Summary
Short bowel syndrome is an acquired condition in which the length of the small intestine is insufficient to perform its normal absorptive function. Current therapies are limited as the developmental mechanisms that normally regulate elongation of the small intestine are poorly understood. Here, we identify Fgf9 as an important epithelial to mesenchymal signal required for proper small intestinal morphogenesis. Mouse embryos that lack either Fgf9 or the mesenchymal receptors for Fgf9 contained a disproportionately shortened small intestine, decreased mesenchymal proliferation, premature differentiation of fibroblasts into myofibroblasts and significantly elevated Tgf-β signaling. These findings suggest that Fgf9 normally functions to repress Tgf-β signaling in these cells. In vivo, a small subset of mesenchymal cell expressed phospho-Erk and the secreted Tgf-β inhibitors, Fst and Fstl1, in an Fgf9-dependent fashion. The p-Erk/Fst/Fstl1 expressing cells were most consistent with intestinal mesenchymal stem cells (iMSCs). We found that isolated iMSCs expressed p-Erk, Fst and Fstl1 and could repress the differentiation of intestinal myofibroblasts in co-culture. These data suggest a model in which epithelial-derived Fgf9 stimulates iMSCs that in turn regulate underlying mesenchymal fibroblast proliferation and differentiation at least in part through inhibition of Tgf-β signaling in the mesenchyme. Taken together, the interaction of FGF and TGF-β signaling pathways in the intestinal mesenchyme could represent novel targets for future short bowel syndrome therapies.
Keywords: Fibroblast growth factor 9 (Fgf9); gut development, Tgf-β signaling; epithelial-mesenchymal cross-talk, mesenchymal stem cells, follistatin
Introduction
The mammalian small intestine undergoes dramatic developmental changes during the last third of embryogenesis. In mice, this period begins at embryonic day (E) 13.5, when the small intestine is lined on its inner surfaced by a pseudo-stratified, uniformly proliferative endoderm (Kaufman, 1992). From E15.5 to E18.5, a proximal-to-distal wave of cytodifferentiation converts the epithelium to a simple columnar histology, organized into nascent villi with intervillus regions (Schmidt et al., 1988; Calvert and Pothier, 1990). A less appreciated developmental change that also occurs during this time period is a remarkable cephalo-caudal lengthening of the organ. Early in embryonic development, the lengthening of the small intestine occurs in proportion to total body length. However, in the latter third of embryogenesis, the rate of intestinal lengthening far outpaces that of the body, becoming exponential in both humans and rodents (Sbarbati, 1979; Weaver et al., 1991). As postnatal development commences, the rate of intestinal lengthening slows with a resumption of a linear relationship between the two.
In humans, the issue of intestinal lengthening becomes clinically important in patients with short bowel syndrome (Ladefoged et al., 1996). This condition most often occurs when a section of intestine is surgically removed postnatally for a variety of conditions in which a portion of the small intestine is no longer viable (Nightingale and Lennard-Jones, 1993). Following resection, an adaptive response occurs in which the diameter of the small intestine increases in response to loss of surface area. In mouse models of this process, crypts and villi lengthen due to increased epithelial progenitor proliferation and decreased epithelial turnover (Erwin et al., 2006; Dekaney et al., 2007, Wang et al., 2007). However, overall small intestinal length is never regenerated to any appreciable degree. Because the adaptive response is often inadequate to restore sufficient surface area for absorption, patients with short bowel syndrome are often treated with total parenteral nutrition, which has a 3-year survival rate of 65-80% (Howard et al., 1995). Thus, understanding the exponential lengthening of the small intestine in the mouse during late-stage embryonic development has potential clinical implications.
However, little is known regarding the developmental pathways that regulate small intestinal lengthening during this critical period. Most well-studied major developmental pathways play important roles in the proper patterning of the crypt-villus axis or the development of the outer muscle wall during embryogenesis. The formation of villus structures during the late prenatal period is dependent on an intact Pdgf pathway (Karlsson et al., 2000). The canonical Wnt pathway is essential for the regulation of epithelial stem cell homeostasis (Korinek et al., 1998). The Bmp arm of the Tgf-β superfamily regulates crypt census, and thus indirectly impacts stem cell number in the small intestine (Haramis et al., 2004). The ultimate cell fate decisions of committed stem cell daughters are regulated at least in part by the Notch signaling pathway (van Es et al., 2005; Fre et al., 2005; Vooijs et al., 2007). Finally, the outer muscular layer of the intestine depends upon hedgehog signals for its proper formation (Ramalho-Santos et al., 2000).
More recent studies have begun to elucidate how these disparate developmental pathways interact during intestinal embryogenesis. For instance, Wnt and Notch perform synergistic roles in the maintenance of proliferation and homeostasis of the epithelial stem cell compartment (van Es et al., 2005). Wnt and Indian hedgehog play antagonistic roles in the determination of epithelial cell differentiation in the colon (van den Brink et al., 2004). Finally, the Pten/Akt family interacts with Wnt/β-catenin signaling to regulate epithelial homeostasis (He et al., 2004, 2007). Thus, the interaction of multiple developmental pathways plays a critical role in several aspects of small intestinal development.
Fibroblast growth factors (Fgfs) comprise a large family of polypeptide growth factors that are found in organisms ranging from C. elegans to humans (Ornitz and Itoh, 2008). Specifically, Fgf9 plays a key role in embryonic development of the lung, heart, cecum and testes (Colvin et al., 2001a, 2001b; Lavine et al., 2005; White et al., 2006; Zhang et al., 2006). However, Fgf9 expression is not limited to these organs. We and others have previously shown that this growth factor is also expressed throughout the length of the intestine during late stage embryogenesis (Zhang et al., 2006; Sala et al., 2006). A careful study of the role of Fgf9 in the development of the intestine outside of the cecum has not been undertaken.
Here, by utilizing mice that lack mesenchymal Fgf9 signaling (either through loss of the Fgf9 ligand or its mesenchymal receptors), we show that Fgf9 signals are required for longitudinal growth of the small intestine during late-stage embryogenesis. Fgf9 signaling to the intestinal mesenchyme regulates proliferation of fibroblasts, which in turn appears to drive gut lengthening. Additionally, we found that Fgf9 inhibits the fibroblast to myofibroblast transition in the small intestinal mesenchyme during embryogenesis. Interestingly, this process is regulated by an indirect cellular mechanism that controls the Tgf-β signaling pathway in fibroblasts.
Results
Fgf9 is required for normal small intestinal development during late stage embryogenesis
We examined the gastrointestinal tract of wild type (WT), Fgf9+/- and Fgf9-/- E18.5 embryos. The length of the small intestine of Fgf9-/- embryos was significantly shorter than the length of controls at E14.5, E16.5 and E18.5 but not at E12.5 (Fig. 1A,B; defined as WT and Fgf9+/- littermates). In this and all other parameters examined in this manuscript, WT and Fgf9 +/- embryos were indistinguishable indicating no significant gene dosage effect occurred. Thus, WT and Fgf9 +/- were taken together as controls. No statistically significant differences were noted when comparing the crown-rump lengths of control and Fgf9-/- embryos at E18.5 (data not shown). In addition, the colon length was not significantly different when comparing control and Fgf9-/- embryos (average colon length=1.4±0.2 cm for Fgf9-/- and =1.5±0.3 cm for control). The ratio of small intestinal/colon length was 3.0 in Fgf9-/- and 4.3 in controls (35% reduction). Fgf9 expression was previously shown to be detectable in the epithelium starting at E12.5 (Zhang et al., 2006). Fgf9 expression persists in a similar distribution during late stage embryogenesis (Supp. Fig. 1A).
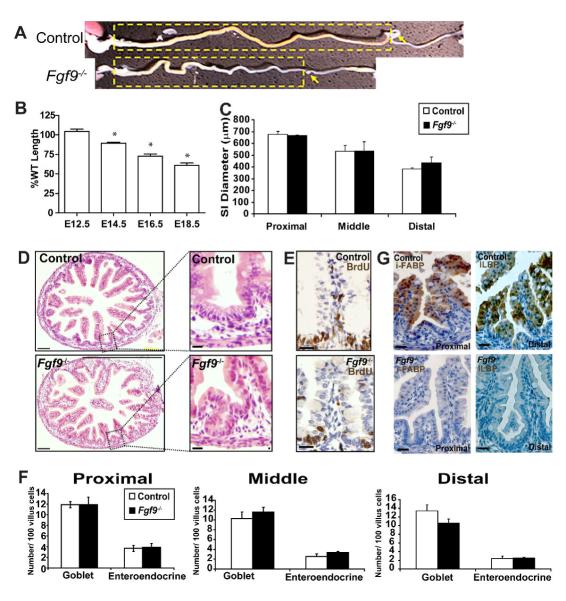
Fig. 1. Late stage Fgf9-/- embryos were disproportionately shortened as compared to littermate controls.
(A) Whole mount photograph of dissected gastrointestinal tracts of E18.5 Fgf9-/- and Fgf9+/- littermates. The small intestines are indicated by the yellow dashed boxes and the site of cecal development by the yellow arrows. The colons are to the right of the small intestines. (B) Quantification of the mean small intestinal lengths (+/-s.d.) of E12.5, E14.5, E16.5 and E18.5 Fgf9-/- embryos normalized to control littermates (=100%; control= WT and Fgf9+/- littermates). N=5-7 embryos/group. An asterisk indicates the difference of the means is statistically significant (as compared to controls of the same time point) by a Student’s t test (P<0.01). (C) Quantification of the mean small intestinal calibers measured at the mid-point of the proximal third, middle third and distal third (+/-s.d.) of E18.5 control and Fgf9-/- embryos. No statistically significant differences were observed between these two groups. (D) H + E stained cross-section of control and Fgf9-/- E18.5 proximal small intestines showing premature formation of crypt-like structures in Fgf9-/- mice that were not observed in controls (insets). (E) No other morphological differences were noted including restriction BrdU-positive cells to the intervillus epithelium (middle insets). (F) Quantification of mean number of goblet and enteroendocrine cells per 100 epithelial cells (+/-s.d.) in proximal, middle, and distal small intestines of E18.5 control and Fgf9-/- mice. No statistically significant differences were observed between these two groups. (G) Immunohistochemically-stained sections of E18.5 control (top) and Fgf9-/- (bottom) littermates showing that I-fabp (left panels) in the proximal small intestines and Ilbp (right panels) in the distal small intestines were expressed at detectable levels in villus enterocytes of controls but not Fgf9-/- embryos. Bars=100 μm (D) and =30 μm (D inset, E and F).
