Abstract
Familial adenomatous polyposis (FAP) is an autosomal-dominant disease characterized by the development of hundreds of adenomatous polyps of the colorectum. Approximately 80% of FAP patients can be shown to have truncating mutations of the APC gene. To determine the cause of FAP in the other 20% of patients, MAMA (monoallelic mutation analysis) was used to independently examine the status of each of the two APC alleles. Seven of nine patients analyzed were found to have significantly reduced expression from one of their two alleles whereas two patients were found to have full-length expression from both alleles. We conclude that more than 95% of patients with FAP have inactivating mutations in APC and that a combination of MAMA and standard genetic tests will identify APC abnormalities in the vast majority of such patients. That no APC expression from the mutant allele is found in some FAP patients argues strongly against the requirement for dominant negative effects of APC mutations. The results also suggest that there may be at least one additional gene, besides APC, that can give rise to FAP.
Presymptomatic diagnosis of patients with a family history of cancer is the first clinical application of the genetic revolution in cancer research. Though many issues about genetic testing remain unresolved, such testing can dramatically improve the medical and psychological management of affected patients and their families and can even be life-saving when performed prudently (1).
Familial adenomatous polyposis (FAP) provides an excellent example of both the potential and the problems inherent in genetic diagnosis for cancer predisposition. FAP is an autosomal dominant disease in which affected patients develop hundreds to thousands of adenomatous polyps in their colorectum beginning in their teenage years. If left untreated, colorectal cancer will be diagnosed at a median age of 40 in such patients (2). Truncating mutations in the APC gene cause approximately 80% of all cases of FAP (3–5). Before the advent of APC genetic testing, it was necessary that all members of FAP kindreds be screened for the presence of disease by using colonoscopy or related clinical methods. Genetic testing can spare a significant number of first- and second-degree relatives the need for frequent colonoscopies and also can alleviate anxiety associated with the uncertainty of their genetic state. Though the only definitive treatment for FAP historically has been colectomy, chemopreventive agents currently are showing promise, and genetic testing should allow such agents to be used before disease onset (6, 7).
Experience with FAP also illustrates the technical problems associated with genetic testing. APC is a large gene, encoding a protein of 2,843 aa contained within 15 exons (8, 9). Sequencing the entire gene, including introns, untranslated, and promoter regions, is impractical. Fortunately, all confirmed FAP-causing mutations detected to date result in truncations of the protein (4, 5). One major consequence of these truncating mutations is the disruption of APC’s ability to inhibit the function of β-catenin (10, 11). It is believed that APC normally binds to β-catenin (12, 13) and promotes its degradation (14), thereby preventing activation of growth-promoting genes, such as c-myc (15), by a β-catenin/Tcf-4 transcription complex (16, 17). This mutation spectrum has stimulated the development of testing approaches that can reveal truncated APC proteins. In particular, the most commonly used test [called in vitro synthesized protein (IVSP) or protein truncation test (PTT)] involves in vitro transcription and translation of APC PCR products. Gel electrophoretic analysis of the translated polypeptides reveals truncated proteins indicative of mutations (3, 18).
Extensive analyses of FAP kindreds with IVSP and direct or indirect DNA sequencing methods have been used to identify more than 200 different mutations (4, 5). The frequency of APC mutations detected among FAP kindreds varies with the technique used, but in no case has it been more than 80%. The basis for the inability to identify mutations in a substantial proportion of such kindreds is unclear. One possibility involves the existence of APC mutations that are difficult to detect by standard mutational analyses. Indeed, the patient whose analysis originally led to the chromosome 5 localization of APC had a large deletion that would have been impossible to detect with any standard sequencing or IVSP assay (19). Furthermore, some FAP patients without truncating APC mutations appeared to express significantly reduced levels of APC transcript from one allele (3). A second possibility is that some cases of FAP are caused by mutations in genes other than APC.
