Abstract
We performed a genomic study combining single-cell mRNA differential display and RNA subtractive hybridization to elucidate CD8 T-cell quiescence/ignorance. By comparing actively maintained quiescent CD8 T cells from liver tumour tumour-infiltrating lymphocytes (TILs) with quiescent T cells at the single-cell level, we identified differentially expressed candidate genes by high-throughput screening and comparative analysis of expressed sequence tags (ESTs). While genes for the T-cell receptor, tumour necrosis factor (TNF) receptor, TNF-related apoptpsis inducing ligand (TRAIL) and perforin were down-regulated, key genes such as Tob, transforming growth factor (TGF)-β, lung Krüpple-like factor (LKLF), Sno-A, Ski, Myc, Ets-2 repressor factor (ERF) and RE1-silencing transcription factor (REST/NRSF) complex were highly expressed in the quiescent TIL CD8 cells. Real-time polymerase chain reaction (PCR) further confirmed these results. A regulation model is proposed for actively maintained quiescence in CD8 T cells, including three components: up-regulation of the TGF-β pathway, a shift in the MYC web and inhibition of the cell cycle.
Keywords: CD8, gene expression profiling, quiescence, T cells, tumour-infiltrating lymphocytes (TILs)
Introduction
Quiescent T lymphocytes are a group of T cells that display no spontaneous proliferation and a low metabolic rate. In quiescent T cells, quiescence reduces consumption of the resources (energy and cell size) required to maintain the vast repertoire of T cells. Only a small fraction of native lymphocytes will be clonally selected by antigens during the lifetime of the host. Based on the characteristics of T-cell quiescence, six different mechanisms for quiescence of T lymphocytes have been proposed: (i) thymic negative selection; (ii) peripheral clonal deletion; (iii) peripheral induced anergy; (iv) T-cell ignorance/indifference; (v) T-cell suppression; and (vi) T-cell senescence (exhaustion).1
Of particular interest is T-cell quiescence/ignorance in many solid tumours, such as sarcomas and carcinomas, which express tumour-specific antigens but evade immune surveillance. The quiescent state of the T lymphocyte has been defined as a phenotype lacking direct cytotoxicity and cell proliferation ex vivo and is thought to be attributable to a lack of activation signals.2,3 However, recent studies have indicated that quiescence in CD8 T cells is an actively maintained state rather than a default state in the absence of stimulated signals.4 To date, several proteins have been indicated to be involved in the regulation of T-cell quiescence. For example, the lung Krüpple-like factor (LKLF), a zinc finger-containing transcription factor, plays a critical role in maintaining T-cell quiescence.5 Another nuclear protein, Tob, is also necessary for regulation of quiescence of T lymphocytes.6 Recent microarray studies have demonstrated that reactivation of quiescent T lymphocytes is associated with increased gene expression promoting cell growth, as well as decreased gene expression maintaining T-cell quiescence.7 Although these studies have suggested the factors involved in the T-cell immune reaction, the molecular mechanisms underlying the quiescent status of CD8 T cells of TILs remain unclear because of technological difficulties in analysing the small number of T cells present within cancer tissues.
Here we successfully applied a genomic approach, at the single-cell level, to analysis of the gene expression profiles of quiescent CD8 T cells obtained from liver cancer patients. Our results demonstrate that inactivation of CD8 T cells involves increased expression of active genes. Quantitative real-time polymerase chain reaction (rtPCR) further confirmed these gene expression changes in quiescent CD8 T cells. The combination of our genomic and molecular approaches represents a promising strategy to characterize important factors involved in the maintenance of T-cell quiescence. Application of gene expression profiling provides a more efficient and comprehensive approach to the study of the genes responsible for actively maintaining the quiescent status of CD8 T cells.
Materials and methods
CD8 cells isolated from TILs
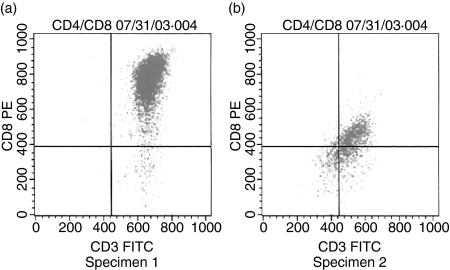
Here, we give an overview of the processes of isolation, validation, genomic analysis and functional assay of the quiescent CD cells. CD8 cells from TILs obtained from two liver cancer patients were isolated as described in our previous reports.8 Briefly, freshly isolated tumour tissue was washed in phosphate-buffered saline (PBS), cut into small pieces, digested in 0·25 mg/ml of collagenase IV at 4° for 24 hr and centrifuged in a Ficoll–Hypaque solution at 500 g for 15 min. The TILs were recovered from the interface of the cell suspension. After staining with fluorescein isothiocyanate (FITC)-labelled anti-CD8 monoclonal antibodies (mAbs) (BD Biosciences, San Jose, CA), each single CD8 cell was manually harvested under fluorescent microscopy in a 0·6-ml PCR tube (1 cell/μl) by single-cell manipulation as described for embryonic stem cells (ESCs).9 This harvesting process ensures pure CD8 cell collections. These individually harvested CD8 cells were then used for differential display and quantitative rtPCR. However, a large number of CD8 T cells from the same TILs were needed to determine their quiescent status using cell proliferation and cytotoxicity assays. These CD8 cells were isolated from the same TILs using magnetic anti-CD8 microbeads [magnetic antibody cell sorting (MACS) technology; Miltenyi Biotech, Foster City, CA] according to the manufacturer's recommendations. As a control, peripheral blood mononuclear cells (PBMC) and therein natural quiescent CD8 cells were prepared similarly as described above. CD8 cells incubated with interleukin (IL)-2 were used as activated CD8 cells. The purities of all CD8 cells were confirmed by fluorescence-activated cell sorting (FACS) after staining with FITC-labelled anti-CD8 mAb (Figs 1b,c). The quiescence of CD8 cells was measured by cell proliferation and cytotoxicity assays as previously described.10
Figure 1.
