Abstract
Innate immunity plays a role in systemic lupus erythematosus (SLE). Our objective was to determine the levels of defensins, which are antimicrobial and immunomodulatory polypeptides, in SLE. Sera from SLE patients and healthy controls were tested for pro-inflammatory human β-defensin 2 (hBD-2) and for α-defensin human neutrophil peptide 1 (HNP-1). hBD-2 could not be detected by enzyme-linked immunosorbent assay (ELISA) and its mRNA levels were low in SLE patients and similar to those found in controls. In contrast, the mean α-defensin level in the sera of all SLE patients (11·07 ± 13·92 ng/μl) was significantly higher than that of controls (0·12 ± 0·07 ng/μl). Moreover, 60% of patients demonstrated very high serum levels (18·5 ± 13·36 ng/μl) and 50% showed elevated gene expression in polymorphonuclear cells. High α-defensin levels correlated with disease activity, but not with neutrophil count. Thus, activation and degranulation of neutrophils led to α-defensin secretion in SLE patients. Given the immunomodulatory role of α-defensins, it is possible that their secretion may activate the adaptive immune system leading to a systemic response.
Keywords: defensin, human neutrophil peptide, systemic lupus erythematosus
Introduction
Systemic lupus erythematosus (SLE) is an autoimmune disorder that involves every organ system, although the joints, skin, blood cells (red blood cells, white blood cells and platelets), kidneys, brain and serosa are most frequently affected.1,2 The precise aetiology of SLE is unknown, but hormonal,3 genetic,4 viral,5 and environmental factors6 appear to play a role in the development and course of the disease. SLE is characterized by the production of a variety of autoantibodies and impaired function of B and T cells.7,8 Previous studies also demonstrated roles for cytokines,9 apoptosis,10 and dysfunction of regulatory T cells11 in the pathogenesis of murine and human SLE. Recently, it has been shown that the innate immune system also plays a role in the pathogenesis of SLE.12,13
The innate immune system confers broad protection against pathogens and most multicellular organisms depend upon it to combat microbial infections. One mechanism of innate immunity is the secretion of broad-spectrum antibacterial substances, such as small cationic polypeptides (3–5 kDa) named defensins.14–16α-Defensins, which are particularly abundant in neutrophils and Paneth cells of the small intestine, are small polypeptides with 29–35 residues and a six-cysteine motif forming three intramolecular disulphide bonds. β-Defensins, which are found in skeletal muscles, the airways, the oesophagus, the tongue, the trachea, the intestine and the liver, differ from α-defensins in size (38–42 amino acid residues) and cysteine pairing.16–18
Some defensins are produced constitutively, and some in response to microbial products or pro-inflammatory cytokines.17,18 All defensins identified to date have the capability to kill and/or inactivate a spectrum of bacteria, fungi and some enveloped viruses.19 This ability to kill micro-organisms is ascribed to their ability to disrupt membrane integrity and function.17,19 In addition to exerting direct antimicrobial effects, defensins facilitate and amplify subsequent innate and adaptive immune responses, such as activation and degranulation of mast cells, interleukin and tumour necrosis factor production, and maturation of dendritic cells.17,19–21 Thus, defensins provide the first line of defence against colonization of pathogens and play a crucial and indispensable role in both the innate and adaptive immune responses.
Recently, using oligonucleotide microarrays, the α-defensin gene DEFA-3 was shown to be up-regulated in SLE patients.22,23 In addition, a high correlation was found between high SLE disease activity and expression of the formyl peptide receptor-like 1 protein that mediates the chemotactic activities of defensins.22 Also, increased levels of anti-defensin antibodies were found in sera of patients with SLE compared with those of normal controls. Higher levels of anti-defensin antibodies were detected in both patients with active SLE and those with inactive SLE, but the level in patients with active SLE was significantly higher. These high levels of anti-defensin antibodies decreased after therapy with high doses of corticosteroids.24 These recent findings suggest a role for defensins in the pathogenesis of SLE. Therefore, we set out to study α- and β-defensin levels in the blood of SLE patients at both the mRNA and protein levels.