We have described previously a mutation detection approach called monoallelic mutation analysis (MAMA). Chromosomes from an affected individual are isolated in hybrid cells formed from fusion of the patient’s cells with a suitable rodent recipient (20). Because each allele can be examined independently, mutations are not obscured by the wild-type product from the normal allele, as can occur with standard analyses of patients’ cells. For example, mutations in promoter regions, which decrease expression from the corresponding allele in cis fashion, are very difficult to detect with routine assays. However, because patterns of expression are faithfully preserved in such hybrids (20, 21), mutations that affect expression can be detected as easily as those that affect the structure of the gene. We now have used MAMA to analyze nine FAP patients in whom no APC mutations could be detected with standard methods. The results show that more than 95% of FAP patients have inactivating mutations in APC and that a combination of MAMA and standard genetic testing can identify APC abnormalities in the vast majority of FAP patients. Additionally, the results suggest that there may be at least one other gene besides APC that can give rise to FAP.
MATERIALS AND METHODS
Cell Culture.
Lymphoblastoid lines were established by Epstein–Barr virus infection of peripheral blood leukocytes from patients diagnosed with FAP who had no evidence of APC mutation upon IVSP analysis. These lines were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and grown at 37°C and 5% CO2. The UCW-56 hamster cell line (22) was grown in DMEM supplemented with 10% fetal bovine serum and 60 μg/ml l-proline and grown at 32°C and 5% CO2.
Cell Fusions.
Fusions initially were performed with polyethylene glycol as described (20). In later experiments, electrofusion was employed because a greater yield of viable fusion clones could be obtained. UCW-56 cells were combined with lymphoblastoid cells in FS (0.3 M mannitol/0.1 mM MgCl2/0.1 mM CaCl2) at a ratio of 3:1. The cells were washed and centrifuged three times in FS before resuspending in FS at a final concentration of 5 × 107 cells/ml. Thirty microliters of this solution then was mixed and pipetted into a 0.5-mm gap microfusion plate (BTX Microslide 450; BTX, San Diego). Fusions were performed by using a BTX Electrocell Manipulator, ECM 200. The settings that yielded the greatest number of fusion clones were 15 V (AC) for 10 sec followed by two 100-V (DC) pulses of 30 μsec each.
The cells from one 30-μl fusion were plated into four wells of a 48-well plate (Costar) in DMEM supplemented with 10% fetal bovine serum and 60 μg/ml l-proline, and grown at 32°C. To obtain a sufficient number of clones, we generally performed three fusions from each line, employing a total of 1.8 × 107 UCW-56 cells and 6 × 106 lymphoblastoid cells. After 24 hr, the medium was changed and the 48-well plates were transferred to 39°C. Medium was changed every 4 days for approximately 2 weeks, at which time individual clones were genotyped and expanded for immunoblotting (20). Immunoblotting was performed as described (23) except that cells were lysed in 8 M urea/2 M thiourea/0.05 M Tris base/0.075 M DTT/3% SDS/0.004% bromophenol blue and adjusted to a final pH of 6.8 (24), and APC was detected with an antibody (catalog no. OP47L; Oncogene Science) reactive with the C terminus of APC (23).
RESULTS
Generation of Monoallelic Clones.
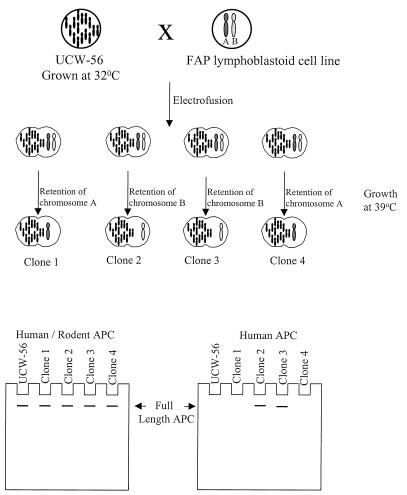
Isolation of human chromosome 5 alleles was accomplished by fusing lymphoblastoid cells from FAP patient lines with the UCW-56 hamster cell line (Fig. 1). UCW-56 has a temperature-sensitive tRNA-leucyl synthetase mutation, which prevents growth at 39°C unless complemented by the human tRNA-leucyl synthetase gene on chromosome 5. Because lymphoblastoid cells are nonadherent, clones derived by fusion are identified easily after growth at 39°C. In the original MAMA technique, fusions were performed with polyethylene glycol. We subsequently found that electrofusion was considerably more efficient at generating fusion clones. After electrofusion with a total of 1.8 × 107 UCW-56 cells and 6 × 106 lymphoblastoid cells, an average of six independent clones could be obtained (see Materials and Methods). Clones were visible approximately 2 weeks after fusion and were expanded for genotyping. More than 175 hybrids derived by electrofusion between UCW-56 and the lymphoblastoid cells of 10 different patients were examined by genotyping. Every hybrid identified had at least one human chromosome 5, documenting a negligible, nonspecific background of the fusion procedure.
