruijn JA: Expression of surfactant protein-C, S100A8, S100A9, and B cell markers in renal allografts: Investigation of the prognostic value. J Am Soc Nephrol 16: 3771–3786, 2005 [DOI] [PubMed] [Google Scholar]
- 10.Kirk AD, Jacobson LM, Heisey DM, Radke NF, Pirsch JD, Sollinger HW: Clinically stable human renal allografts contain histological and RNA-based findings that correlate with deteriorating graft function. Transplant 68: 1578–1582, 1999 [DOI] [PubMed] [Google Scholar]
- 11.Hidalgo LG, Einecke G, Allanach K, Halloran PF: The transcriptome of human cytotoxic T cells: similarities and disparities among allostimulated CD4(+) CTL, CD8(+) CTL and NK cells. Am J Transplant 8: 627–636, 2008 [DOI] [PubMed] [Google Scholar]
- 12.Famulski KS, Einecke G, Reeve J, Ramassar V, Allanach K, Mueller T, Hidalgo LG, Zhu L-F, Halloran PF: Changes in the transcriptome in allograft rejection: IFN-γ induced transcripts in mouse kidney allografts. Am J Transplant 6: 1342–1354, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Famulski KS, Broderick G, Hay K, Cruz J, Sis B, Mengel M, Halloran PF: Transcriptome analysis reveals heterogeneity in the injury response of kidney transplants. Am J Transplant 7: 2483–2495, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Einecke G, Broderick G, Sis B, Halloran PF: Early loss of renal transcripts in kidney allografts: Relationship to the development of histologic lesions and alloimmune effector mechanisms. Am J Transplant 7: 1121–1130, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Mueller TF, Einecke G, Reeve J, Sis B, Mengel M, Jhangri G., Bunnag S, Cruz J, Wishart D, Meng C, Broderick G, Kaplan B, Halloran PF: Microarray analysis of rejection in human kidney transplants using pathogenesis-based transcript sets. Am J Transplant 7: 2712–2722, 2007 [DOI] [PubMed] [Google Scholar]
- 16.Einecke G, Melk A, Ramassar V, Zhu LF, Bleackley RC, Famulski KS, Halloran PF: Expression of CTL associated transcripts precedes the development of tubulitis in T-cell mediated kidney graft rejection. Am J Transplant 5: 1827–1836, 2005 [DOI] [PubMed] [Google Scholar]
- 17.Torrealba JR, Colburn M, Golner S, Chang Z, Scheunemann T, Fechner JH, Roenneburg D, Hu H, Alam T, Kim HT, Kanmaz T, Oberley T, Knechtle SJ, Hamawy MM: Selenium-binding protein-1 in smooth muscle cells is downregulated in a rhesus monkey model of chronic allograft nephropathy. Am J Transplant 5: 58–67, 2005 [DOI] [PubMed] [Google Scholar]
- 18.Surendran K, Simon TC, Liapis H, McGuire JK: Matrilysin (MMP-7) expression in renal tubular damage: Association with Wnt4. Kidney Int 65: 2212–2222, 2004 [DOI] [PubMed] [Google Scholar]
- 19.Trevillian P, Paul H, Millar E, Hibberd A, Agrez MV: αvβ6 Integrin expression in diseased and transplanted kidneys. Kidney Int 66: 1423–1433, 2004 [DOI] [PubMed] [Google Scholar]
- 20.Einecke G, Mueller TF, Famulski KS, Ramassar V, Sis B, Halloran PF: Cytotoxic T cells, interferon-γ and the renal response: Pathogenesis-based transcript sets have high diagnostic value in human kidney allograft rejection. Am J Transplant 6 (Suppl 12): 401, 2006 [Google Scholar]
- 21.Racusen LC, Solez K, Colvin RB, Bonsib SM, Castro MC, Cavallo T, Croker BP, Demetris AJ, Drachenberg CB, Fogo AB, Furness P, Gaber LW, Gibson IW, Glotz D, Goldberg JC, Grande J, Halloran PF, Hansen HE, Hartley B, Hayry PJ, Hill CM, Hoffman EO, Hunsicker LG, Lindblad AS, Marcussen N, Mihatsch MJ, Nadasdy T, Nickerson P, Olsen TS, Papadimitriou JC, Randhawa PS, Rayner DC, Roberts I, Rose S, Rush D, Salinas-Madrigal L, Salomon DR, Sund S, Taskinen E, Trpkov K, Yamaguchi Y: The Banff 97 working classification of renal allograft pathology. Kidney Int 55: 713–723, 1999 [DOI] [PubMed] [Google Scholar]
- 22.Hidalgo LG, Einecke G, Allanach K, Mengel M, Sis B, Mueller TF, Halloran PF: The transcriptome of human cytotoxic T cells: Measuring the burden of CTL-associated transcripts in human kidney transplants. Am J Transplant 8: 637–646, 2008 [DOI] [PubMed] [Google Scholar]
- 23.Famulski KS, Sis B, Billesberger L, Halloran PF: Interferon-γ and donor MHC class I control alternative macrophage activation and activin expression in rejecting kidney allografts: A shift in the Th1-Th2 paradigm. Am J Transplant 8: 547–556, 2008 [DOI] [PubMed] [Google Scholar]
- 24.Einecke G, Reeve J, Mengel M, Sis B, Bunnag S, Mueller TF, Halloran PF: Expression of B cell and immunoglobulin transcripts is a feature of inflammation in late allografts. Am J Transplant 8: 1434–1443, 2008 [DOI] [PubMed] [Google Scholar]
- 25.Gentleman R, Carey VJ, Huber W, Irizarry R, Dudoit S: Bioinformatics and Computational Biology Solutions Using R and Bioconductor, XII edn. Springer, New York, pp 229–248, 2005
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.