Materials and methods
Patients
All patients were diagnosed with SLE according to four or more of the American College of Rheumatology (ACR) revised criteria.25 Disease activity was defined using the SLE Disease Activity Index (SLEDAI).26 Twenty age- and sex-matched healthy volunteers served as the control group. All participants (SLE patients and healthy controls) signed an informed consent form prior to the initiation of the study. The study was approved by the Kaplan Medical Center ethic committee and was conducted according to all good clinical practice (GCP) rules. Sera obtained from SLE patients (1–11 samples; mean 4·87 ± 2·04 samples per patient) and healthy controls were frozen at −20° prior to the performance of the enzyme-linked immunosorbent assay (ELISA). For RNA analyses, whole blood was collected in Tempus blood RNA tubes (Applied Biosystems, Foster City, CA).
Enzyme-linked immunosorbent assay (ELISA)
Microtitre plates were coated with 5 μl of serum in 50 mm carbonate buffer, pH 9·6, and incubated for 1 hr at room temperature. In parallel, a standard curve of purified human neutrophil peptide 1 (HNP-1) or human β-defensin 2 (hBD-2) (Sigma, Rehovot, Israel) was obtained. Wells were then blocked with blocking solution [phosphate-buffered saline (PBS) containing 1% bovine serum albumin (BSA)] and incubated for 1 hr at room temperature. Blocking solution was then removed and wells were washed once with PBS. Goat anti-HNP-1 or rabbit anti-hBD-2 polyclonal antibody (Chemicon, Billerica, MA) diluted in blocking solution according to the manufacturer's instructions was added and the plates were incubated for 1 hr at room temperature. The plates were washed three times with PBS containing 0·05% Tween-20 and then incubated with alkaline phosphatase-conjugated rabbit anti-goat immunoglobulin G (IgG) or mouse anti-rabbit IgG (Santa Cruz, Santa Cruz, CA) for 1 hr at room temperature. The plates were washed three times with PBS containing 0·05% Tween-20 and then p-nitrophenyl phosphate alkaline phosphatase substrate (PNPP) (Chemicon) was added. The plates were read at 405 nm using a Tristar LB-941 luminometer (Berthold Technologies, Bad Wildbad, Germany). The concentration of defensins in the serum was calculated according to the standard curve. The upper normal level for α-defensins was defined as the mean + 2 standard deviations (SDs) of levels determined for the 20 healthy controls.
RNA extraction and quantitative real-time polymerase chain reaction (PCR)
Total RNA was extracted from whole blood from SLE patients using Tempus blood RNA tubes according to the manufacturer's instructions (Applied Biosystems). Total RNA was DNAse I-treated using RQ1 DNAse (Promega, Madison, WI) for 2 hr at 37°, as was previously described.27,28 Then 2 μg of DNAse I-treated RNA was reverse-transcribed using Moloney murine leukaemia virus (MMuLV) reverse transcriptase (Promega) and random hexamers. A 1/20 aliquot of the reaction was then subjected to quantitative real-time PCR using the Sybr Green Master kit (Applied Biosystems) and the ABI Prism 7300 Sequence Detection System (Applied Biosystems). Primers for HNP-1 (HNP-1-F 5′-tgcatgggacgaaagcttg-3′ and HNP-1-R 5′-catgtttttccttgagcctgg-3′) and hBD-2 (hBD-2-F 5′-tgatgcctcttccaggtgttt-3′ and hBD-2-R 5′-ggcaggtaacaggatcgcc-3′) were tested alongside the normalizing gene glyceraldehyde 3 phosphate dehydrogenase (GAPDH) (GAPDH-F 5′-catgttcgtcatgggtgtgaa-3′ and GAPDH-R 5′-tgcaggaggcattgctgat-3′).