Figure 1.
Schematic of MAMA. The hamster cell line, UCW-56, is fused to an FAP lymphoblastoid line, and clones are subsequently selected at 39°C. Only clones that retain human chromosome 5 can grow at this temperature. During the expansion process, hybrids retain at least one human chromosome 5 and usually lose the other copy. Clones then are genotyped to determine which chromosome 5 they contain. Proteins from the fusion clones are used for Western blotting with antibodies reactive with the amino or carboxyl ends of the APC protein. The N-terminal antibody (Human/Rodent APC) reacts with human and rodent APC protein and serves as a loading control. The C-terminal antibody (Human APC) reacts against human but not hamster APC and is used to determine whether full-length APC expression occurs.
MAMA in a Patient with a Known APC Mutation.
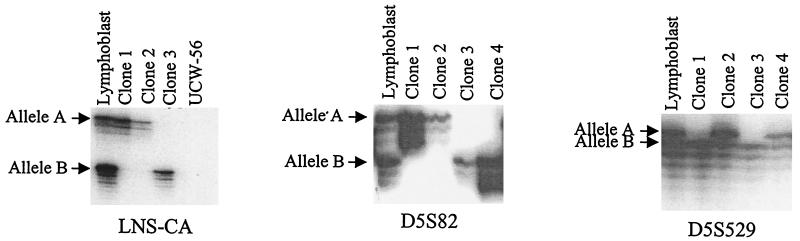
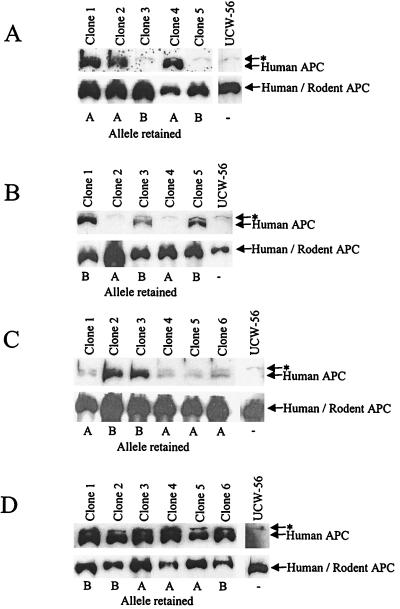
FAP patient C1 was tested by IVSP and found to have a truncating mutation of APC resulting in the removal of ≈2,000 aa from the C terminus. DNA genotyping was performed on clones derived from the fusion of lymphoblasts of patient C1 with UCW-56. Using a polymorphism at the LNS-CA locus, just telomeric of APC, it was found that fusion clones contained either one of two individual chromosomal 5 alleles arbitrarily designated “A” and “B” (examples in Fig. 2). These clones were expanded further and protein was isolated for immunoblotting with a human specific antibody reactive against the C terminus of APC. All clones containing chromosome “B” failed to produce full-length APC protein, whereas those with chromosome “A” produced a protein of the expected wild-type size (Fig. 3A). Thus, the germ-line mutation in this patient resided on the “B” chromosome. Using an N-terminal antibody, the cells with chromosome “B” revealed no truncated protein of the expected size. This is not unusual, because the mRNA or protein from APC genes with truncating mutations often is unstable and thereby undetectable by Western blot assays (23).
Figure 2.
DNA typing of MAMA clones. Primers found to be polymorphic in the germ line of the patient are used to genotype the hybrids. Hybrids found to be monoallelic are propagated and genotyped. Shown are three such examples from three separate patients. In each case the patient is polymorphic and each clone is monoallelic for chromosome “A” or “B.” An overexposure of the autoradiograph is made to confirm that no residual chromosomes containing the “lost” allele remain in the hybrids.
Figure 3.