Interestingly, the defect in small intestinal length in the Fgf9-/- embryos was not accompanied by an alteration in its caliber. We measured caliber at the same relative position along the length of each individual small intestine (Fig. 1C). Thus, the Fgf9-/- small intestine was not a miniaturized version of controls, but instead was growth altered in only one dimension (length). In addition, more severe developmental defects such as small intestinal atresia, duplication or malrotation were never observed as have been previously noted as frequent malformations in other gene knockouts such as Shh-/- and Ihh-/- (Ramalho-Santos et al., 2000).
Histological analysis revealed that Fgf9-/- small intestines were phenotypically similar to littermate controls. Specifically, Fgf9-/- small intestines contained no obvious abnormalities of villus morphology such as bifurcation and fusion, or ectopic crypt placement/polyposis (Fig. 1D). The only morphologic abnormality that we observed in all E18.5 Fgf9-/- small intestines was premature crypt-like structures in the proximal small intestine (Fig. 1D, insets). Normally, crypt formation begins after birth and coincides with the appearance of Paneth cells in this structure. However the premature crypt-like structures in the Fgf9-/- mice did not express markers for Paneth cells such as lysozyme (data not shown).
Analysis of the intestinal epithelium provides a sensitive readout for interaction of Fgf9 with several major developmental pathways. Proliferative epithelial progenitors (as indicated by the presence of BrdU-positive cells) were only present in the intervillus epithelium of control and Fgf9-/- mice (Fig. 1E), indicating that the balance of Wnt and Bmp signaling was not significantly perturbed. The proper allocation of secretory and absorptive lineages is controlled by Notch signaling (van Es et al., 2005). The small intestinal epithelium of Fgf9-/- embryos contained similar proportions of goblet cells, enteroendocrine cells and enterocytes as those seen in their control littermates at E18.5 (Fig. 1F) indicating no severe defects were present in this pathway.
However, we found that Fgf9-/- small intestines showed a subtle defect in enterocytic differentiation at E16.5 and E18.5. These cells expressed lower amounts of intestinal fatty acid binding protein (I-fabp) (Cohn et al., 1992) in the proximal small intestine as well as lower levels of ileal lipid binding protein (Ilbp) in the distal small intestine (Fig. 1G; data not shown for E16.5). However, the enterocytes displayed no obvious alterations in polarity, apical junctions or size in either location of the small intestine.
Proliferation of small intestinal mesenchymal fibroblasts is driven by Fgf9 during late stage embryogenesis
Cell proliferation in the small intestine was quantified by BrdU labeling. We analyzed the three longitudinal layers of this organ (the intervillus epithelium, its underlying mesenchyme, and the muscularis propria) in control and Fgf9-/- embryos as one or more of these layers could drive intestinal elongation (Fig. 2A). We determined the proliferative index (PI) for each compartment as defined by the number of nuclei incorporating BrdU divided by the total number of nuclei in that compartment. We performed this analysis in both the proximal and distal small intestine at three developmental time points (E14.5, E16.5, and E18.5). The average PI for the intervillus epithelium and for the mucularis propria was not significantly different when comparing control and Fgf9-/- embryos at any location or timepoint (Fig. 2B and C shows data for the proximal small intestine at all timepoints). However, the PI for the mesenchyme was significantly reduced (P<0.01) in Fgf9-/- embryos as compared to their littermate controls in both the proximal and distal small intestine at all time points between E14.5 and E18.5 (Fig. 2C). To examine the effects of Fgf9 signaling loss on cell death, TUNEL stained sections of control and Fgf9-/- small intestines were examined. We observed no significant differences in apoptosis in any cellular compartment at any time point (E14.5-E18.5) during late-stage embryogenesis (data not shown).
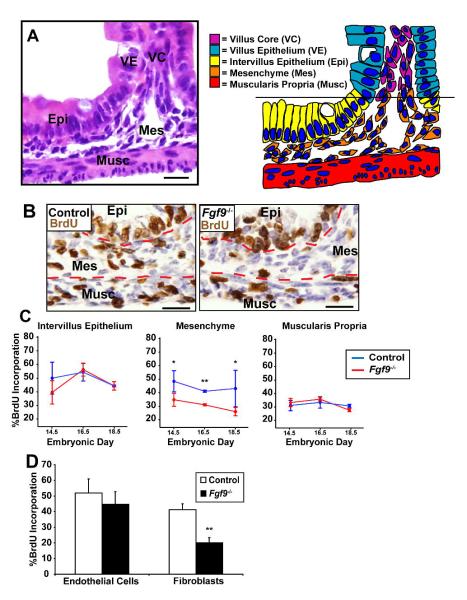
Fig. 2. Fgf9 regulates mesenchymal fibroblast proliferation.
(A) H + E stained section (left) and color-coded cartoon (right) showing micro-anatomical regions of a control E18.5 small intestine (VE=villus epithelium, VC=villus core, IE=intervillus epithelium, Mes=mesenchyme and Musc=muscularis propria). (B) Immunohistochemistry of BrdU stained sections of control (left) and Fgf9-/- (right) E18.5 proximal small intestines. The red dashed lines divide the epithelium, mesenchyme and muscle layers. (C) Quantification of the means (+/s.d.) of the proportion of S-phase cells (as indicated by BrdU incorporation) in the Epi, Mes and Musc for E14.5, E16.5 and E18.5 for control and Fgf9-/- embryos. Statistically significant differences between the two groups (N=4 embryos/group) were determined by a Student’s t test (double asterisk indicates P<0.01 and the single asterisk indicates P<0.05). (D) Quantification of the mean percent BrdU incorporation (+/-s.d.) in fibroblasts (prolyl 4-hydroxylase positive) and endothelial cells (Pecam-1 positive) from the mesenchyme of E18.5 control and Fgf9-/- small intestines. Statistically significant differences between the two groups were determined by comparison of SEMs using a Student’s t test; two asterisks denotes P<0.01; N=4 mice/group). Bars=30 μm.
The embryonic intestinal mesenchyme contains multiple cell types that may increase proliferation in response to Fgf9. The vast majority of the mesenchyme in E18.5 control small intestines was composed of prolyl 4-hydroxylase-positive, alpha smooth muscle actin (α-SMA)-negative fibroblasts (Powell et al., 2005). Two other smaller populations included Pecam1-positive endothelial cells and CD45-positive, F4/80-positive monocyte-derived cells. As expected, the monocyte-derived cells did not proliferate to any appreciable levels in the mesenchyme of these embryos (data not shown). In the absence of Fgf9, fibroblasts exhibited a significant decrease in proliferation (P<0.01; Fig. 2D) while endothelial cells maintained a proliferative rate similar to controls. These findings suggested that mesenchymal fibroblast proliferation contributed to small intestinal elongation during embryogenesis.
Fgf9 signals to mesenchymal Fgfr1 and Fgfr2 to drive proliferation in the small intestinal mesenchyme
Fgfr1c and Fgfr2c show functional redundancy in the development of several mouse organ systems (Lavine et al., 2005; White et al., 2006; Yu and Ornitz, 2008). To examine non-epithelial targets for Fgf9 and potential redundancy among Fgfrs, we conditionally inactivated floxed alleles of both Fgfr1 and Fgfr2 (Pirvola et al., 2002; Herbert et al., 2003; Trokovic et al., 2003; White et al., 2006) using the Dermo1-Cre allele (Yu et al., 2003). To define the targeted lineages in the small intestine, Dermo1-Cre mice were mated with ROSA26 reporter mice (Soriano, 1999). At E14.5, the mesenchyme and muscularis propria expressed Dermo1-Cre, as indicated by positive X-Gal staining in these regions (Fig. 3A). All mesenchymal lineages including fibroblasts, endothelial cells and macrophages contained detectable β-galactosidase while all epithelial cells were negative (Fig. 3A,B).
Fig. 3. Fgf9 signals through mesenchymal receptors to drive small intestine development/elongation.