Flow cytometry analysis on day 30 of CD8 cell culture. (a, b) Flow cytometry analysis on day 30 of CD8 cell culture. The x-axis represents CD3 fluorescein isothiocyanate (FITC) and the y-axis represents CD8 FITC. Approximately 97·57% in the upper right quadrant and 2·33% in the lower right quadrant in specimen 1, and approximately 53·57% in the upper right quadrant and 24·26% in the lower right quadrant in specimen 2. PE, phycoerythrin.
CD8 cell cDNA libraries
Establishment of cDNA libraries from CD8 cells. A cDNA library was generated using a protocol that has been previously reported.10 Briefly, eight CD8+ cells from TILs were lysed in 8 μl of DNA digestion buffer with DNase I (Sigma, St Louis, MT). Two microlitres of DNA digestion solution was added to a cocktail mixture containing 1 μl of dNTP containing 5-methy-dCTP for protection, 1 μl of 50 mm 3′ anchor primer containing a HindIII restriction site [5′-CTCTAAGCTT(T)11-3′], 2 μl of MgCl2, 1 μl of 10× buffer, 0·25 μl of RNasin, 0·25 μl of AMV reverse transcriptase and 4·5 μl of sterile ddH2O (Promega, Fitchburg, WI). First-strand synthesis was performed at 25° for 10 min, 42° for 1 hr and 95° for 5 min. We used our previous design for the arbitrary primers in the CD8 cell differential display.11–13 The cDNA was analysed by PCR with four arbitrary 5′ primers containing an EcoRI restriction site and one 3′ primer containing a HindIII site as shown in Table 1 in a 25-μl volume using AmpliTaq Gold from Perkin Elmer (Washington, DC). After 35–40 cycles of PCR and double digestion by EcoRI and HindIII, the library was ligated to the pUC19 vector and transformed into the DH5α strain. The strains containing the CD8 cell library were stored at 4° for further study.
Table 1.
CD8 cDNA library primers
| Primer | Sequence |
|---|---|
| 5′-terminals1 | 5′-CTCTGAATTCCTGATCCATG-3′ |
| 5′-CTCTGAATTCCTTCATTGCC-3′ | |
| 5′-CTCTGAATTCCTGCTCTCAT-3′ | |
| 5′-CTCTGAATTCTCTGGAGGCA-3′ | |
| 3′-terminals2 | 5′-CTCTAAGCTT(T)11-3′ |
| pUC19 forward3 | 5′-AAACGACGGCCAGTGAATTC-3′ |
| pUC19 reverse3 | 5′-ATGACCATGATTACGCCAA-3′ |
Arbitary polymerase chain reaction (PCR) primers with CTCT protective base pairs and EcoRI restriction site.
Oligo-T terminal with CTCT protective base pairs and HindIII restriction site.
pUC19 primers for determining PCR product sizes.
Removal of the non-specific CD8 library and PCR detecting representatives. The removal of the non-specific TIL library was performed as reported previously.11 Briefly, after denaturation, neutralization and prehybridization, the nylon membrane replicas blotted from library plates were hybridized using a reference cDNA library obtained from pairing of PBMC CD8+ cells and prepared using murine Moloney leukaemia virus (MMLV) reverse transcriptase (Promega) with an oligoT primer. Digoxigenin-dUTP was incorporated into the reference cDNA library using reverse transcription PCR, and then the reference library was hybridized onto the nylon membranes. Anti-digoxigenin-AP and X-phosphate with nitroblue tetrazolium dye (NBT) were used to indicate positive colonies (DIG nucleic acid detection kit; Boehringer Mannheim Biochemica, Indianapolis, IN). The colonies demonstrating negative signals (which contained the CD8-specific genes) were picked out for PCR analysis. Briefly, the negative colonies were eluted into SM buffer. Eluted colonies (5 μl) were amplified by PCR in 50 μl of SM buffer with 10× Taq buffer and 1 μl of Taq polymerase. Primers flanking pUC 19 terminals (Table 1) were used to amplify clone pools and PCR cycles were run in 96-well thermal cycling plates (Hybaid, Waltham, MA). If the sizes of PCR products were similar, one of the products was chosen as a representative for DNA sequencing.