Results
SLE patients
Fifty-two SLE patients (45 female and seven male) and 20 age- and sex-matched healthy controls participated in our study. The mean ± SD age of the patients at the time of SLE diagnosis was 36·2 ± 16·4 (range 7–73) years. The interval from the time of diagnosis to the initiation of the present study was 11·5 ± 9·9 (range 1–58) years. The mean follow-up period of the present study was 44·3 ± 24·5 (range 1–96) months. The main clinical manifestations in the histories of our 52 patients were arthritis (69%), lymphopenia, leucopenia and thrombocytopenia (60, 48 and 27%, respectively), renal manifestations (31%) and dermatological manifestations (29%) (Table 1). Eight patients (15%) had serositis (pericarditis and/or pleuritis). All patients had antinuclear antibodies in their sera and 81% of them revealed anti-dsDNA autoantibodies (Table 1). The mean polymorphonuclear cell count at the time of the study was 4069·9 ± 2032 (range 760–10 340; normal range 1800–8800) cells/mm3. The mean serum creatinine level was 0·85 ± 0·49 (range 0·38–3·9; normal range < 1·2) mg/dl (Table 1). The treatment of our patients during the period of the study included corticosteroids (71%), plaquenil (54%) and cytotoxic agents, mainly cyclophosphamide (37%).
Table 1.
Clinical and laboratory manifestations for 52 systemic lupus erythematosus (SLE) patients with and without high serum α-defensin levels
| All patients | Patients with high α-defensin serum levels1 | Patients with normal α–defensin serum levels2 | P-value | |
|---|---|---|---|---|
| Number (%) | 52 | 31 (60) | 21 (40) | |
| Female (%) | 45 (86) | 28 (62) | 17 (37) | 0·185 |
| Male (%) | 7 (14) | 3 (42) | 4 (58) | 0·185 |
| Arthritis (%)3 | 36 (69) | 23 (74) | 13 (62) | 0·184 |
| Renal involvement (%) | 16 (31) | 11 (35) | 5 (24) | 0·186 |
| Neurological involvement (%) | 11 (21) | 6 (19) | 5 (24) | 0·356 |
| Malar rash (%) | 10 (19) | 7 (20) | 3 (14) | 0·251 |
| Discoid lesions (%) | 5 (10) | 4 (13) | 1 (0·5) | 0·153 |
| Serositis (%) | 8 (15) | 8 (26) | 0 (0) | 0·001 |
| Lymphopenia (%) | 31 (60) | 18 (58) | 13 (62) | 0·314 |
| Leucopenia (%) | 25 (48) | 15 (48) | 10 (48) | 0·479 |
| Thrombocytopenia (%) | 14 (27) | 10 (32) | 4 (19) | 0·143 |
| Haemolytic anaemia (%) | 12 (23) | 9 (29) | 3 (14) | 0·101 |
| ANAs (%) | 52 (100) | 31 (100) | 21 (100) | 1 |
| Anti-dsDNA antibodies (%) | 42 (81) | 28 (90) | 14 (67) | 0·027 |
| Hypocomplementaemia (C3 and/or C4) (%) | 22 (42) | 15 (48) | 9 (33) | 0·143 |
| Creatinine (mg/dl) (mean ± SD) | 0·85 ± 0·49 | 0·79 ± 0·21 | 0·94 ± 0·72 | 0·190 |
| Corticosteroid treatment (%) | 37 (71) | 26 (84) | 11 (52) | 0·019 |
High serum α-defensin levels were defined as levels above 0·26 ng/μl at at least one time-point during the follow-up period.
α-Defensin serum levels were below 0·26 ng/μl throughout the follow-up period.
All clinical manifestations were defined according to the American College of Rheumatology (ACR) revised criteria definitions.25
ANAs, antinuclear antibodies; SD, standard deviation.