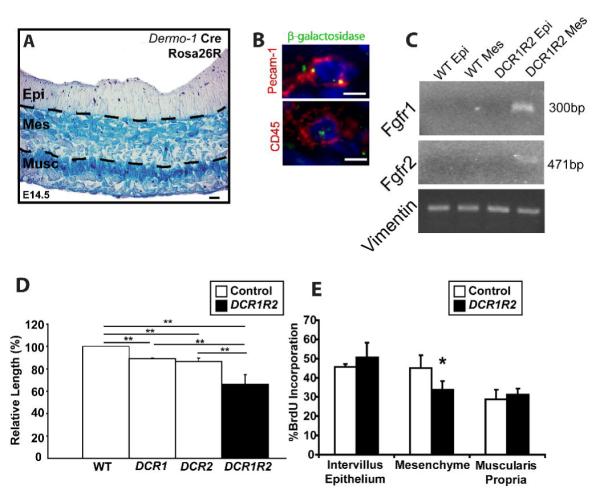
(A) Section of an E14.5 Dermo1-cre; Rosa26R small intestine stained with X-Gal. The epithelium, mesenchyme and muscularis propria are indicated. Blue staining showed regions of Dermo1-cre activity in all cells in the mesenchyme and muscularis propria. The epithelial compartment showed no detectable staining. (B) Section of an E18.5 Dermo1-cre; Rosa26R small intestine stained with anti-β-galactosidase and anti-Pecam (red, top) or CD45 (red, bottom). Both cells show co-localization of punctuate β-galactosidase with the cell lineage markers. No detectable β-galactosidase staining was detected in the epithelium. (C) PCR products of DNA that was procured from the epithelium and mesenchyme of control and DCR1R2 E18.5 small intestines. The recombined alleles of Fgfr1 and Fgfr2 were detected in the mesenchyme of DCR1R2 mice but not in the epithelium. PCR of vimentin is shown as a control for the isolation of the DNA. (D) Quantification of the means of the small intestinal lengths (+/- s.d. normalized to WT length; N=4-5 mice/group) in mice with loss of mesenchymal Fgfr1, (DCR1), Fgfr2 (DCR2) and both Fgfr1 and Fgfr2 (DCR1R2). Loss of both Fgfrs was additive. Two asterisks indicate statistically significant differences when comparing any two groups (Student’s t test P<0.01). (E) Quantification of S-phase cells in three cellular compartments (Intervillus epithelium, Mes and Musc) of E18.5 control and DCR1R2 small intestines. Means +/- s.d. for each group are plotted. Statistically significant differences in the means are denoted by an asterisk (P<0.05 by Student’s t test). Bars=10 μm (A), =5 μm (B).
Dermo1-Cre; Fgfr1+/-: Fgfr2+/- mice were mated with mice containing two floxed alleles of both Fgfr1 and Fgfr2 to generate mice with the genotype Dermo1-Cre; Fgfr1loxP/-;Fgfr2loxP/- (DCR1R2). This breeding scheme also produced Dermo1-Cre Fgfr1loxP/- Fgfr2LoxP/+ (DCR1), Dermo1-Cre; Fgfr1loxP/+; Fgfr2loxP/- (DCR2) and control mice that lack Dermo1-Cre. We confirmed by immunofluorescence using Fgfr1 and Fgfr2 antisera that DCR1R2 mice contained neither Fgfr1 nor Fgfr2 expression in mesenchymal cells of the small intestine while the levels of expression in the epithelium were not perceptibly changed (data not shown). In addition, we performed PCR of laser capture microdissected epithelium and mesenchyme from control and DCR1R2 mice. We could only detect PCR products that indicted the presence of a recombined allele for Fgfr1 and Fgfr2 in the mesenchyme of DCR1R2 mice (Fig. 3C).
Late stage DCR1R2 embryos contained shortened small intestines as compared to littermate controls (P<0.01; Fig. 3D) while the crown-rump length was not significantly altered (data not shown). The gross morphology, histologic analysis, and epithelial differentiation showed defects similar to those described in the Fgf9-/- mouse (data not shown). DCR1 and DCR2 embryos both showed milder but significant reductions in gut length (P<0.01; Fig. 3D). The effects of loss of Fgfr1 and Fgfr2 were additive as the small intestinal lengths of DCR1R2 mice were significantly different than both DCR1 and DCR2 (P<0.01; Fig. 3D). Proliferation data for the DCR1R2 mice showed that these embryos displayed a similar phenotype as Fgf9-/- embryos. Only the mesenchyme contained a significant (P<0.01) decrease in proliferation (Fig. 3E). These data suggest that Fgf9 acts primarily through both Fgfr1 and Fgfr2 in the mesenchyme of the developing small intestine to drive mesenchymal proliferation and small intestinal lengthening.
Fgf9 regulates Tgfβ signaling to control mesenchymal differentiation
As the small intestine transitions to the postnatal period, the majority of the mesenchymal fibroblasts are replaced by myofibroblasts, which are characterized by elevated expression of α-SMA and their lack of detectable desmin expression (Powell et al., 2005). To examine mesenchymal differentiation, sections of the small intestine of control and Fgf9-/- embryos were stained for α-SMA and desmin (Fig. 4A-F and Supp. Fig. 1B, C). Interestingly, at E14.5, E16.5 and E18.5, Fgf9-/- small intestines displayed a profound increase in α-SMA expression in the mesenchyme when compared to the same region in their littermate controls. These α-SMA positive cells did not express detectable levels of desmin, consistent with myofibroblast differentiation (Supp. Fig. 1B, C).
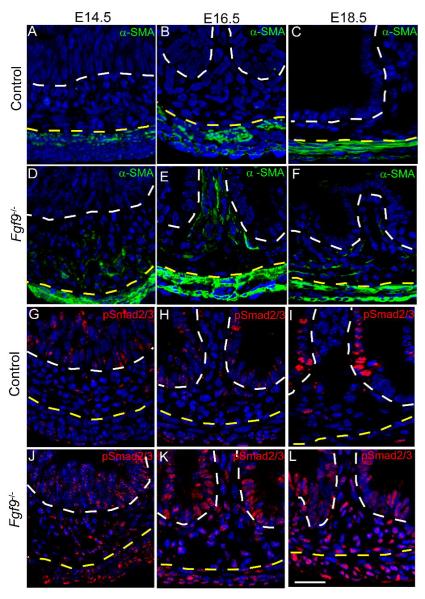
Fig. 4. Fgf9 prevents the mesenchymal fibroblast-myofibroblast transition by inhibition of Tgf-β signaling.
Immunofluorescence-stained sections of E14.5, E16.5 and E18.5 small intestines from (A-C,G-I) control and (D-F, J-L) Fgf9-/- embryos. Sections were stained with antisera directed against α-SMA (A-F; green) and p-Smad2/3 (G-L; red). At all three time points, the mesenchyme from control embryos showed no reactivity to α-SMA and only scattered p-Smad2/3 positive cells were present in this region. In contrast, α-SMA in Fgf9-/- embryos displayed numerous positive staining cells in the mesenchyme. Nearly all mesenchymal cells in Fgf9-/- embryos were positive for pSmad2/3. The white dashed lines indicate the epithelial-mesenchymal borders. The yellow dashed lines indicate the mesenchyme-muscularis propria borders. Bar=30 μm.
Tgf-β signals are well known to drive fibroblast to myofibroblast transitions in other model systems, such as wound repair in skin (Powell et al., 2005). Smad2 and Smad3 serve as intermediate molecules in the Tgf-β signaling cascade that are phosphorylated in response to active binding of Tgf-β and activin ligands to their receptors, while Bmp signaling phosphorylates and activate Smads1, 5 and 8 (Waite et al., 2003). To assess the level of Tgf-β signaling in intestinal mesenchyme of control and Fgf9-/- mice, we examined phospho (p)-Smad2/3 expression using antibodies specific for the active, phosphorylated forms. At E14.5, E16.5 and E18.5, both Fgf9-/- and control embryos showed high level expression of p-Smad2/3 in the nuclei of villus epithelial cells (Fig. 4G-L). At all three of these time points, Fgf9-/- small intestines showed robust p-Smad2/3 staining in nearly all mesenchymal cells while control mice showed only scattered p-Smad2/3-positive cells. In comparison, absence of Fgf9 did not similarly alter the levels of p-Smad1/5/8 in the mesenchyme (Supp. Fig. 1D, E). The staining pattern of p-Smad1/5/8 in control embryos corresponded to previously shown patterns in adult small intestines (Haramis et al., 2004).
Mesenchymal cells located at the villus base expressed p-Erk1/2 and the Tgf-β inhibitors, follistatin (Fst) and follistatin-like 1 (Fstl1)
Fgf9 signaling in the small intestine ultimately targets proliferation, differentiation, and Tgf-β signaling of mesenchymal fibroblasts. A commonly used method for examining Fgf signaling activity is to evaluate the phosphorylation state of Erk1/2 (Thisse and Thisse, 2005; Que et al., 2007). If the mechanism of Fgf9 action was exclusively direct towards fibroblasts, then we would predict that p-Erk1/2 stained sections should label these cells. However, stained sections of E18.5 small intestines from control mice showed that only a small subset of mesenchymal cells showed detectable p-Erk1/2 staining (Fig. 5A, denoted as high p-Erk1/2). We found that these high p-Erk1/2 cells were located closer to the epithelium than the muscle wall and were typically clustered at the base of villi. Significantly fewer of these high p-Erk1/2 mesenchymal cells were detected in Fgf9-/- small intestines (Fig. 5B, C) as well as in DCR1R2 mice (Supp. Fig. 2A). In control small intestines, the high p-Erk1/2 mesenchymal cells did not express well-characterized markers for either of the less abundant mesenchymal lineages (CD45-positive macrophages and Pecam-1-positive endothelial cells; Fig. 5D, E). In addition, the p-Erk1/2 high cells were neither Brdu-positive (Fig. 5F) nor prolyl 4-hydroxylase-positive (data not shown) in control small intestines suggesting these cells were distinct from the proliferative fibroblasts in the mesenchyme.
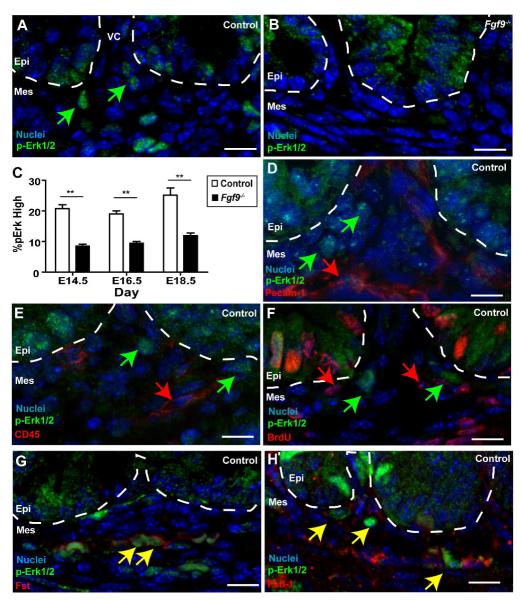
Fig. 5. A subset of small intestinal mesenchymal cells showed a robust response to Fgf9 and expressed Tgf-β inhibitors.
