Abstract
We analysed seven genetic polymorphisms that are candidates to explain individual variations in human endurance phenotypic traits, at least in Caucasian people (ACE Ins/Del, ACTN3 Arg577Ter, AMPD1 Gln12Ter, CKMM 1170 bp/985 + 185 bp, HFE His63Asp, GDF-8 Lys153Arg and PPARGC1A Gly482Ser) in 46 world-class endurance athletes and 123 controls (all Spanish Caucasians). Using the model developed by Williams & Folland we determined (1) the ‘total genotype score’ (TGS, from the accumulated combination of the seven polymorphisms, with a maximum value of ‘100’ for the theoretically optimal polygenic score) in the non-athlete (control) group, in the athlete group and in the total Spanish population, and (2) the probability for the occurrence of Spanish individuals with the ‘perfect’ polygenic endurance profile (i.e. TGS = 100). The probability of a Spanish individual possessing a theoretically optimal polygenic profile for up to the seven candidate genetic polymorphisms we studied was very small, i.e. ∼0.07% (or 1 in 1351 Spanish individuals). The mean TGS was higher in athletes (70.22 ± 15.58) than in controls (62.43 ± 11.45) and also higher than predicted for the total Spanish population (60.80 ± 12.1), suggesting an overall more ‘favourable’ polygenic profile in the athlete group. However, only three of the best Spanish endurance athletes (who are also amongst the best in the world) had the best possible score for up to six genes and none of them had the optimal profile. Other polymorphisms yet undiscovered as well as several factors independent of genetic endowment may explain why some individuals reach the upper end of the endurance performance continuum.
The combined influence of several genetic variants, each with a significant contribution, as well as the complex interaction of genetic variants (with or without an individual contribution) may help to explain individual variations in human endurance performance. Williams & Folland (2008) recently determined the probability for the occurrence of humans with the ‘perfect’ polygenic endurance profile. Such optimal profile was obtained from the theoretically best accumulated combination of 23 genetic polymorphisms, each of which is a candidate to individually influence human variability in one or more endurance phenotypic traits. These variants were chosen based on previous association studies on overall Caucasian populations. For instance, the optimal genotype for the Gly482Ser variant in the peroxisome proliferator-activated receptor γ coactivator 1α gene (PPARGC1A) is Gly/Gly; accordingly, it was given an individual maximum genotype score of ‘2’ (vs. ‘1’ and ‘0’ for Gly/Ser and Ser/Ser). Three candidate genes (PPARGC1A, hereditary haemochromatosis gene (HFE) and the gene encoding the skeletal muscle (M) isoform of adenosine monophosphate deaminase (AMPD1)) were selected by Williams & Folland based on previous data from our laboratory, showing differences in allelic/genotype frequencies between Caucasian (Spanish) world-class endurance male athletes and controls (Chicharro et al. 2004; Lucia et al. 2005a; Rubio et al. 2005).
Williams & Folland (2008) predicted that the probability of a Caucasian individual existing in the planet with all 23 optimal genetic variants is extremely low (0.0005%), which indicates that there would only be three such individuals in the United Kingdom (60 million population). The authors also predicted that the distribution of the polygenic endurance profile in the planet is leptokurtic – that is, clustered towards the middle. Such a clustered distribution would limit the potential for existing humans with a theoretically ‘perfect’ or ‘near perfect’ polygenic endurance profile. Although more research is necessary, the polygenic profile approach was recently incorporated into clinical medicine to predict the risk of cardiovascular events. Kathiresan et al. (2008) computed a polygenic profile of nine validated genotypes that are associated with modulation in levels of LDL or HDL cholesterol. They showed that the polygenic profile was an independent risk factor for incident cardiovascular disease.
The report of Williams & Folland (2008) presents a very attractive model, as it provides a quantitative way of combining existing genotype data to predict a complex phenotype. This model is, however, limited by the fact that the genotype frequency data used in their calculations originated from heterogeneous sources. Heterogeneity was due to (1) between-studies differences in the geographical origin of subjects, and (2) the fact that previous studies did not use a ‘polygenic approach’ for each subject, i.e. they mostly focused on a single genetic variant. Based on Williams & Folland's original approach, our main goal was to compare actual individual polygenic data obtained from a sample of elite Spanish endurance athletes and non-athletic Spanish controls. A secondary purpose was to determine the probability for the occurrence of Spanish individuals with the ‘perfect’ polygenic endurance profile. While recognizing the possibility that several polymorphisms yet to be identified might play a more important role, we used the seven candidate polymorphisms (six studied by Wiliams & Folland) that we believed to be more important at present (at least in Caucasians) for explaining individual variations in endurance sports performance.
