Abstract
Muscle protein synthesis and mTORC1 signalling are concurrently stimulated following muscle contraction in humans. In an effort to determine whether mTORC1 signalling is essential for regulating muscle protein synthesis in humans, we treated subjects with a potent mTORC1 inhibitor (rapamycin) prior to performing a series of high-intensity muscle contractions. Here we show that rapamycin treatment blocks the early (1–2 h) acute contraction-induced increase (∼40%) in human muscle protein synthesis. In addition, several downstream components of the mTORC1 signalling pathway were also blunted or blocked by rapamycin. For instance, S6K1 phosphorylation (Thr421/Ser424) was increased post-exercise 6-fold in the control group while being unchanged with rapamycin treatment. Furthermore, eEF2 phosphorylation (Thr56) was reduced by ∼25% post-exercise in the control group but phosphorylation following rapamycin treatment was unaltered, indicating that translation elongation was inhibited. Rapamycin administration prior to exercise also reduced the ability of raptor to associate with mTORC1 during post-exercise recovery. Surprisingly, rapamycin treatment prior to resistance exercise completely blocked the contraction-induced increase in the phosphorylation of ERK1/2 (Thr202/Tyr204) and blunted the increase in MNK1 (Thr197/202) phosphorylation. However, the phosphorylation of a known target of MNK1, eIF4E (Ser208), was similar in both groups (P > 0.05) which is consistent with the notion that rapamycin does not directly inhibit MAPK signalling. We conclude that mTORC1 signalling is, in part, playing a key role in regulating the contraction-induced stimulation of muscle protein synthesis in humans, while dual activation of mTORC1 and ERK1/2 stimulation may be required for full stimulation of human skeletal muscle protein synthesis.
Resistance exercise is an excellent means to manipulate the size and force production of skeletal muscle in mammals (Widrick et al. 2002). The mammalian target of rapamycin (mTOR) has received considerable attention as an integral control point for muscle cell growth and atrophy (Bodine et al. 2001). In skeletal muscle, mTOR is found in two distinct complexes: mTORC1 and mTORC2. Three known proteins make up the mTORC1 complex (G-protein β subunit-like protein (GβL), proline-rich Akt substrate 40 and raptor) while the mTORC2 complex is comprised of GβL and rictor. The mTORC1 complex plays an important regulatory role during the hypertrophy process of skeletal muscle cells (Bodine et al. 2001). Less is known about the regulation of mTORC2 although it is not thought to be involved in the regulation of translation initiation and elongation (Sarbassov et al. 2006).
Phosphorylation of mTORC1 occurs within minutes following an anabolic stimulus (Bolster et al. 2003). An increase in mTORC1 kinase activity results in the phosphorylation of two key downstream effectors: ribosomal S6 kinase 1 (S6K1) and eukaryotic initiation factor 4E binding protein, 4E-BP1 (Bolster et al. 2003; Karlsson et al. 2004; Cuthbertson et al. 2006; Dreyer et al. 2006). S6K1, when activated, phosphorylates the ribosomal protein (rp) S6 (Roux et al. 2007) and eukaryotic elongation factor 2 (eEF2) kinase (Wang et al. 2001), subsequently enhancing translation initiation and elongation. Phosphorylation of rpS6 has been reported to enhance cell size and cell proliferation (Ruvinsky et al. 2005) by an unknown mechanism although the role of rpS6 has been heavily debated (Ruvinsky & Meyuhas, 2006). Reduced phosphorylation of eEF2 (due to eEF2 kinase inhibition) results in mediation of the ribosome to the mRNA after each amino acid is added to the growing peptide chain (Browne & Proud, 2002). mTORC1 phosphorylation of 4E-BP1 inhibits the binding of eIF4E to 4E-BP1, thereby promoting eIF4E to complex with eIF4G, further enhancing the formation of the translation initiation complex (Wang & Proud, 2006).