Sections of E18.5 small intestines from (A,D,E, F,G,H) control and (B) Fgf9-/- embryos stained with antisera directed against p-Erk1/2 (green) and co-stained with antisera against (D) Pecam-1, (E),CD45 (F) BrdU, (G) Fst and (H) Fstl1 (In D-H co-stains are each red). In all panels, the mesenchyme at the base of a single villus is shown. The dashed white lines denote the epithelial-mesenchymal boundaries. The green arrows indicate high p-Erk1/2 positive mesenchymal cells near the base of villi. The red arrows denote mesenchymal cells that do not co-localize with p-Erk1/2 cells for a given marker (e.g. CD45, Pecam-1 and BrdU). The yellow arrows denote mesenchymal cells that co-localize with a given marker (e.g. Fst and Fstl1). (C) Quantification of the mean percentage (+/- s.d.) of mesenchymal cells with detectable p-Erk in control and Fgf9-/- embryos. The double asterisks indicate a statistically significant difference when comparing Fgf9-/- embryos versus controls as determined by a Student’s t test (P<0.01). Bars=10 μm.
The enrichment of the Fgf9-responsive, high p-Erk1/2 mesenchymal cells suggested a mechanism of action whereby Fgf9 indirectly inhibits Tgf-β signaling in mesenchymal fibroblasts. This model would suggest that high p-Erk1/2 cells function as a signaling intermediary that inhibits Tgf-β signaling. Known secreted inhibitors of Tgf-β signaling include follistatin (Fst), a homologue, follistatin-like 1 (Fstl1), gremlin and noggin (Smith, 1999). We screened for mRNA expression of these candidates in the whole small intestines of WT mice by RT-PCR. We detected expression of Fst and Fstl (data not shown). Both of these genes were previously identified using expression profiles as preferentially expressed in the mesenchyme (versus the epithelium) of E15.5 small intestines (Li et al., 2007). We then evaluated expression of Fst and Fstl1 in control embryonic small intestines by immunohistochemistry. At E14.5 we found scattered mesenchymal Fst and Fstl1 positive cells (e.g. Supp. Fig. 2B). At later times in embryonic development, we observed clusters of both Fst and Fstl1-positive mesenchymal cells near the base of villi of control (Fig. 5G, H) but not Fgf9-/- mice. Double-label immunofluorescence for p-Erk1/2 with either Fst or Fstl1 showed that the subset of stromal cells that expressed high levels of pErk1/2 showed co-localization with both of these proteins (Fig. 5G, H). In addition, Fgfr2 colocalized with both p-Erk and Fst indicating that epithelial Fgf9 signals through mesenchymal receptors in cells that contain high levels of Fst and Fstl1 (Supp. Fig. 2C, D).
The p-Erk/Fst/Fstl1-expressing cells near the villus base are mesenchymal stem cells
One mesenchymal cell type that cannot, as yet, be identified on tissue sections with a single marker is mesenchymal stem cells (MSC). Though these cells were originally considered to function as the source of many different types of mesenchymal lineages, recent work has suggested that they may play a role in signaling, particularly in injury repair (Gnecchi et al., 2006). Most mouse tissues from either adults (da Silva Meirelles et al., 2006; Wang et al., 2006) or embryos (Lee and Tarantal, 2006) contain MSCs that can be readily isolated by well-established protocols (da Silva Meirelles et al., 2006). Therefore, we isolated MSCs from late stage WT embryonic small intestines using these same protocols.
The resulting embryonic small intestinal MSC (iMSC) lines displayed characteristics of previously described MSCs from other tissues. The iMSCs could be maintained in culture for at least 40 passages. The cell surface marker expression matched previous studies of MSCs from other tissues (iMSCs were positive for CD29, CD44, CD54, CD105 and CD106 and negative for CD45 and Pecam-1; Fig. 6A) (da Silva Meirelles et al., 2006; Wang et al., 2006). Lastly, the iMSCs differentiated into either adipocytes or osteocytes when cultured under inductive conditions (Supp. Fig. 3A, B) (da Silva Meirelles et al., 2006).
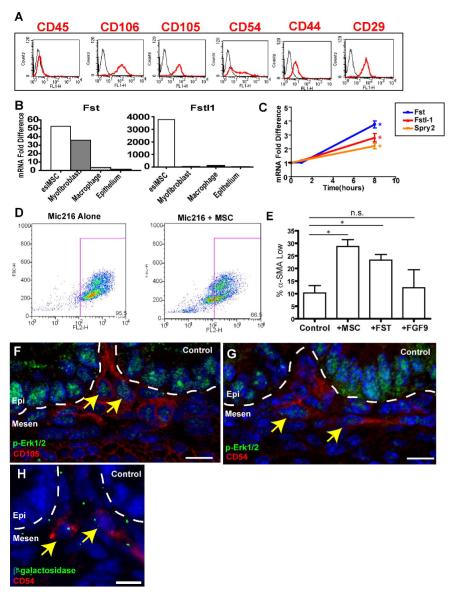
Fig. 6. The p-Erk1, 2/Fst/Fstl1-positive mesenchymal cells were embryonic small intestinal (esi)MSCs.
(A) Fluorescence profiles (X-axis) for E18.5 WT iMSCs (passage 5) stained with antigen specific antibodies (red) and isotype-matched negative controls (black) that showed the majority of these cells (Y-axis=cell counts) were CD29-, CD44-, CD54-, CD105-, and CD106- positive and CD45 negative. (B) Cultured iMSCs expressed genes encoding secreted Tgfβ inhibitors including follistatin (Fst) and follistatin-like 1 (Fstl-1). Quantification of relative mRNA amounts of Fst and Fstl1 in multiple cell lines as determined by qRT-PCR. Expression of Fst and Fstl1 in iMSCs was compared to myofibroblasts (Mic216), macrophages (RAW 264.7) and gut epithelial cells (AGS). (C) Expression of mRNAs encoding Fst (blue), Fstl-1 (orange) and Spry-2 (a known transcriptional target of Fgf9, red) in iMSCs increased with Fgf9 treatment. A single asterisk indicates that the mean values for the 8h time points were statistically significantly different from 0h (P<0.05 by Student’s t test). (D) FACS analysis of intracellular α-SMA of MIC-216 (intestinal myofibroblast line) cells either cultured alone or in a transwell co-culture with iMSCs. The X-axis is fluorescence intensity for α-SMA and the Y-axis is forward scatter (an indication of cell size). The box indicates cell population defined as α-SMA high. (E) Quantification of the percentage of α-SMA low Mic216 cells cultured alone, in the presence of iMSCs , Fst of Fgf9 (N=3 experiments/condition). (F,G) Sections of control E18.5 small intestines stained with p-Erk (green) and (I) CD54 (red) and (J) CD105 (red). The latter two markers co-localize with p-Erk1/2 positive mesenchymal cells (yellow arrows). (H) Section of an E14.5 Dermo1-cre; Rosa26R small intestine stained with anti-β-galactosidase and anti-CD54. Bars =10 μm.
The iMSCs also displayed similar characteristics as the high p-Erk/Fst/Fstl1 small intestinal mesenchymal cells. When cultured alone in 10% serum, the iMSCs expressed much higher relative amounts of mRNA encoding Fst (ranging from 10-128 fold higher than mRNAs from epithelial, macrophage and myofibroblast cell lines) and Fstl1 (ranging from 47-3500 fold higher than the same set of cell lines; Fig. 6B). Importantly, the iMSCs responded in vitro to Fgf9 stimulation in a time- and dose-dependent manner by increasing p-Erk1/2 expression (Supp. Fig. 3C and data not shown). We found that Fst was co-expressed with p-Erk1/2 in the iMSCs in vitro (Supp. Fig. 3D). Furthermore, exogenous Fgf9 increased expression of both Fst and Fstl1 in these iMSCs (Fig. 3E).
To test the hypothesis that the iMSCs could affect myofibroblast differentiation, we co-cultured them with a line of well characterized intestinal myofibroblasts (Mic216) (Plateroti et al., 1998) using a transwell system. We found that the presence of the iMCSs resulted in a de-differentiation of myofibroblasts towards fibroblasts, based on decreased expression of α-SMA in the Mic216 cells (Fig. 6D, E). This finding was partially recapitulated by the incubation of Mic216 cells with Fst (Fig. 6E). Mic216 cells did not significantly alter their αSMA expression in response to Fgf9 (Fig. 6E). Lastly, we performed double-label immunofluorescence on sections of WT small intestines with p-Erk1/2 and markers of iMSCs. We found that iMSC markers co-localized with the high p-Erk1/2 mesenchymal cells (Fig, 6F, G and data not shown). iMSCs are also targeted by Dermo1-Cre as shown by the co-localization of β-galactosidase with markers of this cell type (e.g. Fig. 6H). Taken together, these data support the hypothesis that the p-Erk/Fst/Fstl1-expressing small intestinal stromal cells are iMSCs.