Methods
Subjects
The studied population comprised 123 healthy male non-athletic (sedentary) controls and 46 male endurance athletes (14 endurance runners and 32 professional road cyclists) who previously participated as subjects in published studies (see Table 1). All controls and athletes were of the same Caucasian (Spanish) descent for at least three generations. The athlete sample size is limited as we wanted to ensure that all of these individuals were in the absolute upper end of the human endurance performance continuum. All runners had participated in at least one Olympic final (5 km to marathon), including two world champions (Marathon) and two track-and-field European champions (5 and 10 km) (maximal oxygen uptake 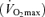 : 77.2 ± 5.2 ml O2 kg−1 min−1). All cyclists were (i) Tour de France finishers and (ii) stage winners or top-5 finishers in at least one edition of this race, or winners of other major events (tour races, one-day classics, etc) (
: 77.2 ± 5.2 ml O2 kg−1 min−1). All cyclists were (i) Tour de France finishers and (ii) stage winners or top-5 finishers in at least one edition of this race, or winners of other major events (tour races, one-day classics, etc) (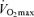 : 73.2 ± 4.2 ml O2 kg−1 min−1).
: 73.2 ± 4.2 ml O2 kg−1 min−1).
Table 1.
Studied polymorphisms in Spanish population and Spanish endurance elite athletes
| Symbol | Gene | Polymorphism | Genotypes (2 =‘optimal’ endurance genotype) | Frequency in Spanish population (%) | Frequency in Spanish endurance elite athletes (%) | Reference |
|---|---|---|---|---|---|---|
| ACE | Angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | 287 bp Ins(I)/Del(D) | 0 = DD, 1 = ID, 2 = II | 35, 46, 19 | 48, 22, 30 | (Lucia et al. 2005b; Muniesa et al. 2009) |
| ACTN3 | Actinin, α3 | Arg(R)577Ter(X) (rs1815739) | 0 = RR, 1 = RX, 2 = XX | 28, 54, 18 | 22, 54, 24 | (Lucia et al. 2008; Muniesa et al. 2009) |
| AMPD1 | Adenosine monophosphate deaminase 1 (isoform M) | Gln(Q)12Ter(X) (rs17602729) | 0 = XX, 1 = QX, 2 = QQ | 1, 17, 82 | 0, 4, 96 | (Rubio et al. 2005; Muniesa et al. 2009) |
| CKMM | Creatine kinase, muscle | NcoI RFLP 1170 bp/985 + 185 bp | 0 = 1170/1170, 1 = 985 + 185/1170, 2 = 985 + 185/985 + 185 | 12, 53, 35 | 11, 43, 46 | (Lucia et al. 2005b; Muniesa et al. 2009) |
| HFE | Hereditary Haemochromatosis | His(H)63Asp(D) (rs1799945) | 0 = HH/HH, 1 = HD/HD, 2 = HD/HH | 67, 9, 24 | 51, 7, 42 | (Chicharro et al. 2004) |
| GDF-8 | Myostatin (growth and differentiation factor) | Lys(K)153Arg(R) (rs1805086) | 0 = RR, 1 = RK, 2 = KK | 0, 10, 90 | 0, 13, 87 | (Muniesa et al. 2009) |
| PPARGC1A | Peroxisome proliferator-activated receptor-γ, coactivator 1, α | Gly(G)482Ser(S) (rs8192678) | 0 = S/S, 1 = G/S, 2 = G/G | 11, 51, 38 | 4, 39, 57 | (Lucia et al. 2005a; Muniesa et al. 2009) |
Written consent was obtained from each subject. The study protocol was approved by the institutional ethics committee (Universidad Europea de Madrid, Spain) and was in accordance with the Declaration of Helsinki for human research of 1974 (last modified in 2000).
Genotypes
From the report by Williams & Folland (2008), we only studied those genetic polymorphisms (see below) for which an association with endurance performance was previously shown in publications using real world-class endurance Caucasian athletes as subjects. We also included the myostatin gene given its crucial role on muscle function (Table 1).