Protein analysis of fusion clones. The Western blots in A were derived from a patient who had a truncating mutation of APC detected with the IVSP assay. Clones 1, 2, and 4 retained allele A and expressed full-length APC whereas clones 3 and 5 retained allele “B” and did not express full-length APC. The blots in B–D were derived from patients in whom no truncating proteins could be detected with IVSP assays. The blots in B were derived from patient 4, who had no full-length APC expression from allele “A” (clones 2 and 4), but who expressed full-length protein from allele “B” (clones 1, 3, and 5). The blots in C were from patient 7, who had reduced expression of APC protein from all fusion clones containing allele “A” compared with those containing allele “B.” The blots in D were derived from patient 9, who had comparable amounts of full-length APC protein expressed from clones retaining either allele “A” or “B.” The asterisk (∗) represents a cross-reactive band present in all hamster cells that is detected with the C-terminal antibody.
MAMA in Patients Without APC Mutations Identifiable by IVSP.
Nine unrelated FAP patients in whom no APC mutations could be identified by IVSP were analyzed by MAMA. Each patient had at least 100 adenomas (verified by pathologic records), and each patient had a family history of FAP, ruling out germ-line mosaicism as a cause of the negative IVSP results (Table 1). Upon fusion to UCW-56, hybrids were expanded and genotyped with LNS-CA and other chromosome 5 polymorphic markers to identify monoallelic clones. Of 165 clones analyzed, 101 were found to contain one allele, whereas 64 were found to contain both maternal and paternal alleles. Monoallelic clones then were expanded and their proteins were used for immunoblotting. A minimum of two clones representing each allele from each patient was used to exclude clonal variability as an explanation for failure to detect full-length protein. Analysis of the nine patients revealed three classes of expression; no detectable APC expression from one allele (six patients; Fig. 3B), reduced APC expression from one allele (one patient, Fig. 3C), or equal APC expression from both alleles (two patients, Fig. 3D).
Table 1.
Clinical phenotype of FAP patients tested by MAMA
| Patient no. | MAMA result | IVSP result | Age at diagnosis | No. of polyps | No. of affected family members | Extracolonic features in the family |
|---|---|---|---|---|---|---|
| 1 | No expression | Negative | 54 | >100 | 2 | Cysts |
| 2 | No expression | Negative | 13 | >100 | 5 | Cysts, pancreatic CA |
| 3 | No expression | Negative | 44 | >100 | 32 | Cysts, osteoma |
| 4 | No expression | Negative | 16 | >100 | 2 | Desmoids |
| 5 | No expression | Negative | 29 | >100 | 11 | None |
| 6 | No expression | Negative | 18 | >100 | 4 | Pancreatic CA |
| 7 | Reduced expression | Negative | NA | >100 | 37 | NA |
| 8 | Full-length expression | Negative | 13 | >100 | 3 | None |
| 9 | Full-length expression | Negative | 27 | >100 | 3 | Cysts |
NA, not available.
Confirmatory Studies.
There was little doubt about the mutational basis of FAP in patients with no detectable expression of APC. However, we were less confident about the patient with detectable but reduced expression. This patient was from a reasonably large kindred, and linkage analysis was carried out with markers closely surrounding APC. Two significant results were evident from this linkage analysis. First, a multipoint logarithm of odds (lod) score of 2.06 was obtained at a θ of 0.1 by using markers D5S82, LNS-CA, and MCC (25), strongly suggesting that the disease was caused by an APC mutation in this family. Second, the haplotype that was associated with disease in the linkage analysis was also the haplotype of the chromosome that showed reduced expression. Haplotypes are extremely easy to define with MAMA and can be used as diagnostic tools for genetic testing of other members of the family; only the proband needs to be tested by MAMA.
In the two cases in which full-length APC protein was expressed by both alleles, it was possible that missense mutations or small in-frame insertions or deletions of APC were responsible for the disease. To evaluate this possibility, the complete coding region of APC was sequenced directly from appropriate PCR products. No mutations were observed in either patient, strongly suggesting that both APC alleles were normal in these two patients. No other members of these two families were available to compare haplotypes or to exclude linkage to APC.
DISCUSSION
Several important conclusions can be made on the basis of this study. First, it is now clear that at least 95% of FAP patients have mutations in APC. In other studies (ref. 3 and unpublished data) we have observed that ≈80% of FAP patients have APC mutations detectable with IVSP. In the other 20% of patients, our new results show that seven of nine (78%) contain mutations detectable with MAMA. In total, it can be estimated that 95% of FAP patients harbor APC mutations. These figures are consistent with linkage analyses showing that disease in FAP kindreds generally is linked to chromosome 5q markers (26). Most previous failures to detect APC mutations in FAP kindreds therefore are likely to have been a result of the insensitivity of the assays used rather than the absence of APC mutations.