Discussion
In this report, we demonstrated that Fgf9 is essential for small intestinal development during late stage embryogenesis. Loss of function models for both the Fgf9 ligand and its mesenchymal receptors showed that epithelial Fgf9 signals to the underlying mesenchyme to regulate elongation of the small intestine. A key cellular target of Fgf9 was ultimately mesenchymal fibroblasts. In the absence of Fgf9 signaling, these fibroblasts showed diminished proliferation, premature differentiation into myofibroblasts and increased Tgf-β signaling, a pathway well known to drive such cellular transitions (Powell et al., 2005). Surprisingly, the response to Fgf9 signaling in the small intestinal mesenchyme did not occur uniformly. Instead the response to Fgf9 signaling occurred robustly in a select subset of mesenchymal cells that also expressed high levels of Tgf-β inhibitors, Fst and Fstl1. We found that these cells most closely resembled iMSCs and that these cells appeared to act as a cellular intermediary in the signaling of Fgf9 to intestinal mesenchymal fibroblasts (Fig. 7).
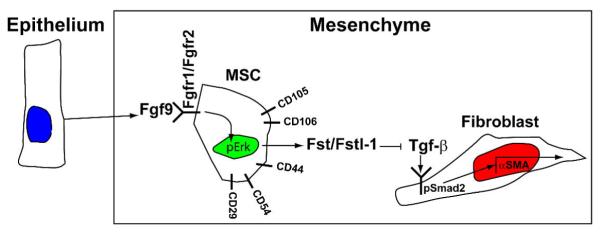
Fig. 7. Proposed cellular mechanism for the control of small intestinal fibroblast differentiation by Fgf9.
Epithelially-expressed Fgf9 interacts with its receptors (Fgfr1 and Fgfr2) on mesenchymal stem cells. These cells in turn express inhibitors of Tgf-β signaling that interact with mesenchymal fibroblasts to inhibit their expression of α-SMA.
An important feature of Fgf9 signaling in the small intestine is that it occurs across an epithelial-mesenchymal boundary to regulate elongation of this organ during late-stage embryogenesis. This type of crosstalk is a common mechanism of action for fibroblast growth factors. For example, epithelial to mesenchymal Fgf9 signaling, with reciprocal mesenchymal to epithelial Fgf10 signals, is known to be essential for formation of a cecum (Burns et al., 2004; Zhang, et al., 2006). In the lung, epithelial to mesenchymal and mesothelial to mesenchymal Fgf9 and reciprocal Fgf10 signaling is required for airway branching (Bellusci et al., 1997; Colvin et al., 2001a; White et al., 2006). Finally, it has recently been shown that epithelial to mesenchymal Fgf9 is necessary for myocardial growth and proper coronary vascular development (Lavine et al., 2005). The function of Fgf9 in the small intestine is somewhat distinct from these other organs, as no branching of a targeted structure (epithelium or blood vessel) occurs. Thus far, we have not uncovered a functional reciprocal Fgf partner that is produced in the intestinal mesenchyme that signals back to the epithelium. Further studies will be ongoing to determine if this occurs.
Our report suggests that a single molecule (Fgf9) targets the mesenchyme to affect the elongation of the entire organ during development. Importantly, communication between the three functional layers of the intestine is absolutely required, as proper functioning of the organ as a whole requires each layer to be properly oriented to each other. Thus, extensive communication across epithelial-mesenchymal boundaries is necessary for proper intestinal development and adult homeostasis. An unanticipated interaction of Fgf9 was with Tgf-β but not the Bmp signaling pathways. Previous studies have demonstrated that development of the mouse small intestine utilizes mesenchymal to epithelial signaling to control epithelial proliferation and differentiation during development. Knockout mice for the transcription factors Nkx2.3 (Pabst et al., 1999) and Foxl1 (Kaestner et al., 1997) each show diminished expression of Bmps in the intestinal mesenchyme. In turn, loss of Bmp signaling has several effects including increased epithelial proliferation and dysmorphic villi. In addition, Wang et al., (2006) have shown that expression of epimorphin in mesenchymal myofibroblasts is required for proper epithelial maintenance in adult animals, at least in part by mesenchymal modulation of the Bmp pathway. The relationship between Fgf9 signaling and Nkx2.3, Foxl1 and epimorphin is currently unknown but they appear to be distinct pathways as Fgf9 does not appear to affect Bmps based on the pattern of expression of p-Smad1/5/8 as well as the absence of epithelial/villus defects in Fgf9-/- mice. The antagonistic relationship of Fgf9 with Tgf-β that we observed in the developing mouse small intestine is the opposite of the synergistic interaction that has been extensively documented for Fgfs with Tgf-β in other systems, most notably the early development of Xenopus and zebrafish during gastrulation (e.g. Cornell and Kimelman, 1994; Mathieu, et al., 2004). Antagonistic interactions of Fgf and Tgf-β signaling have been observed much later in development such as during melanocyte formation and maturation (Stocker et al., 1991).
Interestingly, we show that an important downstream function of Fgf9 is the inhibition of Tgf-β signaling that we hypothesize occurs through an indirect mechanism utilizing iMSCs as cellular intermediaries. It is known that Erk, a downstream target of the Fgf receptor, and other MAP kinases, have the ability to directly modulate the activity of the Tgf-β superfamily through the targeting of Smad1 molecules for degradation by phosphorylation and later ubiquination (Kretzschmar et al., 1997). Interestingly, this ability of Erk to target Smad for degradation has been shown to be a property of Smads associated with the Bmp signaling pathway (Smads 1, 5, and 8), and not the Tgfβ and activin specific Smads 2 and 3. However, this is still a possible mechanism as both Smad2 and Smad3 have the ability to be ubiquitinated and degraded following prolonged activity (Lo and Massagué, 1999). Here, we propose an indirect mechanism that involves Fgf9 regulation of secreted inhibitors of Tgf-β. The overall effect of Fgf9 on the small intestinal mesenchyme may involve other mechanisms whereby Fgf9 targets all fibroblasts and additionally inhibits Tgf-β signaling in a cell autonomous manner. Such mechanisms need not be mutually exclusive. Indeed, considering the importance of Tgf-β signaling in GI tract development and the multiple levels through which Tgf-β is known to be regulated, such multifaceted control is very likely.
MSCs are most likely present in all adult and fetal organs and share the ability to differentiate into several mesenchymal cell types ex vivo (McTaggert and Atkinson, 2007). However, their exact role in development is still unclear. Fgfs are known to modulate the behavior of MSCs, serving to increase proliferation and suppress differentiation of bone marrow derived MSCs in vitro (Tsutsumi et al., 2001; Farré et al., 2007). We propose that iMSCs play a role in development by regulating the differentiation of adjacent mesenchymal cells (fibroblasts) through expression of secreted Tgf-β inhibitors. The location of the iMSCs near the base of villi may be important for this function as the range of signaling for Tgf-β is very limited (Reilly and Melton, 1996).
This potential function of MSCs is in stark contrast to the traditional role of stem cells, which is to divide only occasionally in order to produce daughter cells which, through repeated divisions via transit amplification, produce committed lineages of mature, differentiated cells. However, growing evidence suggests that MSCs have the ability to function outside of this paradigm. For example, it has been recently shown that MSCs have the potential to play a role in disease by functioning as a nidus of paracrine signaling (Gnecchi et al., 2006; Karnoub et al., 2007). Here, we show that this idea of MSCs as paracrine signaling centers can be extended to developmental biology, and that these cells play an important role in gut development by functioning as cellular transceivers. In this role, iMSCs preferentially receive an active Fgf9 signal, and in turn communicate to the surrounding mesenchyme by secreting Tgf-β superfamily inhibitors, thereby preventing premature mesenchymal differentiation. In this manner, they create a permissive environment through which mesenchymal expansion occurs, which we propose contributes to the lengthening of the intestine.
It has been reported that the mesenchymal response to Fgf in other organ systems, particularly the trachea, occurs in a scattered population similar to what we observe in this study (Que et al., 2007). Based on our observations in the intestine, we would speculate that these scattered mesenchymal cells represent mesenchymal stem cells residing in these organs. Thus, the concept of Fgf preferentially signaling to MSC populations might extend to other organ systems. Understanding the regulation of the signaling function of MSCs in response to Fgfs may be informative in our understanding of their normal physiologic roles in development and disease.
Materials and Methods
Mice
All animal experiments were performed in accordance with approved protocols from the Washington University School of Medicine Animal Studies Committee. Mice used in this study were housed in microisolator cages, in a specified pathogen-free barrier facility, under a strict 12h light cycle. Mice were fed a standard irradiated chow diet (PicoLab Rodent Chow 20, Purina Mills Inc.) ad libitum. Fgf9-/-, Fgfr1fl/+, Fgfr2fl/+ and Dermo1-Cre were previously generated (Colvin et al., 2001a; Trokovic et al., 2003; White et al., 2006; Yu et al., 2003) and all are on a C57Bl/6 background.
Histochemistry and BrdU labeling
For embryo collections, time-mated females were sacrificed at E14.5-E18.5, 1 hour after an intraperitoneal injection of bromodeoxyuridine (BrdU, 120μg/gm, Sigma). The GI tract was isolated from embryos, pinned on black wax, fixed in 2% paraformaldehyde for 4 hours and processed as previously described (Stappenbeck and Gordon, 2000). Tissue samples were embedded in paraffin and 5 μm thick serial sections were prepared. Sections were then stained with hematoxylin and eosin. Adjacent, unstained sections were used for immunohistochemical studies (see below).
Paraffin sections were processed for immunohistochemistry using an antibody to BrdU (BD bioscience). Sections were counterstained with Hematoxylin. Proliferation was measured in three cellular compartments in the proximal and distal small intestines as illustrated in Fig. 2 by scoring the number of BrdU-labeled nuclei to the number of total nuclei in each compartment.