The 287 bp Ins/Del polymorphism of the angiotensin converting enzyme gene (ACE) (location: 17q23.3) is related to cardiovascular and skeletal muscle function, training response of muscle efficiency and hypertrophy (Jones & Woods, 2003; Yang et al. 2007);
The Arg577Ter polymorphism (rs1815739) of the actinin α3 gene (ACTN3) (location: 11q13.1), which encodes for the synthesis of α-actinin-3 in skeletal muscle fibres, is involved in muscles’ ability to produce fast contractions while avoiding damage originated by eccentric muscle contractions such as those involved in running (Yang et al. 2007; Lucia et al. 2008);
The Gln12Ter polymorphism (rs17602729) of the adenosine monophosphate deaminase1 gene (AMPD1) (location: 1p13) is involved in the salvage of adenine nucleotides and regulation of muscle glycolysis during intense exercise (Rubio et al. 2005);
The NcoI RFLP 1170 bp/985 + 185 bp polymorphism of the muscle-specific creatine kinase gene (CKMM) (location: 19q13.2-q13.3) is related to energy buffering in skeletal muscle fibres and to tolerance to skeletal muscle damage (Rivera et al. 1997);
The His63Asp polymorphism (rs1799945) in the hereditary haemochromatosis gene (HFE) (location: 6p21.3) relates to iron storage capacity in response to iron supplementation with no deleterious health effects (Chicharro et al. 2004);
The Lys153Arg polymorphism (rs1805086) in the myostatin (growth and differentiation factor) gene (GDF-8) (location: 2q32.2) is associated with muscle strength (Ferrell et al. 1999; Seibert et al. 2001; Huygens et al. 2004);
The Gly482Ser (rs8192678) polymorphism in the PPARGC1A gene (location: 4p15.1) is associated with mitochondrial biogenesis and skeletal muscle fibre-type conversion (i.e. II→I) (Lucia et al. 2005a).
Additionally, we genotyped a control polymorphism that is not associated with endurance phenotypes, i.e. the L/S (long and short allele, respectively) polymorphism of the serotonin (5-HTT) transporter protein (or solute carrier family 6, member 4 gene (SLC6A4)) (location: 17q11.1-q12) (Bray et al. 2009). The L allele is associated with higher levels of 5-HTT expression than the S allele (Heils et al. 1996).
Genotyping methods have been described elsewhere (Gonzalez-Freire et al. 2009; Muniesa et al. 2009) and were in accordance with stringent recommendations for replicating human genotype–phenotype association studies (Chanock et al. 2007). Briefly, we extracted genomic DNA in all subjects from peripheral EDTA treated anti-coagulated blood according to standard phenol–chloroform procedures followed by alcohol precipitation. Sequences corresponding to each mutation of the different genes were amplified by the polymerase chain reaction (PCR). The resulting PCR products were genotyped (in the Genetics Laboratory of Universidad Europea de Madrid, Spain) by single base extension (HFE and GDF-8), restriction fragment length polymorphisms (ACTN3, AMPD1, CKMM and PPARGC1A) or electrophoresis through agarose gel (ACE and SLC6A4).
Probability of having an ‘optimal’ polygenic profile for endurance performance in the Spanish (Caucasian) population
We calculated the probability of any given Spanish (Caucasian) individual possessing every ‘optimal’ genotype for from one up to all seven polymorphisms by using the typical frequency of ‘optimal’ genotypes observed in a Spanish (Caucasian descent for ≥3 generations) population (Muniesa et al. 2009) (Table 1). The studied polymorphisms were ranked in alphabetical succession based on their official symbols. Based on the observed typical frequencies of the ‘optimal’ genotypes, we produced a scale indicating the probability of possessing a ‘perfect’ genetic profile as the number of polymorphisms included in the profile increases.