DNA sequencing of representatives and dot blot hybridization. DNA sequences were determined using a sequencing kit (Sequencing Kit; USB Inc., Cleveland, OH) as shown previously.8 After DNA sequencing, the probes chosen from the known sequence representatives and labelled using a DIG nucleic acid detection kit (Boehringer Mannheim Biochemica) were hybridized onto nylon membranes, on which DNA from different colonies containing similar sizes of PCR products had been immobilized.
Bioinformatics search and UniGene comparison. Similarity searches of the DNA sequence representatives were performed with the BLAST algorithm at the National Center for Biotechnology Information (NCBI), using the blastn program and, when necessary, the blastx program (http://www.ncbi.nlm.nih.gov/blast). Sequence comparisons for the expressed gene were carried out against all entries in the non-redundant (nr) database for Homo sapiens, and, when necessary, using the dbEST database (http://www.ncbi.nlm.nih.gov/dbEST/index.html). As described previously,8 for assignment of putative identities, an E value of < 10−10 and a blastxE value of < 10−5 were used. Subsequently, UniGene (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene) was used to determine the similarity between T-cell UniGene cluster and TIL gene expression. Approximately 7–9% of sequences did not have significant hits against Homo sapiens sequences, suggesting contamination with DNA from bacteria. In a total of 312 and 288 colonies from specimens 1 and 2, respectively, sequence analyses indicated that nine and 13 colonies were probably bacterial, and 13 and 11 colonies, respectively, did not have significant hits according to our E value requirement. These sequences were effectively removed using our E-value cut-offs.
CD8 cell profile expression confirmed by rtPCR. The rtPCR assay was performed in triplicate for each gene. The 50-μl PCR reaction mixture contained 25 μl of 2× SYBR Green (BioRad, Cleveland, OH), 500 nm of each primer, RNA extract and 1 μl of iScript reverse transcriptase™ (Promega). Using appropriate primer conditions and following the manufacturer's recommendations, one-step rtPCR was performed for 10 min at 50° and 5 min at 95°, followed by 45 cycles of denaturation for 10 seconds at 95° and annealing/extension for 30 seconds at 55°. The SYBR fluorescence signal intensities were recorded and analysed during the PCR in an ABI Prism 7700 sequence detector system (Applied Biosystems, Foster City, CA) using sds (version 1.91) software.14
Kinetic change in CD8 cell gene expression
To study the kinetic change in gene expression, CD8 T cells were divided into four groups (100 cells per group). Group 1, at day 0, was isolated by single-cell manipulation and used for cDNA library construction and rtPCR confirmation as described above, and groups 2, 3 and 4 were isolated by MACS and then by single-cell manipulation after culture at 37° in a 5% CO2 atmosphere until days 2, 7 and 30, respectively, to determine CD8 T-cell activation status. The culture medium contained 10% [volume/volume (v/v)] heat-inactivated human AB serum (Sigma) along with recombinant IL-2 (rIL-2) at 2000 U/ml culture medium (Sigma). To study the expression kinetics of the genes in the activated state, an rtPCR assay was performed similarly to that described above using the cultured CD8 T cells incubated with IL-2 on days 2, 7 and 30.
Results
Library screen
We first determined the status of CD8 cells isolated from TILs (Fig. 1). No activated T cells were detected using cell proliferation and cytotoxicity assays. We previously reported a strategy for screening a cDNA library using CD3 cells.8 Using the same strategy, here we examined quiescent CD8 cell gene expression in the same liver cancer specimens. The cDNA library of CD8 cells was amplified using rtPCR and PCR, and then cloned using a prokaryotic vector. The non-specific TIL library was removed using the reference cDNA library obtained from the pairing of PBMC quiescent CD8 cells. In total, 312 and 288 colonies from two liver tumour specimens (1 and 2), respectively, were picked out as library representatives. After sequencing and bioinformatics analyses of the selected colonies, the combined results demonstrated that expression of at least five and seven genes, respectively, in CD8 T cells was different in these two tumour specimens as compared with normal specimens.
cDNA sequence analyses
After sequencing, five and seven differentially expressed cDNAs in these two tumour specimens, respectively, were analysed using a blastn search of the nr (non-redundant) nucleotide and dbEST Homo sapiens databases, and, when necessary, a blastx search of a GenBank Homo sapiens protein database. Interestingly, by comparison with reported human genomic sequences in GenBank and dbEST, we found four genes expressed in both specimens 1 and 2, namely Myc, Tob, Ets-2 repressor factor (ERF) and REST/NRSF. Genes expressed in only one library were TGF-β for specimen 1 and LKLF, Ski and Sno-A for specimen 2, as shown in Table 2.