Immunohistochemistry
Slide preparations for immunohistochemistry varied depending on primary antibody utilized. For antibodies directed against α-SMA (clone 1a4; Sigma-Aldrich; 1:2,000 dilution), prolyl 4-hydroxylase (clone ER-TR7; BMA Biomeidcal; 1:100), desmin (clone ZC18; Zymed; 1:100), I-Fabp, Fgfr1 (Santa Cruz,; 1:50), Fgfr2 (Santa Cruz; 1:100), Fst (Santa Cruz; 1:10), Fstl-1 (Santa Cruz; 1:10), Pecam-1 (rat anti-mouse CD31; clone MEC13.3, BD Pharmingen; 1:100), p-Smad1/5/8 (Cell Signaling; 1:100), and p-Smad2/3 (Cell Signaling; 1:100), embryonic small intestines were embedded in paraffin and sectioned at 5μm prior to staining. Sections were then deparaffinized in 3 washes of xylene, and rehydrated in isopropanol. Antigen retrieval was performed by varying methods depending on primary antibody used. For I-Fabp, Fst, Fstl-1, p-Smad2/3, and p-Smad1/5/8 slides were heated at 95°C for 20 minutes in 10 mM sodium citrate buffer, pH=6.0. Slides were then washed in ddH20 for 5 minutes prior to washing in blocking buffer (composed of 1% BSA, 0.1% Triton in PBS) for 20 minutes. For primary antibody to Pecam-1, antigen retrieval was done by 10 minute incubation with a 1% chymotrypsin solution, followed by a 10 minute wash in ddH20 prior to application of blocking buffer. Antibodies to α-SMA, prolyl 4-hydroxylase, Fgfr2, and desmin required no antigen retrieval.
For antibodies to p-Erk1/2 (Cell Signaling; 1:100), CD29 (BD Biosciences; 1:100), CD44 (BD; 1:100), CD45 (BD; 1:100), CD54 (BD; 1:100), CD105 (BD; 1:100), and CD106 (BD; 1:100), embryonic small intestines were embedded in OCT compound (Sakura) and immediately frozen in liquified Cytocool (Richard-Allan Scientific). Frozen sections of 5 μm were then fixed for 5 minutes in 4% paraformaldehyde, rinsed in PBS, and then immersed in blocking buffer for 20 minutes prior to incubation with primary antibody. Primary antibodies were incubated at 4°C overnight, followed by secondary antibody incubation for 1 hour at 24°C using a 1:500 dilution. Slides were then counterstained with bis-benzimide and coversliped with a 1:1 glycerol/PBS solution. Sections were viewed with a Zeiss Axiovert 200 with Axiocam MRM camera and Apotome optical sectioning slider.
Isolation of embryonic small intestinal mesenchymal stem cells
A single WT E18.5 small intestine was minced with a razor blade followed by treatment with collagenase (Gibco) in Dulbeccos modified eagle medium (DMEM) containing 10mM HEPES buffer for 1 hour at 37°C. After passage through a 70 μm filter, the cell suspension was centrifuged for 10 minutes at 400g. The cell pellet was then suspended in DMEM containing 10% FBS and plated on standard plastic tissue culture plates. After incubation for 2 hours at 37°C, the growth media containing nonadherent cells was removed and discarded. Adherent cells were replenished with fresh media and passaged once weekly.
To differentiate MSCs into adipocytes, isolated cultured embryonic small intestinal MSCs were treated with 10−8 M dexamethasone, 5 μg/ml insulin, for 21 days. Cells were then stained with Oil Red O to verify presence of lipid stores. To differentiate MSCs into osteocytes, MSCs were treated with 10−8 M dexamethasone, 5 μg/ml ascorbic acid 2-phosphate, 10 mM β-glycerophosphate for 21 days. Cells were then stained with alizarin red to verify presence of calcium deposits in the extracellular matrix.
FACS analysis
Cultured cells were treated for 5 minutes with a trypsin/EDTA solution. Cells were pelleted and fixed in 2% formaldehyde in PBS for 10 minutes at 37°C. Cells were then chilled at 4°C for 1 minute, and pelleted by centrifugation at 250g for 5 minutes. Cells were then permeabilized by 30 minute incubation in 90% methanol at 4°C. Cells were then washed twice with PBS, and then re-suspended in 1% BSA in PBS, containing the primary antibody for 1 hour at 24°C. The cells were then washed twice in PBS before incubation in fluorescently-conjugated secondary antibody (diluted 1:500 in 1% BSA in PBS for 1 hour). Cells were then rinsed in PBS, before finally suspending in 1% BSA in PBS for flow cyotometric analysis. All analysis was performed on a FACSCalibur flow cytometer and analyzed with FlowJo software.
Immunoblotting
MSCs were serum starved for 2 days in DMEM containing 0.5% FCS. Following starvation, 10 ng/ml Fgf9 (Peprotech) was added to the growth media. Following treatment, cells were lysed in RIPA buffer for 5 minutes on ice and then centrifuged at 14000g for 10 minutes. Protein content of the supernatant was quantified using a Bio-Rad Dc colorimetric protein assay (Bio-Rad). 10μg of protein was then reduced and loaded into 10% Bis-Tris gels and electrophoresed.Samples were then transferred onto PVDF membranes and blocked in a solution containing 4% BSA (gels and membranes from Invitrogen), 0.1% Tween in TBS for 1 hour at 24°C. Blots were then incubated in primary antibody overnight, washed in TBST, and then probed with HRP-conjugated secondary antibody (Bio-Rad) for one hour at 24°C before development with the SuperSignal West Pico chemiluminescent kit (Pierce).
qRT-PCR Analysis
Cells were serum starved and treated with Fgf9 as above. Total cellular RNA was collected and purified using a Qiagen RNeasy mini kit. cDNAs were then synthesized using Superscript III reverse transcriptase (Invitrogen) and random primers according to the manufacturer’s protocol. qRT-PCR that was performed in triplicate for each sample using SYBR-green master mix (Invitrogen). The following primers were used: 18S (5′-CATTCGAACGTCTGCCCTATC and 5′-CCTGTGCCTTCCTTGGA), Fst (5′-TACTGTGTGACCTGTAATCGG-3′ and 5′-TGATACACTTTCCCTCATAGGC-3′), Fstl1 (5′-CACGGCGAGGAGGAACCTA-3′ and 5′-TCTTGCCATTACTGCCACACA-3′.
Co-culture experiments
A rat myofibroblast line (Mic216) (Plateroti et al., 1998) was co-cultured with MSCs using a transwell system (Corning). 5.0 × 104 MSCs were plated on the upper chamber, which was separated from the lower chamber by a membrane containing 0.4 μm pores. The lower chamber was seeded with 1.0 × 104 myofibroblasts. Experiments were performed was done in DMEM containing 0.5%FCS. When confluent (∼3 days) myofibroblasts were collected for FACS analysis as described above. Fst was obtained from R+D systems for treatment of Mic216.
Laser capture microdissection
10,000 cells were separately procured from the mesenchyme and epithelium of control and DCR1R2 E18.5 embryonic small intestines as previously described (Pull et al., 2005). DNA was isolated by incubating the captured cells with proteinase K in TE buffer at 42°C for 10 hours. DNA was amplified by 40 cycles of PCR using primers as previously described (Yu et al., 2003).
Supplementary Material
Supplemental Figure 1. Fgf9, desmin and p-SMAD 1/5/8 expression in embryonic mouse small intestines. A) Whole mount small intestine from a WT E14.5 mouse utilized for in situ hybridization using a probe for Fgf9. The staining is consistent with epithelial expression as was demonstrated at earlier time points (Zhang et al., 2006).
B-E) Immunofluorescence-stained sections of E18.5 small intestines from (B, D) control and (C, E) Fgf9-/- embryos. Sections were stained with antisera directed against desmin (B, C; green) and p-Smad1/5/8 (D, E; red). No differences in desmin or p-Smad 1/5/8 expression when comparing small intestines from control and Fgf9-/- animals. Bars=30μm.
Supplemental Figure 2. p-Erk positive mesenchymal cells are diminished in DCR1R2 embryos and express Fst and Fgfr2. (A) Quantification of the mean percentage (+/- s.d.) of mesenchymal cells with detectable p-Erk in control and DCR1R2 embryos (E18.5). The double asterisk indicates a statistically significant difference when comparing DCR1R2 embryos versus controls as determined by a Student’s t test (P<0.01). Sections from control (B) E14.5 and (C,D) E18.5 small intestines stained with anti-Fgfr2 and (C) anti-pErk1/2 (red) and (D) anti-Fst (red). The white dashed lines in B-D indicate the epithelial/mesenchymal junction. The yellow dashed line in B indicates the mesenchymal/muscularis propria border. The cells circled by the yellow dashed lines in C and D show mesenchymal cells that are positive for both Fgfr2 and p-Erk1/2 (C) and Fst (D). Bars= 5μm.
Supplemental Figure 3. iMSCs are multi-potent, Fgf9 responsive and co-express Fst and pErk1/2. (A) Osteogenic induction of cultured iMSCs stained with alizarin red. The widespread positive cells indicated that iMSCs can differentiate into osteocytes. (B) Adipogenic induction of iMSCs stained with Oil Red O. The widespread positive cells indicated that iMSCs can differentiate into adipocytes. (C) Cultured, serum starved iMSCs phosphorylated Erk1/2 in response to treatment with 10 ng/ml Fgf9. Immunoblots at various time points post treatment are shown. (D) Cultured iMSC stained with antisera directed against p-Erk1/2 (green) and Fst (red). Arrow indicates the cell nucleus. Bars=50 μm (A, B). Bar=5 μm (D).
Acknowledgements
We thank Dr. Deborah Rubin for her helpful comments on the manuscript and Sarah Brown for technical assistance. This work was supported by the Pew Foundation and DK071619 (TSS) and a grant from the March of Dimes Foundation (DMO). MJG was supported by an MSRT training grant T32DK0713034.