Polygenic potential for endurance performance of the Spanish (Caucasian) population
We computed the combined influence of all the seven studied polymorphisms following the procedure of Williams & Folland (2008). First, we scored each genotype within each polymorphism (Table 1). We assigned a genotype score (GS) of 2 to the ‘optimal’ or preferable endurance genotype, whereas a GS of 0 was assigned to the less optimal genotype. Second, we summed the GS of each single genotype (GSACE+ GSACTN3+ GSAMPD1+ GSCKMM+ GSHFE+ GSGDF-8+ GSPPARGC1A). Third, the score was transformed to the scale of 0–100 for easier interpretation, namely total genotype score (TGS), as follows:
where 14 is the result of multiplying 7 (number of studied polymorphisms) by 2, which is the score given to the ‘optimal’ or preferable endurance genotype. As indicated by Williams & Folland (2008), a TGS of 100 represents a ‘perfect’ polygenic profile for endurance – that is, that all GS are 2. In contrast, a TGS of 0 represents the ‘worst’ possible profile for endurance – that is, that all GS are 0. Fourth, we created a data set of 50 000 hypothetical Spanish individuals, each with a randomly generated polygenetic profile (for all 7 polymorphisms) based on the frequency of each genotype for the Spanish population observed in our laboratory (Muniesa et al. 2009) (Table 1). Finally, we examined the distribution of TGSs within this virtual population. We calculated the mean and kurtosis statistics using the Statistical Package for Social Sciences (SPSS, v. 16.0 for Windows; SPSS Inc., Chicago, IL, USA).
Polygenic potential for endurance performance of a Spanish (Caucasian) population of non-athletes (controls) and top level endurance athletes
We also computed the polygenic profile for each control subject and each endurance elite athlete following the procedures as described above to examine (1) the nature of the distribution of the TGSs in a highly selected group of Spanish endurance elite athletes, and (2) differences between the athlete group, the non-athlete group, and the general Spanish population.
Theoretical TGS cut-off value to predict elite endurance status
We evaluated the ability of the TGS to correctly distinguish potential elite endurance athletes from non-athletes (0 = non-athlete, 1 = elite athlete) by receiver operating characteristic (ROC) curves (Zweig & Campbell, 1993). We calculated the area under the ROC curve (AUC) and 95% confidence intervals (95% CI). Finally, we used binary logistic regression to study the relationship between TGS and athletic status.
Distribution of elite endurance athletes and controls with an ‘optimal’ endurance genotype for one up to seven polymorphisms
We compared frequencies of having an ‘optimal’ endurance genotype for one up to seven polymorphisms between athletes and non-athletes (controls) using a χ2 test with α set at 0.05.
Distribution of the SLC6A4 polymorphisms in athletes and controls
Genotype frequencies of the S/L polymorphism in the SLC6A4 gene were compared between athletes and non-athletes (controls) using a χ2 test with α set at 0.05.
Results
The typical frequency of ‘optimal’ genotype of each polymorphism in the Spanish (Caucasian) population is shown in Table 2. Typical genotypes frequencies ranged from 19% for the II genotype of the ACE gene to 90% for the KK genotype of the GDF-8 gene.
Table 2.
Probability of possessing perfect genetic profile by number of polymorphisms in the Spanish population
| Polymorphisms influencing endurance performance | New gene included at each stage | Typical frequency (%) of ‘optimal’ genotype in Spanish population | Probability of possessing a ‘perfect’ profile |
|
|---|---|---|---|---|
| % chance | Approximate odds ratio | |||
| 1 | ACE | 19 | 19.000 | 1:5 |
| 2 | ACTN3 | 18 | 3.434 | 1:29 |
| 3 | AMPD1 | 82 | 2.806 | 1:36 |
| 4 | CKMM | 35 | 0.992 | 1:101 |
| 5 | HFE | 24 | 0.240 | 1:417 |
| 6 | GDF-8 | 90 | 0.240 | 1:417 |
| 7 | PPARGC1A | 38 | 0.074 | 1:1351 |
The probability of a given Spanish (Caucasian) individual possessing an ‘optimal’ polygenic profile was 19% when considering just one polymorphism (the II genotype for the ACE gene) and it was reduced to ∼3% when adding a second polymorphism (the XX genotype for the ACTN3 gene). When considering all polymorphisms, the probability of possessing an optimal polygenic profile was ∼0.07% (i.e. one in 1351 Spanish individuals).