Table 2.
Gene expression by the specimens
| Specimen | Gene |
|---|---|
| Both specimens 1 and 2 | c-Myc |
| Tob | |
| REST | |
| REST/NRSF | |
| Only specimen 1 | TGF-β |
| Only specimen 2 | LKLF |
| Ski | |
| Sno-A |
LKLF, lung Krüpple-like factor; TGF, transforming growth factor.
Verification of gene expression by rtPCR
The rtPCR assay was performed in triplicate as described in the Materials and methods. Each sample contained 100 quiescent CD8 T cells isolated by single-cell manipulation. All experiments used the β-actin gene as the control. In specimen 1, TGF-β was highly expressed with a threshold of 23·0 ± 4·0 cycles, similar to β-actin, as shown in Table 3. Other genes in specimen 1 were expressed at thresholds ranging from 27·0 ± 4·0 to 30·0 ± 5·0. In specimen 2, Ski and Sno-A were highly expressed at thresholds of 27·0 ± 4·0 and 27·0 ± 5·2, respectively, as indicated in Table 4. Although library screening identified five genes in specimen 1 and seven genes in specimen 2, as mentioned above (with four genes shared and the other four genes expressed differentially in the two specimens), comparison of total eight gene expressions between both specimens were not significantly different using rtPCR (P = 0·081).
Table 3.
Real-time polymerase chain reaction (rtPCR) analysis of gene expression in specimen 1
| Mean ± SD of threshold cycle | ||
|---|---|---|
| Gene | TIL CD8 cells | PB CD8 cells |
| β-actin | 23·0 ± 2·6 | 23·5 ± 2·6 |
| Tob | 28·0 ± 3·5* | 30·2 ± 3·1 |
| Ski | 27·0 ± 4·0 | 28·1 ± 4·1 |
| Sno-A | 30·0 ± 5·2 | 30·9 ± 3·5 |
| TGF-β | 23·0 ± 4·01 | 27·2 ± 3·1 |
| LKLF | 30·0 ± 3·0 | 29·2 ± 2·2 |
| ERF | 26·0 ± 6·01 | 29·3 ± 2·4 |
| REST/NRSF | 28·0 ± 3·0 | 28·9 ± 2·6 |
| c-Myc | 30·0 ± 5·0 | 29·1 ± 3·1 |
For rtPCR, 100 CD8 cells were used.
Mean values more than two threshold cycle differences.
PB, peripheral blood; TIL, tumour-infiltrating lymphocyte; LKLF, lung Krüpple-like factor; TGF, transforming growth factor; SD, standard deviation.
Table 4.
Real-time polymerase chain reaction (rtPCR) analysis of gene expression in specimen 2
| Mean ± SD of threshold cycle | ||
|---|---|---|
| Gene | TIL CD8 cells | PBMC CD8 cells |
| β-actin | 24·0 ± 3·2 | 23·2 ± 2·7 |
| Tob | 29·0 ± 2·81 | 31·0 ± 3·1 |
| Ski | 27·0 ± 4·0 | 28·1 ± 1·9 |
| Sno-A | 27·0 ± 5·2 | 28·0 ± 2·6 |
| TGF-β | 25·0 ± 2·61 | 29·0 ± 2·1 |
| LKLF | 29·0 ± 3·0 | 30·0 ± 1·9 |
| ERF | 27·0 ± 4·5 | 28·1 ± 2·1 |
| REST/NRSF | 26·0 ± 3·41 | 28·1 ± 2·8 |
| c-Myc | 29·0 ± 4·0 | 29·2 ± 3·1 |
For rtPCR, 100 CD8 cells were used.
Mean values are more than two threshold cycle differences.
PBMC, peripheral blood mononuclear cells; TIL, tumour-infiltrating lymphocyte; LKLF, lung Krüpple-like factor; TGF, transforming growth factor; SD, standard deviation.
Change in gene expression kinetics in the activated state
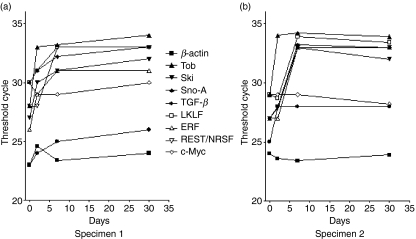
We next compared the gene expression kinetics between the naïve quiescent and in vitro activated states. One hundred CD8 T cells isolated from both specimens were analysed by rtPCR, and the β-actin gene was used as the internal control. As shown in Figs 2(a,b), several categories of genes were identified exhibiting changes in expression kinetics. In both specimens, Ski and Sno-A showed a clear decrease in expression between days 2 and 7 in in vitro culture. However, c-Myc gene expression did not change. The transcription factors LKLF and ERF exhibited reduced expression after in vitro culture but in a different time frame. The decrease started on day 2 in specimen 1 (Fig. 2a) but on day 7 in specimen 2 (Fig. 2b). Interestingly, the Tob gene showed a dramatic reduction from day 0 to day 2, and then remained slightly decreased on days 7 and 30. The last gene identified was TGF-β, whose expression showed a continuous slow reduction throughout the culture in specimen 1; whereas in specimen 2 it decreased markedly between days 0 and 2, and then remained relatively stable until day 30.