Levels of HNP-1 in SLE patients
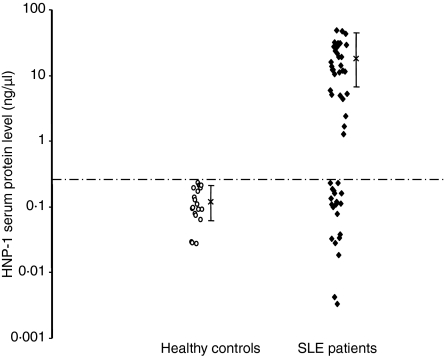
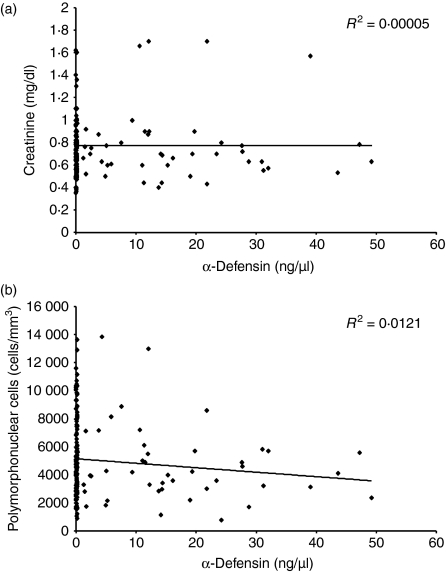
We first set out to determine the levels of the α-defensin HNP-1 in the sera of our SLE patients by ELISA. The mean α-defensin level in the sera of 20 healthy controls was 0·12 ± 0·07 ng/μl (Fig. 1). Thus, we considered 0·26 ng/μl (mean α-defensin level of healthy controls + 2 SD) as the upper normal limit. The mean level of α-defensin in all 52 SLE patients (11·07 ± 13·92 ng/μl) was significantly (Student's t-test, P < 2 × 10−6) higher than that of the healthy controls. Thirty-one of the 52 patients (60%; 28 female and three male) demonstrated high serum levels (18·5 ± 13·36 ng/μl) of α-defensin at at least one time-point during the follow-up period (Fig.1). There was no correlation between the neutrophil count and the levels of α-defensin found in the sera of SLE patients (Fig. 2). Also, creatinine levels did not correlated with the levels of α-defensin found in the sera of SLE patients, suggesting no accumulation of defensin as a result of renal failure (Fig. 2). As 48% of the patients exhibited leucopenia, our results may indicate that either damage to neutrophils or neutrophil degranulation could be the reason for the high α-defensin levels in the sera of SLE patients. However, measurement of the levels of anti-neutrophil cytoplasmic autoantibodies revealed that only 10% of our patients were positive, and this had no correlation with the levels of α-defensins (data not shown). These findings suggest that neutrophil degranulation is characteristic of lupus patients.
Figure 1.
Levels of α-defensin in the sera of 52 systemic lupus erythematosus (SLE) patients and 20 age- and sex-matched healthy controls. Serum α-defensin levels were determined by enzyme-linked immunosorbent assay (ELISA), as described in the Materials and methods section. The means ± SD for SLE patients and healthy controls were 18·5 ± 13·36 and 0·12 ± 0·07 ng/μl, respectively. The mean α-defensin level for 20 healthy controls + 2 standard deviations (0·26 ng/μl) was defined as the upper normal limit.
Figure 2.
Lack of correlation between serum α-defensin levels and the number of polymorphonuclear cells or creatinine levels. α-Defensin levels in the sera of 52 systemic lupus erythematosus (SLE) patients (total of 253 tests; mean ± SD 4·87 ± 2·04 per patient) did not correlate with serum creatinine levels (a) or the number of polymorphonuclear cells (b) determined on the same dates.
Levels of HNP-1 and HNP-3 mRNA in whole blood of SLE patients
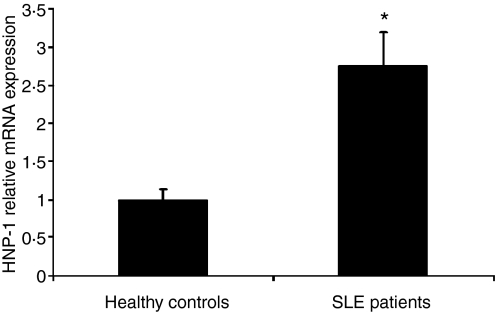
To confirm that neutrophil degranulation rather than damage was the main source of the high α-defensin levels in the sera of SLE patients, we measured α-defensin mRNA in whole blood. We used primers for HNP-1 and HNP-3 in whole blood using quantitative real-time PCR. HNPs are found mainly in neutrophils and analysis of HNP mRNA in whole blood measures almost exclusively HNP in neutrophils. The levels of α-defensin mRNA in whole blood were significantly higher than in controls in 50% of the SLE patients tested (Student's t-test P < 0·0005) (Fig. 3). Eighty per cent of the patients who were positive for α-defensins in serum had increased levels of mRNA. Taken together, these results indicate that high α-defensin levels in the sera of SLE patients probably stem from high α-defensin expression and neutrophil degranulation rather than neutrophil damage.