Rapamycin is a potent inhibitor of mTORC1 activity. As such, rapamycin administration in cell culture and rodent models is a valuable means to investigate mTORC1 function (Bodine et al. 2001). Rapamycin has been shown to complex to the FK-binding protein 12 and then bind to mTORC1 thereby inhibiting its function (Oshiro et al. 2004; Proud, 2007). For example, rapamycin treatment reduces S6K1 phosphorylation and increases eIF4E–BP1 complex formation in cells (Chung et al. 1992; von Manteuffel et al. 1996), while in rodents, activation of mTORC1 and increases in skeletal muscle protein synthesis through nutrients or exercise are blocked with rapamycin administration (Anthony et al. 2000; Kubica et al. 2005). It is not known whether rapamycin administration in humans has a similar effect on skeletal muscle protein synthesis.
Our laboratory has recently demonstrated that the mTORC1 signalling pathway is activated in human skeletal muscle after an acute bout of resistance exercise (Dreyer et al. 2006, 2008; Drummond et al. 2008); however, these studies were largely correlational. The aim of this investigation was to determine whether the increase in human skeletal muscle protein synthesis following a heavy bout of resistance exercise is causally related to mTORC1 activation, i.e. inhibited by rapamycin treatment.
Methods
Subjects
We studied 15 healthy young male subjects, seven of which participated in the rapamycin group while an additional eight subjects participated in the control group (no rapamycin). Age (control: 29 ± 2 years; rapamycin: 28 ± 2 years), height (control: 176 ± 2 cm; rapamycin: 178 ± 3 cm), weight (control: 74 ± 5 kg; rapamycin: 78 ±5 kg), lean mass (control: 57 ± 2 kg; rapamycin: 60 ± 4 kg) and body fat (control: 18 ± 3%; rapamycin: 21 ±2%) were not different between groups. The subjects were not engaged in any regular exercise training at the time of enrollment. Subjects’ screening was performed with clinical history, physical examination and laboratory tests including complete blood count with differential, liver and kidney function tests, coagulation profile, fasting blood glucose and oral glucose tolerance test, hepatitis B and C screening, HIV test, thyroid-stimulating hormone, urinalysis, and drug screening. Upon enrollment, a dual-energy X-ray absorptiometry (DEXA) scan (Hologic QDR 4500 W, Bedford, MA, USA) was performed to measure body composition and lean mass. Subjects were also tested for maximal strength by performing a one-repetition maximum (1RM) on a leg extension machine (Cybex-VR2, Medway, MA, USA) during the initial screening. A second 1RM testing was performed at least 1 week prior to study participation. The starting weight used during the resistance exercise portion of this study was 70% of the subjects’ pre-determined 1RM. All subjects gave informed written consent before participating in the study, which was approved by the Institutional Review Board of the University of Texas Medical Branch (which is in compliance with the Declaration of Helsinki).
Study design
Subjects were admitted to the GCRC of the University of Texas Medical Branch the day prior to the exercise study. The subjects were then fed a standard dinner at 1800 h and a snack at 2200 h and were allowed only water ad libitum afterwards. All subjects were studied following an overnight fast under basal conditions and refrained from exercise for 24 h prior to study participation. The morning of the infusion study, an 18-gauge polyethylene catheter was inserted into a forearm vein for tracer infusion and another catheter was inserted retrogradely in the forearm of the opposite arm which was heated for arterialized blood sampling. After drawing a background blood sample, a primed continuous infusion of l-[ring-2H5] phenylalanine (Cambridge Isotope Laboratories, Andover, MA, USA) was started (time = 0 h) and maintained at a constant rate until the end of the experiment. The priming dose for the labelled phenylalanine was 2 μmol kg−1 and the infusion rate was 0.05 μmol kg−1 min−1. All subjects were studied during the same time of day to avoid potential circadian changes.
A baseline muscle biopsy was taken 150 min following the initiation of the tracer infusion. The biopsy was obtained from the lateral portion of the vastus lateralis using a 5 mm Bergström biopsy needle, under sterile procedure and local anesthesia (1% lidocaine). Once collected, the muscle tissue was immediately blotted and frozen in liquid nitrogen and stored at −80°C until analysis. Following the first biopsy, two blood samples were drawn 30 min apart from each other. One hour after the first biopsy, a second muscle biopsy was taken marking the end of the baseline period. However, under this circumstance, the biopsy needle was inclined at a different angle so that the second biopsy was taken approximately ∼5 cm apart from the first.