Second, these results have significant implications concerning the relationship between specific APC mutations and phenotype. In particular, it has been shown that the precise mutation within APC often is correlated with disease severity (27–30), leading some investigators to propose that dominant negative effects of specific mutations contribute to polyposis. These hypotheses are dependent on the idea that specific mutant gene products interact with either the product of the wild-type allele of APC (31, 32) or with other gene products in the APC pathway (e.g., β-catenin or GSK3B). It has been proposed that such “dominant negative” mutants would lead to more severe disease because they disrupt the APC tumor-suppressor pathway even in the presence of a wild-type allele from the unaffected parent, whereas other mutants are only functionally apparent when the wild-type allele is lost. Our new results provide evidence against the idea that dominant negative effects of specific mutant APC alleles are required for severe disease. In the six patients with no detectable expression of the mutant APC allele, no interaction of the missing gene product is possible. Yet, some of the individuals with these mutations had severe polyposis (Table 1). Our results therefore are more consistent with the idea that the total absence of APC protein can predispose to significant polyposis and that specific mutations which lead to less severe disease may be explained by the partial retention of function. Though our results rule out a requirement for dominant negative effects of mutations in general, they do not exclude the possibility that specific mutations are associated with such dominant negative effects.
Third, the results have significant implications for diagnosis. As noted previously, standard genetic tests “miss” a significant number of APC mutations. These are presumably in promoter or other noncoding regions within or surrounding the gene, and sequencing these very large regions is impractical. As shown here, MAMA can detect the presence of such mutations, and the haplotype analysis that is derived from MAMA can be used straightforwardly to help counsel family members of the proband. Though MAMA clearly is not cost-effective as a screening tool, it is likely to be valuable in the management of FAP patients whose mutations cannot be identified by standard methods. It should be noted in this regard that more than three-quarters of the individuals in the extended families of FAP probands do not inherit a mutant APC allele, yet many must be screened with colonoscopy and some will suffer the emotional consequences of uncertainty. The cost of alternative techniques such as MAMA must be judged against the medical and human costs of having no definitive diagnoses in such cases.
Finally, two of nine FAP patients analyzed harbored alleles from which full-length, completely wild-type sequence was expressed at normal levels. These patients may have contained mutations in noncoding regions of the gene that specifically diminished their expression in colonic stem cells but that did not lead to decreased expression in the hybrids generated upon fusion to UCW-56 cells. Alternatively, the FAP in these two kindreds may be a result of a gene different from APC. Unfortunately, neither of these two patients came from a kindred large enough for linkage analysis, so these possibilities cannot be distinguished at present. It is interesting that a homolog of APC, located on chromosome 19p13.3, recently has been described (33, 34). This homologue as well as the functionally related family of axin genes (35–40) are good candidates for FAP causation in these kindreds. Elucidation of other genes responsible for FAP might shed considerable additional light on the mechanisms through which APC causes polyposis, just as mutations in β-catenin in sporadic cancers have illuminated the pathway (10, 41).
Acknowledgments
Financial support for this work was provided by the Clayton Fund, Grant P01 ES06052; National Institute on Environmental Health Sciences Center Grant P30 ES03819; National Cancer Institute Center Grant P30 CA06973; National Cancer Institute Grants CA43460, CA57345, CA62924, and CA63721; the Lucille P. Markey Foundation in Cellular and Molecular Medicine; and Association for International Cancer Research Grant 94275. Under an agreement between Calbiochem and Johns Hopkins University, K.W.K. and B.V. are entitled to a share of the sales royalty for the OPL47L antibody received by the University from Calbiochem. Under an agreement between Hoffmann–La Roche and Johns Hopkins University, K.W.K. and B.V. are entitled to a share of the sales royalty for the IVSP assay received by the University from Hoffmann–La Roche. The terms of these arrangements are being managed by the University in accordance with its conflict of interest policies.