Figure 3.
mRNA levels of human neutrophil peptide 1 (HNP-1) in the sera of systemic lupus erythematosus (SLE) patients and healthy controls. mRNA levels were analysed by quantitative real-time polymerase chain reaction (PCR), normalized using glyceraldehyde 3 phosphate dehydrogenase (GAPDH) as the reference gene (n = 5 and n = 15 for the healthy controls and the SLE patients, respectively; mean ± standard error of the mean). All experiments were performed in triplicate. Results are presented as fold increase compared with the healthy group. *Student's t-test, P < 0·0005.
Clinical correlation
The levels of α-defensins fluctuated in the sera of SLE patients. The presence of high serum levels of α-defensins, at at least one time-point in the follow-up period, correlated (although without statistical significance) with higher rates of arthritis and renal and dermatological involvement (Table 1). More SLE patients with high levels of α-defensins in their sera were treated with corticosteroids, as compared with patients who did not have elevated α-defensin levels (84% versus 52%; Student's t-test, P = 0·019) (Table 1). Follow-up of the 31 SLE patients with high levels of α-defensins in their sera demonstrated that α-defensin levels correlated significantly (Student's t-test, P = 0·013) with disease activity, as high α-defensin levels were found in more active disease states (mean SLEDAI 7·44 ± 5·06). However, when α-defensin levels decreased to the normal range (< 0·26 ng/μl), an improvement of lupus-related manifestations (mean SLEDAI 5·14 ± 4·82) was observed (data not shown). It is of interest to note that all eight patients with serositis (at one time-point in the course of their disease) had high α-defensin levels (Table 1). It is not known whether there is a link between high α-defensin levels and serositis, and this warrants further study.
Levels of hBD-2 in sera and whole blood of SLE patients
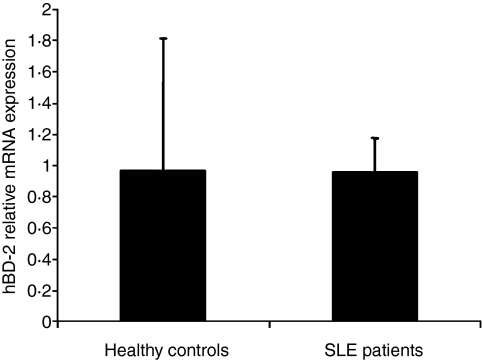
We next analysed the levels of hBD-2, as this β-defensin is expressed in macrophages and is elevated in instances of infection and inflammation.17,21,29–31 We could not detect hBD-2 protein in the sera of our SLE patients by ELISA. In parallel, we examined the levels of hBD-2 in whole blood at the mRNA level using quantitative real-time PCR. Our measurements showed that hBD-2 mRNA levels were barely detectable, confirming the negative results obtained with the ELISA, and not different from those of healthy volunteers (Student's t-test, P = 0·2) (Fig. 4). Taken together, our results show that, although several types of inflammation were manifested in our SLE patients, their hBD-2 levels did not indicate increased expression in blood cells or secretion to serum.
Figure 4.
mRNA levels of human β-defensin 2 (hBD-2) in the whole blood of systemic lupus erythematosus (SLE) patients and healthy controls. mRNA levels were extremely low in both patients and healthy controls. mRNA levels were analysed by quantitative real-time polymerase chain reaction (PCR), normalized using glyceraldehyde 3 phosphate dehydrogenase (GAPDH) as the reference gene (n = 5 and n = 15 for the healthy controls and the SLE patients, respectively; mean ± standard error of the mean). All experiments were performed in triplicate. Results are presented as fold increase compared with the healthy group.