References
- Bellusci S, Grindley J, Emoto H, Itoh N, Hogan BL. Fibroblast growth factor 10 (FGF10) and branching morphogenesis in the embryonic mouse lung. Development. 1997;124:4867–4878. doi: 10.1242/dev.124.23.4867. [DOI] [PubMed] [Google Scholar]
- Burns RC, Fairbanks TJ, Sala F, De Langhe S, Mailleux A, Thiery JP, Dickson C, Itoh N, Warburton D, Anderson KD, et al. Requirement for fibroblast growth factor 10 or fibroblast growth factor receptor 2-IIIb signaling for cecal development in mouse. Dev. Biol. 2004;265:61–74. doi: 10.1016/j.ydbio.2003.09.021. [DOI] [PubMed] [Google Scholar]
- Calvert R, Pothier P. Migration of fetal intestinal intervillus cells in neonatal mice. Anat. Rec. 1990;227:199–206. doi: 10.1002/ar.1092270208. [DOI] [PubMed] [Google Scholar]
- Cohn SM, Simon TC, Roth KA, Birkenmeier EH, Gordon JI. Use of transgenic mice to map cis-acting elements in the intestinal fatty acid binding protein gene (Fabpi) J. Cell Biol. 1992;119:27–44. doi: 10.1083/jcb.119.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colvin JS, White A, Pratt SJ, Ornitz DM. Lung hypoplasia and neonatal death in Fgf9-null mice identify this gene as an essential regulator of lung mesenchyme. Development. 2001a;128:2095–2106. doi: 10.1242/dev.128.11.2095. [DOI] [PubMed] [Google Scholar]
- Colvin JS, Green RP, Schmahl J, Capel B, Ornitz DM. Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell. 2001b;104:875–889. doi: 10.1016/s0092-8674(01)00284-7. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Kimelman D. Activin-mediated mesoderm induction requires FGF. Development. 1994;120:453–62. doi: 10.1242/dev.120.2.453. [DOI] [PubMed] [Google Scholar]
- da Silva Meirelles L, Chagastelles PC, Nardi NB. Mesenchymal stem cells reside in virtually all post-natal organs and tissues. J Cell Sci. 2006;119:2204–2213. doi: 10.1242/jcs.02932. [DOI] [PubMed] [Google Scholar]
- Dekaney CM, Fong JJ, Rigby RJ, Lund PK, Henning SJ, Helmrath MA. Expansion of intestinal stem cells associated with long-term adaptation following ileocecal resection in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;293:G1013–1022. doi: 10.1152/ajpgi.00218.2007. [DOI] [PubMed] [Google Scholar]
- Erwin CR, Jarboe MD, Sartor MA, Medvedovic M, Stringer KF, Warner BW, Bates MD. Developmental characteristics of adapting mouse small intestine crypt cells. Gastroenterology. 2006;130:1324–1332. doi: 10.1053/j.gastro.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Farré J, Roura S, Prat-Vidal C, Soler-Botija C, Llach A, Molina CE, Hove-Madsen L, Cairó JJ, Gòdia F, Bragós R, Cinca J, Bayes-Genis A. FGF-4 increases in vitro expansion rate of human adult bone marrow-derived mesenchymal stem cells. Growth Factors. 2007;25:71–76. doi: 10.1080/08977190701345200. [DOI] [PubMed] [Google Scholar]
- Fre S, Huyghe M, Mourikis P, Robine S, Louvard D, Artavanis-Tsakonas S. Notch signals control the fate of immature progenitor cells in the intestine. Nature. 2005;435:964–968. doi: 10.1038/nature03589. [DOI] [PubMed] [Google Scholar]
- Gnecchi M, He H, Noiseux N, Liang OD, Zhang L, Morello F, Mu H, Melo LG, Pratt RE, Ingwall JS, Dzau VJ. Evidence supporting paracrine hypothesis for Akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. FASEB J. 2006;20:661–669. doi: 10.1096/fj.05-5211com. [DOI] [PubMed] [Google Scholar]
- Haramis AP, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJ, Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- He XC, Zhang J, Tong WG, Tawfik O, Ross J, Scoville DH, Tian Q, Zeng X, He X, Wiedemann LM, et al. BMP signaling inhibits intestinal stem cell self-renewal through suppression of Wnt-beta-catenin signaling. Nat. Genet. 2004;36:1117–1121. doi: 10.1038/ng1430. [DOI] [PubMed] [Google Scholar]
- He XC, Yin T, Grindley JC, Tian Q, Sato T, Tao WA, Dirisina R, Porter-Westpfahl KS, Hembree M, Johnson T, et al. PTEN-deficient intestinal stem cells initiate intestinal polyposis. Nat. Genet. 2007;39:189–198. doi: 10.1038/ng1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert JM, Lin M, Partanen J, Rossant J, McConnell SK. FGF signaling through FGFR1 is required for olfactory bulb morphogenesis. Development. 2003;130:1101–1111. doi: 10.1242/dev.00334. [DOI] [PubMed] [Google Scholar]
- Howard L, Ament M, Fleming CR, Shike M, Steiger E. Current use and clinical outcome of home parenteral and enteral nutrition therapies in the United States. Gastroenterology. 1995;109:355–365. doi: 10.1016/0016-5085(95)90321-6. [DOI] [PubMed] [Google Scholar]
- Kaestner KH, Silberg DG, Traber PG, Schütz G. The mesenchymal winged helix transcription factor Fkh6 is required for the control of gastrointestinal proliferation and differentiation. Genes Dev. 1997;11:1583–1595. doi: 10.1101/gad.11.12.1583. [DOI] [PubMed] [Google Scholar]
- Karlsson L, Lindahl P, Heath JK, Betsholtz C. Abnormal gastrointestinal development in PDGF-A and PDGFR-a deficient mice implicates a novel mesenchymal structure with putative instructive properties in villus morphogenesis. Development. 2000;127:3457–3466. doi: 10.1242/dev.127.16.3457. [DOI] [PubMed] [Google Scholar]
- Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–63. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- Kaufman MH. The atlas of mouse development. Harcourt Brace and Company; London: 1992. [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat. Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Kretzschmar M, Doody J, Massagué J. Opposing BMP and EGF signaling pathways converge on the TGF-β family mediator Smad1. Nature. 1997;389:617–622. doi: 10.1038/39348. [DOI] [PubMed] [Google Scholar]
- Ladefoged K, Hessov I, Jarnum S. Nutrition in short-bowel syndrome. Scand. J. Gastroenterol. Suppl. 1996;216:122–131. doi: 10.3109/00365529609094567. [DOI] [PubMed] [Google Scholar]
- Lavine KJ, Yu K, White AC, Zhang X, Smith C, Partanen J, Ornitz DM. Endocardial and epicardial derived FGF signals regulate myocardial proliferation and differentiation in vivo. Developmental Cell. 2005;8:85–95. doi: 10.1016/j.devcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Lee CCI, Tarantal AF. Comparison of growth and differentiation of fetal and adult rhesus monkey MSCs. Stem Cells and Development. 2006;15:209–220. doi: 10.1089/scd.2006.15.209. [DOI] [PubMed] [Google Scholar]
- Li X, Madison BB, Zacharias W, Kolterud A, States D, Gumucio DL. Deconvoluting the intestine: molecular evidence for a major role of the mesenchyme in the modulation of signaling cross talk. Physiol. Genomics. 2007;29:290–301. doi: 10.1152/physiolgenomics.00269.2006. [DOI] [PubMed] [Google Scholar]
- Mathieu J, Griffin K, Herbomel P, Dickmeis T, Strähle U, Kimelman D, Rosa FM, Peyriéras N. Nodal and Fgf pathways interact through a positive regulatory loop and synergize to maintain mesodermal cell populations. Development. 2004;131:629–641. doi: 10.1242/dev.00964. [DOI] [PubMed] [Google Scholar]
- McTaggert SJ, Atkinson K. Mesenchymal stem cells: Immunobiology and therapeutic potential in kidney disease. Nephrology. 2007;12:44–52. doi: 10.1111/j.1440-1797.2006.00753.x. [DOI] [PubMed] [Google Scholar]
- Nightingale JM, Lennard-Jones JE. The short bowel syndrome: what’s new and old? Dig. Dis. 1993;11:12–31. doi: 10.1159/000171397. [DOI] [PubMed] [Google Scholar]
- Ornitz DM, Itoh N. Functional evolutionary history of the mouse Fgf gene family. Dev. Dyn. 2008;237:18–27. doi: 10.1002/dvdy.21388. [DOI] [PubMed] [Google Scholar]
- Pabst O, Zweigerdt R, Arnold HH. Targeted disruption of the homeobox transcription factor Nkx2-3 in mice results in postnatal lethality and abnormal development of small intestine and spleen. Development. 1999;126:2215–2225. doi: 10.1242/dev.126.10.2215. [DOI] [PubMed] [Google Scholar]
- Pirvola U, Ylikoski J, Trokovic R, Hébert JM, McConnell SK, Partanen J. FGFR1 is required for the development of the auditory sensory epithelium. Neuron. 2002;35:671–680. doi: 10.1016/s0896-6273(02)00824-3. [DOI] [PubMed] [Google Scholar]
- Plateroti M, Rubin DC, Duluc I, Singh R, Foltzer-Jourdainne C, Freund JN, Kedinger M. Subepithelial fibroblast cell lines raised from different levels of the gut axis display regional characteristics. Am. J. Physiol. 1998;274:G945–G954. doi: 10.1152/ajpgi.1998.274.5.G945. [DOI] [PubMed] [Google Scholar]
- Powell DW, Adegboyega PA, Di Mari JF, Mifflin RC. Epithelial cells and their neighbors, role of intestinal myofibroblasts in development, repair and cancer. Am. J. Physiol. Gastrointest Liver Physiol. 2005;289:G2–G7. doi: 10.1152/ajpgi.00075.2005. [DOI] [PubMed] [Google Scholar]