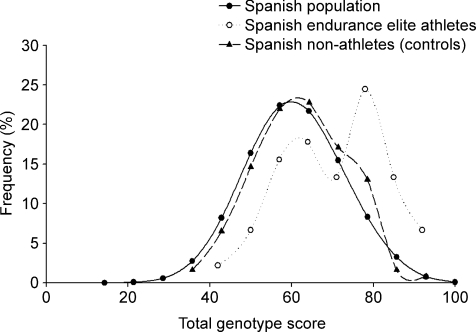
The frequency distributions of TGSs derived from a model sample of 50 000 randomly selected Spanish (Caucasian) individuals and that obtained from 123 Spanish healthy controls and 46 elite endurance athletes are depicted in Fig. 1. In the total Spanish population the predicted mean ±s.d. TGS was 60.80 ± 12.21, kurtosis statistic: –0.167 ± 0.022. In the 123 Spanish controls and the endurance Spanish athletes, mean TGS was 62.43 ± 11.45 (kurtosis statistic: –0.417 ± 0.433) and 70.22 ± 15.58 (kurtosis statistics: 0.791 ± 0.688), respectively. The TGS distribution in the Spanish population was platykurtic. This type of distribution has a lower probability than a normally distributed variable of values near the mean, as well as a lower probability than a normally distributed variable of extreme values. The distribution in athletes was shifted to the right with two ‘peaks’, one notably approaching a TGS of ∼80. Interestingly, none of our athletes exhibited an optimal TGS (100) and only three exhibited an optimal GS for up to six variants (TGS ∼93 in all three cases) (Table 3).
Figure 1.
Frequency distribution of total genotype scores from a model sample of 50 000 randomly selected Spanish individuals, 46 elite athletes and 123 non-athletes (controls)
Table 3.
Distribution [n (%)] of elite endurance athletes and non-athletes (controls) with an ‘optimal’ endurance genotype for 1 up to 7 polymorphisms
| Number of accumulated genotypes with an ‘optimal’ individual score (=2) | Athletes (n= 46) | Controls (n= 123) | P value for the between-group comparison |
|---|---|---|---|
| P= 0.006 | |||
| 1 | 2 (4.3%) | 1 (0.8%) | |
| 2 | 6 (13.0%) | 28 (22.8%) | |
| 3 | 11 (23.9%) | 48 (39.0%) | |
| 4 | 13 (28.3%) | 34 (27.6%) | |
| 5 | 11 (23.9%) | 11 (8.9%) | |
| 6 | 3 (6.5%) | 1 (0.8%) | |
| 7 | 0 (0%) | 0 (0%) |
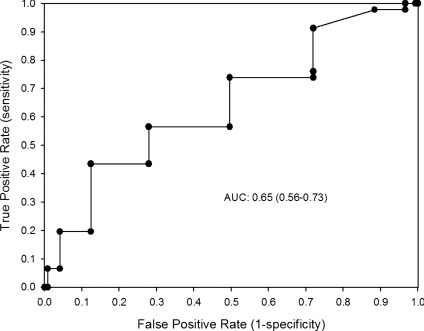
ROC analysis showed a significant discriminating accuracy of TGS in identifying an elite endurance athlete (AUC = 0.65; 95% CI: 0.56–0.73; P= 0.001; sensitivity = 0.435, specificity = 0.875) (Fig. 2). The corresponding TGS value at this point was 74.71. Logistic regression analysis showed that subjects with TGS above 74.71 had an increased odds ratio (OR) of being an elite endurance athlete when compared to those with TGSs below this value (OR: 5.411; 95% CI: 3.019–9.699; P < 0.001).
Figure 2. Receiver operating characteristic curve (ROC) summarizing the ability of total genotype score (TGS) to classify potential elite endurance athletes from non-athletes.
AUC indicates the area under the curve (95% confidence intervals).
SLC6A4 genotype distributions were similar (P= 0.881) in athletes and non-athletes (Table 4).
Table 4.
Genotype distribution (n, %) in elite endurance athletes and non-athletes (controls) of a polymorphism (S/L in solute carrier family 6, member 4 gene (SLC6A4)) not related to endurance phenotypes
| Athletes (n= 46) | Controls (n= 123) | P value | |
|---|---|---|---|
| SLC6A4 | — | — | 0.881 |
| S/S | 15 (32.6%) | 39 (31.7%) | — |
| L/S | 18 (39.1%) | 53 (43.1%) | — |
| L/L | 13 (28.3%) | 31 (25.2%) | — |
Discussion
This is a novel report using actual data on the polygenic endurance profile of athletes with the same ethnic origin who are at the upper end of the human endurance performance continuum. Although several candidate genes have been proposed to individually explain between-people differences in human endurance phenotypic traits (e.g. 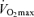 ), results are often conflicting and/or difficult to extrapolate to actual competition performance. One potential source of controversy comes when trying to combine results of different studies. Most studies report data on just one polymorphism and the populations of endurance athletes are frequently from mixed sporting disciplines and different ethnic/geographical origins. Our report itself is not without limitations as we only studied seven polymorphisms. Numerous genetic variants that we did not include in the model are likely to appear in the foreseeable future that can also explain individual variations in the potential for attaining the status of being an endurance athletic champion.