Discussion
In this study, we measured the levels of defensins in SLE patients. To the best of our knowledge, this is the first report in which high α-defensin levels have been found in the blood of SLE patients. Although high levels of hBD-2 have been shown to correlate with inflammation, we detected only low levels of hBD-2, and levels were similar in healthy controls and SLE patients (Fig. 4). However, high levels of the α-defensin HNP-1 were found in the sera of 60% of our SLE patients (Fig. 1). The fact that more SLE patients with high levels of α-defensins in their sera were treated with corticosteroids suggests a correlation between high α-defensin levels and disease severity. Indeed, high α-defensin levels were observed in more active disease states and their decrease correlated with the improvement of lupus-related manifestations. These results are supported by recent findings in which higher levels of anti-defensin antibodies were detected in both patients with active SLE and those with inactive SLE, but levels in the former were significantly higher than in the latter.24 The high levels of anti-defensin antibodies are consistent with our finding that high defensins levels are found in the blood of SLE patients. These high levels of anti-defensin antibodies decreased after therapy with high doses of corticosteroids.24 As anti-defensin antibodies in patient sera may mask defensin detection by ELISA, we cannot rule out the possibility that an even higher percentage of patients had above-normal α-defensin levels. Thus, high α-defensin levels in sera of SLE patients could serve as a marker for disease activity.
The high α-defensin levels found for SLE patients could stem from neutrophil damage or degranulation. Although the difference was not statistically significant, we noted that, at the time-points at which patients were positive for α-defensins in serum, neutrophil counts were lower compared with the time-points at which they were negative for α-defensins (4705 versus 5152 cells/mm3). However, defensin mRNA analysis ruled out the possibility that neutrophil damage could be the sole factor leading to the elevated α-defensin levels in the sera of SLE patients. The high levels of α-defensin mRNA in whole blood suggest that high expression and degranulation could be, at least in part, the reason for the high α-defensin levels. Degranulation of neutrophils has been shown to occur when neutrophils are activated by bacterial patterns32 or beta-chemokines, such as macrophage inflammatory protein 1 (MIP-1)-α.33 In addition, cross-linking of Fc gamma receptor II a (FcγRIIa) and FcγRIIIb by autoantibodies could lead to neutrophil activation and α-defensin release.34 The inflammatory state and the autoantibodies found in SLE patients could trigger the activation of neutrophils, which could lead to induced expression and secretion of α-defensins. Consequently, secreted α-defensins could recruit the adaptive immune system through their function as potent chemotaxins for mononuclear cells,35 including dendritic cells and CD45RA+ and CD8+ T lymphocytes.36,37 In conclusion, activation of neutrophils in SLE patients could result in the secretion of α-defensins, which, in turn, induce chemotaxis and recruit the adaptive immune response. These events could lead to a concerted activation of the innate and adaptive immune systems, as is indeed manifested in SLE patients.
References
- 1.Hahn BH. An overview of the pathogenesis of systemic lupus erythematosus. In: Wallace DJ, Hahn BH, editors. Dubios’ Lupus Erythematosus. Philadelphia: Lippincott Williams & Wilkins; 2002. pp. 87–92. [Google Scholar]
- 2.Rahman A, Isenberg DA. Systemic lupus erythematosus. N Engl J Med. 2008;358:929–39. doi: 10.1056/NEJMra071297. [DOI] [PubMed] [Google Scholar]