- Pull SL, Doherty JM, Mills JC, Gordon JI, Stappenbeck TS. Activated macrophages are an adaptive element of the colonic epithelial progenitor niche necessary for regenerative responses to injury. Proc Natl Acad Sci U S A. 2005;102:99–104. doi: 10.1073/pnas.0405979102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalho-Santos M, Melton DA, McMahon AP. Hedgehog signals regulate multiple aspects of gastrointestinal development. Development. 2000;127:2763–2772. doi: 10.1242/dev.127.12.2763. [DOI] [PubMed] [Google Scholar]
- Reilly KM, Melton DA. Short-range signaling by candidate morphogens of the TGF beta family and evidence for a relay mechanism of induction. Cell. 1996;86:743–754. doi: 10.1016/s0092-8674(00)80149-x. [DOI] [PubMed] [Google Scholar]
- Sala FG, Curtis JL, Veltmaat JM, Del Moral PM, Le LT, Fairbanks TJ, Warburton D, Ford H, Wang K, Burns RC, Bellusci S. Fibroblast growth factor 10 is required for survival and proliferation but not differentiation of intestinal epithelial progenitor cells during murine colon development. Dev Biol. 2006;299:373–85. doi: 10.1016/j.ydbio.2006.08.001. [DOI] [PubMed] [Google Scholar]
- Sbarbati R. Quantitative aspects of the embryonic growth of the intestine and stomach. J. Anat. 1979;129:795–803. [PMC free article] [PubMed] [Google Scholar]
- Schmidt GH, Winton DJ, Ponder BA. Development of the pattern of renewal in the crypt-villus unit of chimaeric mouse small intestine. Development. 1988;103:785–790. doi: 10.1242/dev.103.4.785. [DOI] [PubMed] [Google Scholar]
- Smith WC. TGF beta inhibitors. New and unexpected requirements in vertebrate development. Trends Genet. 1999;15:2–5. doi: 10.1016/s0168-9525(98)01641-2. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with a ROSA26 Cre reporter strain. Nat. Gen. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Stappenbeck TS, Gordon JI. Rac1 mutations produce aberrant epithelial differentiation in the developing and adult mouse small intestine. Development. 2000;127:2629–2642. doi: 10.1242/dev.127.12.2629. [DOI] [PubMed] [Google Scholar]
- Stocker KM, Sherman L, Rees S, Ciment G. Basic FGF and TGF-beta1 influence commitment to melanogenesis in neural crest-derived cells of avian embryos. Development. 1991;111:635–645. doi: 10.1242/dev.111.2.635. [DOI] [PubMed] [Google Scholar]
- Thisse C, Thisse B. Functions and regulations of Fgf signaling during embryonic development. Dev. Biol. 2005;287:390–402. doi: 10.1016/j.ydbio.2005.09.011. [DOI] [PubMed] [Google Scholar]
- Trokovic R, Trokovic N, Hernesniemi S, Pirvola U, Weisenhorn D. M. Vogt, Rossant J, McMahon AP, Wurst W, Partanen J. FGFR1 is independently required in both developing mid- and hindbrain for sustained response to isthmic signals. Embo J. 2003;22:1811–1823. doi: 10.1093/emboj/cdg169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsutsumi S, Shimazu A, Miyazaki K, Pan H, Koike C, Yoshida E, Takagishi K, Kato Y. Retention of multilineage differentiation potential of mesenchymal cells during proliferation in response to FGF. Biochem. Biophys. Res. Commun. 2001;288:413–429. doi: 10.1006/bbrc.2001.5777. [DOI] [PubMed] [Google Scholar]
- van den Brink, Bleuming SA, Hardwick JC, Schepman BL, Offerhaus GJ, Keller JJ, Nielsen C, Gaffield W, van Deventer SJ, Roberts DJ, Peppelenbosch MP. Indian Hedgehog is an antagonist of Wnt signaling in colonic epithelial cell differentiation. Nat. Genet. 2004;36:277–282. doi: 10.1038/ng1304. [DOI] [PubMed] [Google Scholar]
- van Es JH, van Gijn ME, Riccio O, van den Born M, Vooijs M, Begthel H, Cozijnsen M, Robine S, Winton DJ, Radtke F, Clevers H. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- Vooijs M, Ong CT, Hadland B, Huppert S, Liu Z, Korving J, van den Born M, Stappenbeck T, Wu Y, Clevers H, Kopan R. Mapping the consequence of Notch1 proteolysis in vivo with NIP-CRE. Development. 2007;134:535–544. doi: 10.1242/dev.02733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite KA, Eng C. From developmental disorder to heritable cancer: it’s all in the BMP/TGF-beta family. Nat. Rev. Genet. 2003;4:763–773. doi: 10.1038/nrg1178. [DOI] [PubMed] [Google Scholar]
- Wang X, Hisha H, Taketani S, Adachi Y, Li Q, Cui W, Cui Y, Wang J, Song C, Mizokami T, et al. Characterization of mesenchymal stem cells isolated from mouse fetal bone marrow. Stem Cells. 2006;24:482–493. doi: 10.1634/stemcells.2005-0219. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wang L, Iordanov H, Swietlicki EA, Zheng Q, Jiang S, Tang Y, Levin MS, Rubin DC. Epimorphin(-/-) mice have increased intestinal growth, decreased susceptibility to dextran sodium sulfate colitis, and impaired spermatogenesis. J. Clin. Invest. 2006;116:1535–1546. doi: 10.1172/JCI25442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Tang Y, Rubin DC, Levin MS. Chronically administered retinoic acid has trophic effects in the rat small intestine and promotes adaptation in a resection model of short bowel syndrome. Am. J. Physiol. Gastrointest. Liver Physiol. 2007;292:G1559–G1569. doi: 10.1152/ajpgi.00567.2006. [DOI] [PubMed] [Google Scholar]
- Weaver LT, Austin S, Cole TJ. Small intestinal length: a factor essential for gut adaptation. Gut. 1991;32:1321–1323. doi: 10.1136/gut.32.11.1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AC, Xu J, Yin Y, Smith C, Schmid G, Ornitz DM. FGF9 and SHH signaling coordinate lung growth and development through regulation of distinct mesenchymal domains. Development. 2006;133:1507–1517. doi: 10.1242/dev.02313. [DOI] [PubMed] [Google Scholar]
- Yu K, Ornitz DM. FGF signaling regulates mesenchymal differentiation and skeletal patterning along the limb bud proximodistal axis. Development. 2008;135:483–491. doi: 10.1242/dev.013268. [DOI] [PubMed] [Google Scholar]
- Yu K, Xu J, Liu Z, Sosic D, Shao J, Olson EN, Towler DA, Ornitz DM. Conditional Inactivation of FGF Receptor 2 Reveals an Essential role for FGF Signaling in the Regulation of Osteoblast Function and Bone Growth. Development. 2003;130:3063–3074. doi: 10.1242/dev.00491. [DOI] [PubMed] [Google Scholar]
- Zhang X, Stappenbeck TS, White AC, Lavine KJ, Gordon JI, Ornitz DM. Reciprocal epithelial-mesenchymal FGF signaling is required for cecal development. Development. 2006;133:173–180. doi: 10.1242/dev.02175. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. Fgf9, desmin and p-SMAD 1/5/8 expression in embryonic mouse small intestines. A) Whole mount small intestine from a WT E14.5 mouse utilized for in situ hybridization using a probe for Fgf9. The staining is consistent with epithelial expression as was demonstrated at earlier time points (Zhang et al., 2006).
B-E) Immunofluorescence-stained sections of E18.5 small intestines from (B, D) control and (C, E) Fgf9-/- embryos. Sections were stained with antisera directed against desmin (B, C; green) and p-Smad1/5/8 (D, E; red). No differences in desmin or p-Smad 1/5/8 expression when comparing small intestines from control and Fgf9-/- animals. Bars=30μm.
Supplemental Figure 2. p-Erk positive mesenchymal cells are diminished in DCR1R2 embryos and express Fst and Fgfr2. (A) Quantification of the mean percentage (+/- s.d.) of mesenchymal cells with detectable p-Erk in control and DCR1R2 embryos (E18.5). The double asterisk indicates a statistically significant difference when comparing DCR1R2 embryos versus controls as determined by a Student’s t test (P<0.01). Sections from control (B) E14.5 and (C,D) E18.5 small intestines stained with anti-Fgfr2 and (C) anti-pErk1/2 (red) and (D) anti-Fst (red). The white dashed lines in B-D indicate the epithelial/mesenchymal junction. The yellow dashed line in B indicates the mesenchymal/muscularis propria border. The cells circled by the yellow dashed lines in C and D show mesenchymal cells that are positive for both Fgfr2 and p-Erk1/2 (C) and Fst (D). Bars= 5μm.
Supplemental Figure 3. iMSCs are multi-potent, Fgf9 responsive and co-express Fst and pErk1/2. (A) Osteogenic induction of cultured iMSCs stained with alizarin red. The widespread positive cells indicated that iMSCs can differentiate into osteocytes. (B) Adipogenic induction of iMSCs stained with Oil Red O. The widespread positive cells indicated that iMSCs can differentiate into adipocytes. (C) Cultured, serum starved iMSCs phosphorylated Erk1/2 in response to treatment with 10 ng/ml Fgf9. Immunoblots at various time points post treatment are shown. (D) Cultured iMSC stained with antisera directed against p-Erk1/2 (green) and Fst (red). Arrow indicates the cell nucleus. Bars=50 μm (A, B). Bar=5 μm (D).