ABBREVIATIONS
- MAMA
monoallelic mutation analysis
- FAP
familial adenomatous polyposis
- PTT
protein truncation test
- IVSP
in vitro synthesized protein
References
- 1.Petersen G, Codori A-M. In: The Genetic Basis of Human Cancer. Vogelstein B, Kinzler K W, editors. New York: McGraw–Hill; 1998. pp. 591–599. [Google Scholar]
- 2.Giardiello F M. In: Gastrointestinal Cancers: Biology, Diagnosis, and Therapy. Rustgi A K, editor. Philadelphia: Lippincott–Raven; 1995. pp. 367–377. [Google Scholar]
- 3.Powell S M, Petersen G M, Krush A J, Booker S, Jen J, Giardiello F M, Hamilton S R, Vogelstein B, Kinzler K W. N Engl J Med. 1993;329:1982–1987. doi: 10.1056/NEJM199312303292702. [DOI] [PubMed] [Google Scholar]
- 4.Nagase H, Su Y. Hum Mutat. 1993;2:425–434. doi: 10.1002/humu.1380020602. [DOI] [PubMed] [Google Scholar]
- 5.Laurent-Puig P, Beroud C, Soussi T. Nucleic Acids Res. 1998;26:269–270. doi: 10.1093/nar/26.1.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.DuBois R N, Giardiello F M, Smalley W E. Gastroenterol Clin North Am. 1996;25:773–791. doi: 10.1016/s0889-8553(05)70274-0. [DOI] [PubMed] [Google Scholar]
- 7.Giardiello F M. Gastroenterol Clin North Am. 1996;25:349–362. doi: 10.1016/s0889-8553(05)70251-x. [DOI] [PubMed] [Google Scholar]
- 8.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, et al. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 9.Kinzler K W, Nilbert M C, Su L K, Vogelstein B, Bryan T M, Levy D B, Smith K J, Preisinger A C, Hedge P, McKechnie D, et al. Science. 1991;253:661–665. doi: 10.1126/science.1651562. [DOI] [PubMed] [Google Scholar]
- 10.Morin P J, Sparks A B, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler K W. Science. 1997;275:1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 11.Korinek V, Barker N, Morin P J, van Wichen D, de Weger R, Kinzler K W, Vogelstein B, Clevers H. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 12.Su L K, Vogelstein B, Kinzler K W. Science. 1993;262:1734–1737. doi: 10.1126/science.8259519. [DOI] [PubMed] [Google Scholar]
- 13.Rubinfeld B, Souza B, Albert I, Muller O, Chamberlain S H, Masiarz F R, Munemitsu S, Polakis P. Science. 1993;262:1731–1734. doi: 10.1126/science.8259518. [DOI] [PubMed] [Google Scholar]
- 14.Munemitsu S, Albert I, Souza B, Rubinfeld B, Polakis P. Proc Natl Acad Sci USA. 1995;92:3046–3050. doi: 10.1073/pnas.92.7.3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He T C, Sparks A B, Rago C, Hermeking H, Zawel L, da Costa L T, Morin P J, Vogelstein B, Kinzler K W. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 16.Behrens J, von Kries J P, Kuhl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 17.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destree O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 18.van der Luijt R, Khan P M, Vasen H, van Leeuwen C, Tops C, Roest P, den Dunnen J, Fodde R. Genomics. 1994;20:1–4. doi: 10.1006/geno.1994.1119. [DOI] [PubMed] [Google Scholar]
- 19.Herrera L, Kakati S, Gibas L, Pietrzak E, Sandberg A. Am J Med Genet. 1986;25:473–476. doi: 10.1002/ajmg.1320250309. [DOI] [PubMed] [Google Scholar]
- 20.Papadopoulos N, Leach F S, Kinzler K W, Vogelstein B. Nat Genet. 1995;11:99–102. doi: 10.1038/ng0995-99. [DOI] [PubMed] [Google Scholar]
- 21.Gabriel J M, Higgins M J, Gebuhr T C, Shows T B, Saitoh S, Nicholls R D. Proc Natl Acad Sci USA. 1998;95:14857–14862. doi: 10.1073/pnas.95.25.14857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson D, Carnright D V. Somatic Cell Genet. 1977;3:483–495. doi: 10.1007/BF01539120. [DOI] [PubMed] [Google Scholar]