Figure 2.
Changes in gene expression in specimens 1 and 2. Real-time polymerase chain reaction (PCR)-based assays were used to validate gene expression during the in vitro culture of CD8 cells. (a) Expression changes in specimen 1; (b) expression changes in specimen 2. ERF, Ets-2 repressor factor; LKLF, lung Krüpple-like factor; TGF, transforming growth factor.
Change in T-cell receptor (TCR) expression
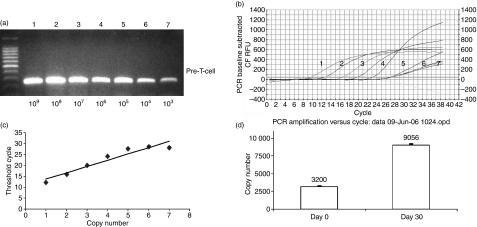
Regarding T-cell function, data obtained in our previous study demonstrated that gene expression profiles varied in isolated TIL CD3 cells, when cultured in vitro for 15–30 days.8 Genes that are related to T-cell cytotoxicity, such as perforin, FAS ligand and granzyme B, were identified. In the current study, we compared gene expression profiles between quiescent and activated stages in TIL CD8 cells. In addition to perforin, FAS ligand and granzyme B, we also examined gene expression for TCR, TNF-R, and TRAIL, which are known to be involved in cell–cell interaction or communication. As indicated in Fig. 3, TCR expression was up-regulated at day 30 compared with the resting level at day 0. TNF-R, perforin and TRAIL were also up-regulated compared with levels on day 0. The magnitude of these increases in gene expression level varied between two- and fourfold (data not shown).
Figure 3.
Normalized expression results for the T-cell receptor obtained using real-time polymerase chain reaction (PCR). (a) Pre-T-cell receptor copy number related to band densities: lane 1, 109; lane 2, 108; lane 3, 107; lane 4, 106; lane 5, 105; lane 6, 104; lane 7, 103. (b) PCR baseline subtracted relative fluorescence units (RFU) curve related to T-cell receptor sample copy numbers: lane 1, 109; lane 2, 108; lane 3, 107; lane 4, 106; lane 5, 105. (c) Relationship between threshold cycle and pre-T-cell receptor copy number: lane 1, 109; lane 2, 108; lane 3, 107; lane 4, 106; lane 5, 105; lane 6, 104; lane 7, 103. (d) Quantitative change in T-cell receptor expression on days 0 and 30.
Discussion
T cells (CD3 cells) comprise CD8 T lymphocytes [cytotoxic T lymphocytes (CTLs)] and CD4 T cells (helper T cells) which can both kill virally infected cells and cancer cells via cytotoxicity. CD8 cells, which can destroy tumour cells, have been studied for over two decades.15–18 Our previous studies characterized the genomic expression profiles of the CD3 cells obtained from TILs.8 In this study, CD8 cells from TILs obtained from the same liver cancer specimens were analysed for gene expression profiles. We addressed the following questions: (i) whether the quiescence of the TIL CD8 cells is an actively maintained state or a default state in the absence of the stimulating signals; (ii) how the CD8 T cells maintain their quiescent status; (iii) which signalling pathways are involved in the quiescent state.
Although several papers have reported CD8 cell gene expression in the activation state, there is a general lack of studies of the quiescent state. This is mainly a consequence of the limited number of CD8 quiescent cells available and/or the mixed cell situation in vivo. The number of quiescent CD8 cells present in TILs is very limited and normally only 5000–50 000 CD8 cells can be harvested in each specimen. Of this limited collection of quiescent CD8 cells, most of them will be used for function assays, such as cell proliferation and cytotoxicity assays, and a portion of them will be used to obtain the activated CD8 cells. Furthermore, although MACS is a quick way to isolate large amounts of CD8 cells, our FACS results showed 100% purity using single-cell manipulation as compared with 89–95% purity using MACS (data not shown). Because CD8 cell purity is crucial for differential display, the quiescent CD8 cells obtained from single-cell manipulation should produce more reliable results as compared with quiescent CD8 cells isolated by MACS. Our integrated strategy was designed to utilize the advantages of both methods: the 100% purity obtained using single-cell manipulation for genomic analyses such as differential display and the collection of large amounts of cells from MACS for functional assays such as cell proliferation and cytotoxicity assays. We have successfully implemented this strategy, combining single-cell mRNA differential display and RNA subtractive hybridization, to study CD3 cells using a single cell or a small number of cells.8 In fact, the CD3 cells obtained from single-cell manipulation produced more reliable results than CD3 cells isolated by MACS. The modified strategy also transforms the gel-band-pattern results of mRNA differential display into colony-pattern results using cloning into vectors. Furthermore, subtractive hybridization is introduced into the modified method so that the artifacts of cDNA amplification from test cells are eliminated by hybridization.