Following the second biopsy, the control group immediately began exercise while the subjects in the rapamycin group consumed 12 mg of rapamycin (1 mg per tablet) (Rapamune/Sirolimus; Wyeth, Madison, NJ, USA) and remained in their hospital bed for two additional hours. During the 2 h rapamycin period, blood was sampled every 15 min for the first hour and at 30 min for the second hour. All subjects (in both groups) were escorted to the Cybex leg extension machine and performed 11 sets of 10 repetitions of two-legged extension exercises set to 70% of their 1RM. The rest period between sets was 3 min. Blood samples were collected immediately after the third, sixth, eighth and tenth sets. All subjects were then transported back to their hospital bed for the remainder of the study. Blood samples were taken every 15 min during the first hour of recovery from exercise. At 1 and 2 h post-exercise, a third and fourth muscle biopsy, respectively, were sampled in a new incision site ∼7 cm proximal to the first incision site and the biopsy needle angled such that each biopsy was ∼5 cm from each other.
Hormone analysis
Insulin (Millipore, Billerica, MA, USA) and cortisol (Calbiotech, Spring Valley, CA, USA) concentrations were measured at select time points via ELISA according to the manufacturers’ instructions.
Rapamycin analysis
Rapamycin was determined from blood samples using a commercially available kit (IMx Sirolimus assay; Abbott Laboratories, Abbott Park, IL, USA). Select samples were analysed during the time course of the current experiment. Due to the sensitivity of the assay, rapamycin (sirolimus) values less than 3 ng ml−1 were undetectable.
Muscle fractional synthetic rate
Muscle tissue samples were ground, and intracellular free amino acids and muscle proteins were extracted as previously described (Dreyer et al. 2006). Blood and muscle intracellular free concentration and enrichment of phenylalanine, and the blood concentrations of the branched-chain amino acids (BCAA; leucine, isoleucine and valine) were determined by gas chromatography–mass spectrometry (GCMS, 6890 Plus GC, 5973N MSD, 7683 autosampler, Agilent Technologies, Palo Alto, CA, USA) using appropriate internal standards (Wolfe & Chinkes, 2005). Mixed muscle protein-bound phenylalanine enrichment was analysed by GCMS after protein hydrolysis and amino acid extraction (Dreyer et al. 2006), using the external standard curve approach (Calder et al. 1992). We calculated the fractional synthetic rate of mixed muscle proteins (FSR) by measuring the incorporation rate of the phenylalanine tracer into the proteins (ΔEp/t) and using the precursor–product model to calculate the synthesis rate:
where ΔEp is the increment in protein-bound phenylalanine enrichment between two sequential biopsies, t is the time between the two sequential biopsies, and EM(1)+EM(2) are the phenylalanine enrichments in the free intracellular pool in the two sequential biopsies. Data are expressed as per cent per hour.
Immunoblot analysis
Details of immunoblotting can be found elsewhere (Dreyer et al. 2006). Briefly, frozen tissue was homogenized, centrifuged for 10 min at 4°C, and supernatant was collected. Total protein concentrations were determined using the Bradford assay (Smartspec Plus, BioRad, Hercules, CA, USA). The supernatant was diluted (1 : 1) in a 2× sample buffer mixture containing 125 mm Tris, pH 6.8, 25% glycerol, 2.5% SDS, 2.5%β-mercaptoethanol and 0.002% bromophenol blue, then boiled for 3 min at 100°C. However, homogenate aliquots used to detect 4E-BP1 were initially boiled at 100°C for 10 min and spun for 30 min at 6000 g before combining the supernatants with the sample buffer. Equal amounts of total protein (50 μg) were loaded into each lane and the samples were separated by electrophoresis (150 V for 60 min) on a 7.5% or 15% polyacrylamide gel as determined by the size of the target protein (Criterion, BioRad). Each sample was loaded in duplicate and each gel contained an internal loading control (rodent skeletal muscle) and molecular weight ladder (Precision Plus, BioRad). Following electrophoresis, protein was transferred to a polyvinylidene difluoride membrane (BioRad) at 50 V for 60 min. Blots were incubated in primary antibody overnight at 4°C (antibody concentrations are described below). The next morning, blots were incubated in secondary antibody for 1 h at room temperature. Chemiluminescent solution (ECL plus, Amersham BioSciences, Piscataway, NJ, USA) was applied to each blot. After a 5 min incubation, optical density measurements were obtained with a phosphoimager (ChemiDoc, BioRad) and densitometric analysis was performed using Quantity One 4.5.2 software (BioRad). Membranes containing phospho-detected proteins were stripped of primary and secondary antibodies using Restore Western blot Stripping Buffer (Pierce Biotechnology, CA, USA). After incubating in stripping buffer, membranes were re-probed for total protein with the specific antibody of interest. Phospho and total density values were normalized to an internal loading control (which was loaded on each respective blot) and the phospho/total protein ratios were determined. Immunoblot data were expressed as phospho divided by total protein and adjusted to represent fold change from basal.