- 23.Smith K J, Johnson K A, Bryan T M, Hill D E, Markowitz S, Willson J K, Paraskeva C, Petersen G M, Hamilton S R, Vogelstein B, Kinzler K W. Proc Natl Acad Sci USA. 1993;90:2846–2850. doi: 10.1073/pnas.90.7.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blough E R, Rennie E R, Zhang F, Reiser P J. Anal Biochem. 1996;233:31–35. doi: 10.1006/abio.1996.0003. [DOI] [PubMed] [Google Scholar]
- 25.Miyoshi Y, Nishisho I, Miki Y, Mori T, Kinzler K W, Vogelstein B, Nakamura Y. Jpn J Cancer Res. 1992;83:10–14. doi: 10.1111/j.1349-7006.1992.tb02344.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mareni C, Stella A, Origone P, Susca F, Montera M P, Lonoce A, Ponz de Leon M, Sassatelli R, Gentile M, Straface A, et al. Hum Genet. 1993;90:545–550. doi: 10.1007/BF00217456. [DOI] [PubMed] [Google Scholar]
- 27.Nagase H, Miyoshi Y, Horii A, Aoki T, Ogawa M, Utsunomiya J, Baba S, Sasazuki T, Nakamura Y. Cancer Res. 1992;52:4055–4057. [PubMed] [Google Scholar]
- 28.Gayther S, Wells D, SenGupta S, Chapman P, Neale K, Tsioupra K, Delhanty J. Hum Mol Genet. 1994;3:53–56. doi: 10.1093/hmg/3.1.53. [DOI] [PubMed] [Google Scholar]
- 29.Caspari R, Olschwang S, Friedl W, Mandl M, Boisson C, Boker T, Augustin A, Kadmon M, Moslein G, Thomas G, et al. Hum Mol Genet. 1995;4:337–340. doi: 10.1093/hmg/4.3.337. [DOI] [PubMed] [Google Scholar]
- 30.Mahmoud N N, Boolbol S K, Bilinski R T, Martucci C, Chadburn A, Bertagnolli M M. Cancer Res. 1997;57:5045–5050. [PubMed] [Google Scholar]
- 31.Joslyn G, Richardson D S, White R, Alber T. Proc Natl Acad Sci USA. 1993;90:11109–11113. doi: 10.1073/pnas.90.23.11109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Su L K, Johnson K A, Smith K J, Hill D E, Vogelstein B, Kinzler K W. Cancer Res. 1993;53:2728–2731. [PubMed] [Google Scholar]
- 33.Nakagawa H, Murata Y, Koyama K, Fujiyama A, Miyoshi Y, Monden M, Akiyama T, Nakamura Y. Cancer Res. 1998;58:5176–5181. [PubMed] [Google Scholar]
- 34.van Es J, Kirkpatrick C, van de Wetering M, Molenaar M, Miles A, Kuipers J, Destree O, Peifer M, Clevers H. Curr Biol. 1999;9:105–108. doi: 10.1016/s0960-9822(99)80024-4. [DOI] [PubMed] [Google Scholar]
- 35.Ikeda S, Kishida S, Yamamoto H, Murai H, Koyama S, Kikuchi A. EMBO J. 1998;17:1371–13784. doi: 10.1093/emboj/17.5.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Behrens J, Jerchow B A, Wurtele M, Grimm J, Asbrand C, Wirtz R, Kuhl M, Wedlich D, Birchmeier W. Science. 1998;280:596–599. doi: 10.1126/science.280.5363.596. [DOI] [PubMed] [Google Scholar]
- 37.Yamamoto H, Kishida S, Uochi T, Ikeda S, Koyama S, Asashima M, Kikuchi A. Mol Cell Biol. 1998;18:2867–2875. doi: 10.1128/mcb.18.5.2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kishida S, Yamamoto H, Ikeda S, Kishida M, Sakamoto I, Koyama S, Kikuchi A. J Biol Chem. 1998;273:10823–10826. doi: 10.1074/jbc.273.18.10823. [DOI] [PubMed] [Google Scholar]
- 39.Hart M J, de los Santos R, Albert I N, Rubinfeld B, Polakis P. Curr Biol. 1998;8:573–581. doi: 10.1016/s0960-9822(98)70226-x. [DOI] [PubMed] [Google Scholar]
- 40.Nakamura T, Hamada F, Ishidate T, Anai K, Kawahara K, Toyoshima K, Akiyama T. Genes Cells. 1998;3:395–403. doi: 10.1046/j.1365-2443.1998.00198.x. [DOI] [PubMed] [Google Scholar]
- 41.Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]