- 3.Sthoeger ZM, Chiorazzi N, Lahita RG. Regulation of the immune response by sex hormones. J Immunol. 1988;141:91–8. I. In vitro effects of estradiol and testosterone on pokeweed mitogen-induced human B cell differentiation. [PubMed] [Google Scholar]
- 4.Sullivan KE. Genetics of systemic lupus erythematosus. Rheum Dis Clin North Am. 2000;26:229–56. doi: 10.1016/s0889-857x(05)70137-x. Clinical implications. v–vi. [DOI] [PubMed] [Google Scholar]
- 5.James JA, Kaufman KM, Farris AD, Taylor-Albert E, Lehman TJ, Harley JB. An increased prevalence of Epstein-Barr virus infection in young patients suggests a possible etiology for systemic lupus erythematosus. J Clin Invest. 1997;100:3019–26. doi: 10.1172/JCI119856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonsen A, Bengtsson AA, Nived O, Truedsson L, Sturfelt G. Gene-environment interactions in the aetiology of systemic lupus erythematosus. Autoimmunity. 2007;40:613–7. doi: 10.1080/08916930701511051. [DOI] [PubMed] [Google Scholar]
- 7.Lu L, Kaliyaperumal A, Boumpas DT, Datta SK. Major peptide autoepitopes for nucleosome-specific T cells of human lupus. J Clin Invest. 1999;104:345–55. doi: 10.1172/JCI6801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagy G, Koncz A, Perl A. T- and B-cell abnormalities in systemic lupus erythematosus. Crit Rev Immunol. 2005;25:123–40. doi: 10.1615/critrevimmunol.v25.i2.30. [DOI] [PubMed] [Google Scholar]
- 9.Horwitz DA, Jacob CO. The cytokine network in the pathogenesis of systemic lupus erythematosus and possible therapeutic implications. Springer Semin Immunopathol. 1994;16:181–200. doi: 10.1007/BF00197516. [DOI] [PubMed] [Google Scholar]
- 10.Emlen W, Niebur J, Kadera R. Accelerated in vitro apoptosis of lymphocytes from patients with systemic lupus erythematosus. J Immunol. 1994;152:3685–92. [PubMed] [Google Scholar]
- 11.Valencia X, Yarboro C, Illei G, Lipsky PE. Deficient CD4+CD25high T regulatory cell function in patients with active systemic lupus erythematosus. J Immunol. 2007;178:2579–88. doi: 10.4049/jimmunol.178.4.2579. [DOI] [PubMed] [Google Scholar]
- 12.Carroll MC. A protective role for innate immunity in systemic lupus erythematosus. Nat Rev Immunol. 2004;4:825–31. doi: 10.1038/nri1456. [DOI] [PubMed] [Google Scholar]
- 13.Truedsson L, Bengtsson AA, Sturfelt G. Complement deficiencies and systemic lupus erythematosus. Autoimmunity. 2007;40:560–6. doi: 10.1080/08916930701510673. [DOI] [PubMed] [Google Scholar]
- 14.Froy O, Gurevitz M. Membrane potential modulators: a thread of scarlet from plants to humans. FASEB J. 1998;12:1793–6. doi: 10.1096/fasebj.12.15.1793. [DOI] [PubMed] [Google Scholar]
- 15.Froy O, Gurevitz M. Arthropod and mollusk defensins – evolution by exon-shuffling. Trends Genet. 2003;19:684–7. doi: 10.1016/j.tig.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Ganz T. Defensins: antimicrobial peptides of innate immunity. Nat Rev Immunol. 2003;3:710–20. doi: 10.1038/nri1180. [DOI] [PubMed] [Google Scholar]
- 17.Lehrer RI, Ganz T. Defensins of vertebrate animals. Curr Opin Immunol. 2002;14:96–102. doi: 10.1016/s0952-7915(01)00303-x. [DOI] [PubMed] [Google Scholar]
- 18.Raj PA, Dentino AR. Current status of defensins and their role in innate and adaptive immunity. FEMS Microbiol Lett. 2002;206:9–18. doi: 10.1111/j.1574-6968.2002.tb10979.x. [DOI] [PubMed] [Google Scholar]
- 19.Yang D, Biragyn A, Kwak LW, Oppenheim JJ. Mammalian defensins in immunity: more than just microbicidal. Trends Immunol. 2002;23:291–6. doi: 10.1016/s1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- 20.Lin PW, Simon PO, Gewirtz AT, Neish AS, Ouellette AJ, Madara JL, Lencer W. Paneth cell cryptdins act in vitro as apical paracrine regulators of the innate inflammatory response. J Biol Chem. 2004;279:19902–7. doi: 10.1074/jbc.M311821200. [DOI] [PubMed] [Google Scholar]