Here, we further applied the genomic approach in using sequence tags to study gene expression in the quiescent CD8 cells obtained from the same in vivo liver cancers. Our results demonstrated that multiple genes (Tob, TGF-β, Sno-A, Ski, LKLF, Myc, ERF and the REST/NRSF complex) were expressed in the actively maintained quiescent state of CD8 cells as compared with the natural quiescent state of T cells.
The Tob gene is a member of a family of anti-proliferative genes, whose protein expression prevents cell proliferation by constraining the cell cycle. Some reports suggested that Tob is involved in the control of G1-S progression through suppression of cyclin D1 expression.19 Our finding of decreased Tob expression after day 2 in in vitro culture sheds light on its important function in TILs at the early quiescent stage. As discussed previously, another possibility is that Tob suppresses transcription of activating cytokines, including IL-2 and interferon (IFN)-γ, as well as cell cycle activators (such as cyclin A and cyclin E).20
The second important gene is TGF-β, which was highly expressed in both specimens. The known function of TGF-β suggests that it is a multifunctional peptide that controls proliferation, differentiation and other functions in many cell types.21–23 Our data demonstrated that TGF-β was highly expressed in the CD8 cell quiescent state. It is known that Ski and Sno-A negatively regulate the TGF-β signalling pathway by recruiting this complex to Smads.24 We also found high levels of expression of Ski and Sno-A, but barely detected Smads expression in the quiescent state. These findings suggest that TGF-β probably maintains cell quiescence depending on the expression levels of Ski and Sno-A in CD8 cells.
Myc protein broadly controls cell growth, including cell proliferation, differentiation, apoptosis and inhibition. Myc protein is known to associate with ancillary factors to control cell growth. Because of its broad control of cell growth, the Myc pathway is also called the MYC web.25 LKLF was reported to play an important role in the establishment and maintenance of the quiescent state in CD4+ and CD8+ T lymphocytes, and it is sufficient to programme T-cell quiescence via a c-Myc-dependent web (pathway).26,27 ERF, which is an ETS domain protein with strong transcriptional repressor activity, has been shown to be able to suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation.28 Consistent with these earlier findings, we detected reduced expression levels of LKLF and ERF after day 2 in in vitro culture. LKLF may play an important role in T-cell quiescence through the MYC web (pathway).
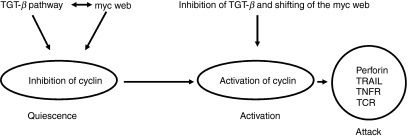
As expression of multiple genes was detected in the CD8 quiescent stage, we propose three possible mechanisms for maintenance of the quiescent state: (i) inhibition of the cell cycle by the Tob protein; (ii) up-regulation of the TGF-β pathway by TGF-β, Sno-A and Ski; and (iii) a shift of the MYC web (pathway) by c-Myc, ERF and LKLF (Fig. 4).
Figure 4.
Hypothesized mechanisms of CD8 quiescence, activation and attack.
Finally, we also compared the expression profiles of genes that play important roles in CD3 cells with those that play important roles in CD8 cells by quantitative rtPCR. In our previous study, six genes (granzyme B, FASL, perforin, TCR, TNF-R and TRAIL) were found to be expressed in quiescent CD3 cells. Here, expression of only four genes (perforin, TCR, TNF-R and TRAIL) was detected in CD8 cells on day 0 and these genes were highly expressed on day 30 as shown in Table 5. Grazyme B and FASL were not detected in CD8 cells on day 0. Our data suggest that the TIL lytic defect might be an acquired property of T cells restrained within the tumour microenvironment. Studies performed by others suggest that TCR-mediated signalling is rapidly blocked after conjugation with cognate tumour targets.29 Our data showed a threefold increase in TCR expression after culture for 30 days. Moreover, high levels of expression of perforin, TNF-R and TRAIL have been suggested to be involved in triggering apoptosis in target cells after activation, thereby providing T cells with cytolytic activity.
Table 5.
Real-time polymerase chain reaction (rtPCR) analysis of CD8 tumour-infiltrating lymphocytes (TILs) on days 0 and 30
| Groups | Fold change (mean ± SD) |
|---|---|
| Pre-TCR | 2·83 ± 0·23 |
| TRAIL | 3·88 ± 0·62 |
| Perforin | 2·63 ± 0·21 |
| TNF receptor | 3·45 ± 0·43 |
TCR, T-cell receptor; TNF, tumour necrosis factor; TRAIL, TNF-related apoptpsis inducing ligand.