), results are often conflicting and/or difficult to extrapolate to actual competition performance. One potential source of controversy comes when trying to combine results of different studies. Most studies report data on just one polymorphism and the populations of endurance athletes are frequently from mixed sporting disciplines and different ethnic/geographical origins. Our report itself is not without limitations as we only studied seven polymorphisms. Numerous genetic variants that we did not include in the model are likely to appear in the foreseeable future that can also explain individual variations in the potential for attaining the status of being an endurance athletic champion.
Compared with the Spanish population, the TGS distribution of our athlete sample was shifted to the right with two ‘peaks’, one notably approaching a score of ∼80. This would suggest the existence of an overall more ‘favourable’ polygenic endowment in athletes. The highest peak corresponds to most of the elite runners. One reason for this finding might lie in the fact that the optimal ACE II genotype (associated with improved cardiovascular function but with less potential for muscle hypertrophy) is more frequent in runners than in cyclists (Lucia et al. 2005b). In runners low leg muscle mass represents a performance advantage as it favours a key phenotypic trait, running economy (Gonzalez-Freire et al. 2009). Interestingly, none of our athletes exhibited an optimal polygenic profile (TGS = 100) and only three exhibited an optimal genotype for up to six variants (TGS∼93). The possibility of finding a Spanish individual with the aforementioned ‘near-optimal’ profile is 0.1%, i.e. there are ∼41 000 Spanish individuals with such profile. A top-3 finisher in the Tour de France had only three optimal individual genotypes, yielding a TGS of ∼57, that is, similar to the mean TGS for the total Spanish population. In other words, and at least for the candidate genes we studied and for the TGS model developed by Williams & Folland, a healthy Spaniard with a normal polygenic endurance profile would not be genetically limited to reach the podium in the Tour de France. This is an important finding as the Tour de France arguably represents the most extreme of challenging endurance events that can be undertaken by humans.
William & Folland obtained a clustered TGS distribution towards the middle, which would limit the potential number of existing humans with a theoretically ‘perfect’ or ‘near perfect’ polygenic endurance profile. In Spain, indeed, there are only ∼3000 individuals (out of a total population of ∼41 million people of Caucasian descent) with the theoretically optimal polygenic endurance profile (TGS = 100) for the seven candidate genes we studied. None of our athletes is amongst these genetically gifted 3000 individuals.
ROC analysis suggested that the theoretical TGS cut-off value that predicts elite endurance status is 74.71. Accordingly, subjects with a TGS above 74.71 had an increased OR (5.411; 95%CI: 3.019–9.699) of being an elite endurance athlete when compared to those with a TGS below this value. Less than half of our elite endurance athletes (43.5%, n= 20) had TGS values above 74.71, and the probability of finding a Spanish individual with TGS above this value is 12.4%– that is, there would be ∼5 804 000 Spanish individuals with the potential to become an elite endurance athlete. However, a critical concept to be kept in mind is that only an extremely small fraction of the planet's population (irrespective of genetic endowment) participates in the artificial selection process (including stringent training regimens since childhood) that ends with the attainment of elite sports performance. Several new candidate polymorphisms will likely appear in the foreseeable future, allowing for more accurate predictions that should also take into account potential complex gene interactions, including those interactions between genetic variants that might not influence endurance performance individually. Nonetheless, even when more adequate genetic data are available, the frequency of top level sports performers will be meaningfully different from predicted by genetic possibility. There are indeed numerous other contributors to the ‘complex trait’ of being an athletic champion that are not likely to be reducible to defined genetic polymorphisms. These include both internal factors (e.g. technique, kinematics, motivation, pain tolerance) and external factors (notably social support, opportunity and economic possibility). On the other hand, future research might determine to what extent the changes that environmental factors can induce in gene expression during critical periods of prenatal and postnatal development (i.e. epigenetic mechanisms) (Waterland & Michels, 2007) can also explain why some individuals reach the upper end of the endurance performance continuum.