- 21.Froy O. Regulation of mammalian defensin expression by Toll-like receptor-dependent and independent signaling pathways. Curr Microbiol. 2005;7:1387–97. doi: 10.1111/j.1462-5822.2005.00590.x. [DOI] [PubMed] [Google Scholar]
- 22.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–23. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishii T, Onda H, Tanigawa A, et al. Isolation and expression profiling of genes upregulated in the peripheral blood cells of systemic lupus erythematosus patients. DNA Res. 2005;12:429–39. doi: 10.1093/dnares/dsi020. [DOI] [PubMed] [Google Scholar]
- 24.Tamiya H, Tani K, Miyata J, et al. Defensins- and cathepsin G-ANCA in systemic lupus erythematosus. Rheumatol Int. 2006;27:147–52. doi: 10.1007/s00296-006-0173-9. [DOI] [PubMed] [Google Scholar]
- 25.Tan EM, Cohen AS, Fries JF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 26.Bombardier C, Gladman DD, Urowitz MB, Caron D, Chang CH. Derivation of the SLEDAI. Arthritis Rheum. 1992;35:630–40. doi: 10.1002/art.1780350606. A disease activity index for lupus patients. The Committee on Prognosis Studies in SLE. [DOI] [PubMed] [Google Scholar]
- 27.Froy O, Gotter AL, Casselman AL, Reppert SM. Illuminating the circadian clock in monarch butterfly migration. Science. 2003;300:1303–5. doi: 10.1126/science.1084874. [DOI] [PubMed] [Google Scholar]
- 28.Froy O, Chapnik N, Miskin R. Mouse intestinal cryptdins exhibit circadian oscillation. FASEB J. 2005;19:1920–2. doi: 10.1096/fj.05-4216fje. [DOI] [PubMed] [Google Scholar]
- 29.Singh PK, Jia HP, Wiles K, et al. Production of β-defensins by human airway epithelia. Proc Natl Acad Sci USA. 1998;95:14961–6. doi: 10.1073/pnas.95.25.14961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Becker MN, Diamond G, Verghese MW, Randell SH. CD14-dependent lipopolysaccharide-induced β-defensin-2 expression in human tracheobronchial epithelium. J Biol Chem. 2000;275:29731–6. doi: 10.1074/jbc.M000184200. [DOI] [PubMed] [Google Scholar]
- 31.Harder J, Meyer-Hoffert U, Teran LM, Schwichtenberg L, Bartels J, Maune S, Schroder JM. Mucoid Pseudomonas aeruginosa, TNF-α, and IL-1β, but not IL-6, induce human β-defensin-2 in respiratory epithelia. Am J Respir Cell Mol Biol. 2000;22:714–21. doi: 10.1165/ajrcmb.22.6.4023. [DOI] [PubMed] [Google Scholar]
- 32.Nilsson M, Sorensen OE, Morgelin M, Weineisen M, Sjobring U, Herwald H. Activation of human polymorphonuclear neutrophils by streptolysin O from Streptococcus pyogenes leads to the release of proinflammatory mediators. Thromb Haemost. 2006;95:982–90. doi: 10.1160/TH05-08-0572. [DOI] [PubMed] [Google Scholar]
- 33.Jan MS, Huang YH, Shieh B, Teng RH, Yan YP, Lee YT, Liao KK, Li C. CC chemokines induce neutrophils to chemotaxis, degranulation, and alpha-defensin release. J Acquir Immune Defic Syndr. 2006;41:6–16. doi: 10.1097/01.qai.0000188336.94090.14. [DOI] [PubMed] [Google Scholar]
- 34.Tanaka S, Edberg JC, Chatham W, Fassina G, Kimberly RP. Fc gamma RIIIb allele-sensitive release of alpha-defensins: anti-neutrophil cytoplasmic antibody-induced release of chemotaxins. J Immunol. 2003;171:6090–6. doi: 10.4049/jimmunol.171.11.6090. [DOI] [PubMed] [Google Scholar]
- 35.Territo MC, Ganz T, Selsted ME, Lehrer R. Monocyte-chemotactic activity of defensins from human neutrophils. J Clin Invest. 1989;84:2017–20. doi: 10.1172/JCI114394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chertov O, Michiel DF, Xu L, et al. Identification of defensin-1, defensin-2 and CAP37/azurocidin as T-cell chemoattractant proteins released from interleukin-8-stimulated neutrophils. J Biol Chem. 1996;271:2935–40. doi: 10.1074/jbc.271.6.2935. [DOI] [PubMed] [Google Scholar]
- 37.Yang D, Chen Q, Chertov O, Oppenheim JJ. Human neutrophil defensins selectively chemoattract naive T and immature dendritic cells. J Leukoc Biol. 2000;68:9–14. [PubMed] [Google Scholar]