In conclusion, we have utilized an optimized and integrated strategy to address questions regarding CD8 T-lymphocyte quiescence and activation in vivo. It is interesting to note the differences in gene expression found between the two liver cancer specimens in the initial screen of cDNA libraries based on differential display and subtractive hybridization. However, all of the gene expression patterns were later confirmed by rtPCR because of its higher sensitivity and these expression patterns did not show significant differences between the specimens. We also repeated cDNA library construction three times using CD3 cells from the same specimen and found that the differences were minimal (data not shown). Therefore, these similar results obtained for different specimens strongly indicate that the expression profiles of the eight genes identified are the generalized common themes of quiescent CD8 cells in TILs. It is possible that, if the CD8 T cells in TILs are not homogenous, our results based on a single cell or a small number of cells reflect the expression pattern in the individual cells and additional single-cell sampling could reveal additional genes. Further investigations will be needed to fully elucidate the mechanisms of blocking and activation of CD8 cells in Th1 CD4 cells to facilitate personalized tumour therapy. Studying gene expression profiles for a small number of cells or at the single-cell level may provide a basis for manipulating the immune response for cancer prevention and therapy through passive and active immunity.
Acknowledgments
Under the support of former Dr H. D. Preisler, we set up the method of analysis of CD3, CD4 and CD8 cells from TILs. The work was supported by National Cancer Institute grant IRG-91-022-09 to BL as principal investigatorand in part by USDA CRIS Project No. 1265-31000-090-00D (to GEL). Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the US Department of Agriculture.
References
- 1.Melero I, Arina A, Cen L. Many Sounds of T lymphocyte Silence. Immunol Res. 2005;33:135–48. doi: 10.1385/IR:33:2:135. [DOI] [PubMed] [Google Scholar]
- 2.Koba F, Akiyoshi T, Tsuji H. Depression of the generation of cell-mediated cytotoxicity in regional lymph nodes of patients with gastric carcinoma. J Clin Lab Immunol. 1987;22:181–4. [PubMed] [Google Scholar]
- 3.Monsurro V, Wang E, Yamano Y, et al. Quiescent phenotype of tumor-specific CD8+ T cells following immunization. Blood. 2004;104:1970–8. doi: 10.1182/blood-2004-02-0525. [DOI] [PubMed] [Google Scholar]
- 4.Marrack P, Mitchell T, Hildeman D, et al. Genomic-scale analysis of gene expression in resting and activated T cells. Curr Opin Immunol. 2000;12:206–9. doi: 10.1016/s0952-7915(99)00075-8. [DOI] [PubMed] [Google Scholar]
- 5.James P, Did S. Lung Krüpple-like factor: a quintessential player in T cell quiescence. Nat Immunol. 2001;2:667–8. doi: 10.1038/90598. [DOI] [PubMed] [Google Scholar]
- 6.Tzachanis D, Freeman GJ, Hirano N, Andre vPAFL, Delfs MW, Alla BLM, Boussiotis VA. Tob is a negative regulator of activation that is expressed in anergic and quiescent T cells. Nat Immunol. 2001;2:1174–82. doi: 10.1038/ni730. [DOI] [PubMed] [Google Scholar]
- 7.Giacomelli L, Nicolini C. Gene expression of human T lymphocytes cell cycle. J Cell Biochem. 2006;2:22–6. doi: 10.1002/jcb.20991. Experimental and bioinformatic analysis. [DOI] [PubMed] [Google Scholar]
- 8.Li B, Perabekam S, Liu G, Yin M, Song S, Larson A. Experimental and bioinformatics comparison of gene expression between T cells from TIL of liver cancer and T cells from UniGene. J Gastroenterol. 2002;37:275–82. doi: 10.1007/s005350200035. [DOI] [PubMed] [Google Scholar]
- 9.Babinet C, Cohen-Tannoudji M. Genome engineering via homologous recombination in mouse embryonic stem (ES) cells: an amazingly versatile tool for the study of mammalian biology. An Acad Bras Cienc. 2001;73:365–83. doi: 10.1590/s0001-37652001000300007. [DOI] [PubMed] [Google Scholar]
- 10.Li B, Ding J, Larson A, Song S. Tumor tissue recycling–a new combination treatment for solid tumors: experimental and preliminary clinical research. In vivo. 1999;13:433–8. [PubMed] [Google Scholar]
- 11.Li B, Chang T, Larson A, Ding J. Identification of mRNAs expressed in tumor-infiltrating lymphocytes by a strategy for rapid and high throughput screening. Gene. 2000;255:273–9. doi: 10.1016/s0378-1119(00)00330-9. [DOI] [PubMed] [Google Scholar]