In summary, the probability of a Spanish individual possessing an optimal polygenic profile for up to the seven candidate genetic polymorphisms we studied (ACE Ins/Del, ACTN3 Arg577Ter, AMPD1 Gln12Ter, CKMM 1170 bp/985 + 185 bp, HFE His63Asp, GDF-8 Lys153Arg and PPARGC1A Gly482Ser) is very small, i.e. ∼0.07% (or 1 in 1351 Spanish individuals). Though the ‘total genotype score’ was slightly higher in athletes than in the general population, only three of the best Spanish endurance athletes (who are also amongst the best in the world) had the best polygenic profile for up to six genes and none of them had the optimal profile. Several genetic polymorphisms and complex genetic interactions that are yet to be identified, as well as factors independent of genetic endowment may also explain why some individuals reach the athletic excellence and some others do not. This is in fact the beauty of sports.
Acknowledgments
This study was funded by the following grants: Spanish Ministry of Education (ref. no. EX-2007-1124), Consejo superior de Deportes (CSD, ref no. UPR10/08) and Fondo de Investigaciones Sanitarias (FIS, ref. no. PI061183).
Glossary
Abbreviations
- ACE
angiotensin-converting enzyme gene
- ACTN3
α-actinin-3 gene
- AMPD1
adenosine monophosphate deaminase1 gene
- AUC
area under the curve
- CKMM
muscle-specific creatine kinase gene
- GDF-8
growth and differentiation factor (myostatin) gene
- HFE
-
hereditary haemochromatosis gene
5-HTT (serotonin)
- PCR
polymerase chain reaction
- PPARGC1A
peroxisome proliferator-activated receptor γ coactivator 1α gene
- ROC
receiver operating characteristic
- SLC6A4
solute carrier family 6, member 4 gene
- TGS
total genotype score
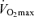
maximal oxygen uptake
Author contributions
JRR: conception and design, analysis and interpretation of data, drafting the article or revising it critically for important intellectual content, final approval of the version to be published; FGG: analysis of data and interpretation of data, drafting the article or revising it critically for important intellectual content, final approval of the version to be published; CS: analysis and interpretation of data, drafting the article or revising it critically for important intellectual content, final approval of the version to be published; MGF: analysis and interpretation of data, drafting the article or revising it critically for important intellectual content, final approval of the version to be published; ZV: analysis and interpretation of data, drafting the article or revising it critically for important intellectual content, final approval of the version to be published; CF: conception and design, drafting the article or revising it critically for important intellectual content, final approval of the version to be published; AL: conception and design, analysis and interpretation of data, drafting the article or revising it critically for important intellectual content, final approval of the version to be published.
References
- Bray MS, Hagberg JM, Pérusse L, Rankinen T, Roth SM, Wolfarth B, Bouchard C. The human gene map for performance and health-related fitness phenotypes: the 2006–2007 update. Med Sci Sports Exerc. 2009;41:35–73. doi: 10.1249/mss.0b013e3181844179. [DOI] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Chicharro JL, Hoyos J, Gomez-Gallego F, Villa JG, Bandres F, Celaya P, Jimenez F, Alonso JM, Cordova A, Lucia A. Mutations in the hereditary haemochromatosis gene HFE in professional endurance athletes. Br J Sports Med. 2004;38:418–421. doi: 10.1136/bjsm.2002.003921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell RE, Conte V, Lawrence EC, Roth SM, Hagberg JM, Hurley BF. Frequent sequence variation in the human myostatin (GDF8) gene as a marker for analysis of muscle-related phenotypes. Genomics. 1999;62:203–207. doi: 10.1006/geno.1999.5984. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Freire M, Santiago C, Verde Z, Lao JI, Oiivan J, Gomez-Gallego F, Lucia A. Unique among unique. Is it genetically determined? Br J Sports Med. 2009;(in press) doi: 10.1136/bjsm.2008.049809. [DOI] [PubMed] [Google Scholar]