- 12.Tian DY, Hayashi M, Yoshida T, Sakakura T, Kawarada Y, Tanaka T. Overexpression of caltractin gene in tumor-infiltrating lymphocytes. Int J Oncol. 1998;13:1135–40. doi: 10.3892/ijo.13.6.1135. [DOI] [PubMed] [Google Scholar]
- 13.Renner C, Trumper L, Pfitzenmeier JP, Loftin U, Gerlach K, Stehle I, Wadle A, Pfreundschuh M. Differential mRNA display at the single-cell level. Biotechniques. 1998;24:720–2. doi: 10.2144/98245bm04. [DOI] [PubMed] [Google Scholar]
- 14.Li B, Phillips BN, Agnes JR, Varda I, Singh R, Jones DN, Haas E, Weiss MA. SRY-directed DNA bending and human sex reversal: reassessment of a clinical mutation uncovers a global coupling between the HMG box and its tail. J Mol Biol. 2006;360:310–28. doi: 10.1016/j.jmb.2006.04.048. [DOI] [PubMed] [Google Scholar]
- 15.Kowalczyk D, Skorupski W, Kwias Z, Nowak J. Activated gamma/delta T lymphocytes infiltrating renal cell carcinoma. Immunol Lett. 1996;53:15–8. doi: 10.1016/0165-2478(96)02605-3. [DOI] [PubMed] [Google Scholar]
- 16.Gervois N, Guilloux Y, Diez E, Jotereau F. Suboptimal activation of melanoma infiltrating lymphocytes (TIL) due to low avidity of TCR/MHC-tumor peptide interactions. J Exp Med. 1996;183:2403–7. doi: 10.1084/jem.183.5.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darmon AJ, Bleackley RC. Proteases and cell-mediated cytotoxicity. Crit Rev Immunol. 1998;18:255–73. doi: 10.1615/critrevimmunol.v18.i3.50. [DOI] [PubMed] [Google Scholar]
- 18.Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, Hengartner H, Golstein P. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science. 1994;265:528–30. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 19.Hiramatsu Y, Kitagawa K, Suzuki T, et al. Degradation of Tob1 mediated by SCFSkp2-dependent ubiquitination. Cancer Res. 2006;66:8477–83. doi: 10.1158/0008-5472.CAN-06-1603. [DOI] [PubMed] [Google Scholar]
- 20.Okochi K, Suzuki T, Inoue JI, Matsuda S, Yamamoto T. Interaction of anti-proliferative protein Tob with poly(A)-binding protein and inducible poly(A)-binding protein: implication of Tob in translational control. Genes. 2005;10:151–63. doi: 10.1111/j.1365-2443.2005.00826.x. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, McKinnon ML, Leong LM, Rusholme SAB, Wang S, Akhurst RJ. Genetic modifiers interact with maternal determinants in vascular development of Tgfb1-/- mice. Hum Mol Genet. 2003;12:1579–89. doi: 10.1093/hmg/ddg164. [DOI] [PubMed] [Google Scholar]
- 22.Thyagarajan T, Sreenath T, Cho A, Wright JT, Kulkarni AB. Reduced expression of dentin sialophosphoprotein is associated with dysplastic dentin in mice overexpressing transforming growth factor-beta-1 in teeth. J Biol Chem. 2001;276:11016–20. doi: 10.1074/jbc.M010502200. [DOI] [PubMed] [Google Scholar]
- 23.Wurdak H, Ittner LM, Lang KS, et al. Inactivation of TGF-beta signaling in neural crest stem cells leads to multiple defects reminiscent of DiGeorge syndrome. Genes Dev. 2005;19:530–5. doi: 10.1101/gad.317405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shinagawa T, Dong HD, Xu M, Maekawa T, Ishii S. The sno gene, which encodes a component of the histone deacetylase complex, acts as a tumor suppressor in mice. EMBO J. 2000;19:2280–91. doi: 10.1093/emboj/19.10.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levens D. Disentangling the MYC web. Proc Natl Aca Sci USA. 2002;99:6274–9. doi: 10.1073/pnas.102173199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carlson CM, Endrizzi BT, Wu JH, et al. Kruppel-like factor 2 regulates thymocyte and T-cell migration. Nature. 2006;442:299–302. doi: 10.1038/nature04882. [DOI] [PubMed] [Google Scholar]
- 27.Haaland RE, Yu WD, Rice AP. Identification of LKLF-regulated genes in quiescent CD4+ T lymphocytes. Mol Immunol. 2005;42:627–41. doi: 10.1016/j.molimm.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 28.Sgouras DN, Athanasiou MA, Beal GJ, Fisher JRJ, Blair DG, Mavrothalassitis GJ. ERF: an ETS domain protein with strong transcriptional repressor activity, can suppress ets-associated tumorigenesis and is regulated by phosphorylation during cell cycle and mitogenic stimulation. EMBO J. 1995;14:4781–93. doi: 10.1002/j.1460-2075.1995.tb00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koneru M, Schae D, Monu N, Ayala A, Frey AB. Defective proximal TCR signaling inhibits CD8+ tumor-infiltrating lymphocyte lytic function. J Immunol. 2005;174:1830–40. doi: 10.4049/jimmunol.174.4.1830. [DOI] [PubMed] [Google Scholar]