- Heils A, Teufel A, Petri S, Stöber G, Riederer P, Bengel D, Lesch KP. Allelic variation of human serotonin transporter gene expression. J Neurochem. 1996;66:2621–2624. doi: 10.1046/j.1471-4159.1996.66062621.x. [DOI] [PubMed] [Google Scholar]
- Huygens W, Thomis MA, Peeters MW, Aerssens J, Janssen R, Vlietinck RF, Beunen G. Linkage of myostatin pathway genes with knee strength in humans. Physiol Genomics. 2004;17:264–270. doi: 10.1152/physiolgenomics.00224.2003. [DOI] [PubMed] [Google Scholar]
- Jones A, Woods DR. Skeletal muscle RAS and exercise performance. Int J Biochem Cell Biol. 2003;35:855–866. doi: 10.1016/s1357-2725(02)00342-4. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Anevski D, Guiducci C, Burtt NP, Roos C, Hirschhorn JN, Berglund G, Hedblad B, Groop L, Altshuler DM, Newton-Cheh C, Orho-Melander M. Polymorphisms associated with cholesterol and risk of cardiovascular events. N Engl J Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- Lucia A, Gomez-Gallego F, Barroso I, Rabadan M, Bandres F, San Juan AF, Chicharro JL, Ekelund U, Brage S, Earnest CP, Wareham NJ, Franks PW. PPARGC1A genotype (Gly482Ser) predicts exceptional endurance capacity in European men. J Appl Physiol. 2005a;99:344–348. doi: 10.1152/japplphysiol.00037.2005. [DOI] [PubMed] [Google Scholar]
- Lucia A, Gomez-Gallego F, Chicharro JL, Hoyos J, Celaya K, Cordova A, Villa G, Alonso JM, Barriopedro M, Perez M, Earnest CP. Is there an association between ACE and CKMM polymorphisms and cycling performance status during 3-week races? Int J Sports Med. 2005b;26:442–447. doi: 10.1055/s-2004-821108. [DOI] [PubMed] [Google Scholar]
- Lucia A, Olivan J, Bravo J, Gonzalez-Freire M, Foster C. The key to top-level endurance running performance: a unique example. Br J Sports Med. 2008;42:172–174. doi: 10.1136/bjsm.2007.040725. discussion 174. [DOI] [PubMed] [Google Scholar]
- Muniesa CA, Gonzalez-Freire M, Santiago C, Lao JI, Buxens A, Rubio JC, Martin MA, Arenas J, Gomez-Gallego F, Lucia A. World-class performance in lightweight rowing: Is it genetically influenced? A comparison with cyclists, runners and non-athletes. Br J Sports Med. 2009;(in press) doi: 10.1136/bjsm.2008.051680. [DOI] [PubMed] [Google Scholar]
- Rivera MA, Dionne FT, Simoneau JA, Perusse L, Chagnon M, Chagnon Y, Gagnon J, Leon AS, Rao DC, Skinner JS, Wilmore JH, Bouchard C. Muscle-specific creatine kinase gene polymorphism and VO2max in the HERITAGE Family Study. Med Sci Sports Exerc. 1997;29:1311–1317. doi: 10.1097/00005768-199710000-00006. [DOI] [PubMed] [Google Scholar]
- Rubio JC, Martin MA, Rabadan M, Gomez-Gallego F, San Juan AF, Alonso JM, Chicharro JL, Perez M, Arenas J, Lucia A. Frequency of the C34T mutation of the AMPD1 gene in world-class endurance athletes: does this mutation impair performance? J Appl Physiol. 2005;98:2108–2112. doi: 10.1152/japplphysiol.01371.2004. [DOI] [PubMed] [Google Scholar]
- Seibert MJ, Xue QL, Fried LP, Walston JD. Polymorphic variation in the human myostatin (GDF-8) gene and association with strength measures in the Women's Health and Aging Study II cohort. J Am Geriatr Soc. 2001;49:1093–1096. doi: 10.1046/j.1532-5415.2001.49214.x. [DOI] [PubMed] [Google Scholar]
- Waterland RA, Michels KB. Epigenetic epidemiology of the developmental origins hypothesis. Annu Rev Nutr. 2007;27:363–388. doi: 10.1146/annurev.nutr.27.061406.093705. [DOI] [PubMed] [Google Scholar]
- Williams AG, Folland JP. Similarity of polygenic profiles limits the potential for elite human physical performance. J Physiol. 2008;586:113–121. doi: 10.1113/jphysiol.2007.141887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, MacArthur DG, Wolde B, Onywera VO, Boit MK, Lau SY, Wilson RH, Scott RA, Pitsiladis YP, North K. The ACTN3 R577X polymorphism in East and West African athletes. Med Sci Sports Exerc. 2007;39:1985–1988. doi: 10.1249/mss.0b013e31814844c9. [DOI] [PubMed] [Google Scholar]
- Zweig MH, Campbell G. Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem. 1993;39:561–577. [PubMed] [Google Scholar]