Quantification of raptor–mTOR complex (mTORC1)
mTOR protein was isolated in whole muscle homogenates using an immunoprecipitation technique as previously characterized (Williamson et al. 2006a). Approximately 30 mg of tissue was homogenized in CHAPS buffer (1 : 9 w/v, pH 7.5). The buffer consisted of 40 mm Hepes, pH 7.5, 120 mm NaCl, 1 mm EDTA, 10 mm pyrophosphate, 10 mmβ-glycerophosphate, 40 mm NaF, 1.5 mm sodium vanadate, 0.3% CHAPS, microcystin, and a Sigma protease inhibitor cocktail (P8340; St Louis, MO, USA). Samples were centrifuged at ∼2500 g for 5 min at 4°C and a total protein assay was conducted as described above. Seven-hundred micrograms of protein was combined with 2 μl of mTOR antibody and then rocked overnight at 4°C. mTOR complexes were isolated using BioMag goat anti-rabbit IgG beads (Qiagen, Valencia, CA, USA) and captured using a magnetic tube rack. Before use, the magnetic beads were washed twice in CHAPS buffer (without protease inhibitors) then re-suspended in CHAPS buffer combined with 1% non-fat dry milk. Five-hundred microlitres of this mixture (beads + buffer) were added to the samples then rocked 1 h at 4°C. Samples were washed two times in CHAPS buffer (without protease inhibitors) and once in a second buffer (Hepes, NaCl, pH 7.5). Sixty microlitres of 2× sample buffer were added to the slurry then boiled for 5 min. The sample was separated from the beads using the magnetic tube rack, then loaded onto a 7.5% gel in duplicate and later probed with anti-raptor and anti-mTOR antibody (diluted in 5% bovine serum albumin in tris-buffered saline (1% Tween)) with an overnight incubation in at 4°C. Blots were then developed and analysed as described above. The amount of raptor bound to mTORC1 was normalized to the total amount of mTOR in the precipitates. The data were expressed as raptor–mTORC1 association.
Antibodies
The phospho and total antibodies used for immunoblotting were purchased from Cell Signaling (Danvars, MA, USA) unless otherwise noted: phospho-Akt (Thr473; 1 : 1000), phospho-mTOR (Ser2448; 1 : 1000), Raptor (1 : 500), phospho-S6K1 (Thr389 and Thr421/Ser424; 1 : 500), phospho-4E-BP1 (Thr37/46; 1 : 1000), phospho-eEF2 (Thr56; 1 : 1000), phospho-rpS6 (Ser235/236 and Ser240/244; 1 : 1000), phospho-ERK1/2 (Thr202/Tyr204; 1 : 1000) phospho-MNK1 (Thr197; 1 : 1000), phospho-eIF4E (Ser208; 1 : 1000), phospho-eIF4G (Ser1108; 1 : 000), and phospho-AMP-activated protein kinase α (AMPKα) (Thr172; 1 : 1000). Phospho-eIF2Bɛ was purchased from GeneTex Inc. (San Antonio, TX, USA) (Ser535; 1 : 1000) and the total eIF2Bɛ antibody was purchased from Abcam (Cambridge, MA, USA). Total protein was detected using an antibody dilution of 1 : 1000. Anti-rabbit IgG horseradish peroxidase-conjugated secondary antibody was purchased from Amersham Bioscience (1 : 2000).
Statistical analysis
Data are reported as mean ±s.e.m. Between and within group differences were tested using a two-way repeated-measures ANOVA. When main effects existed, Bonferonni post hoc tests were conducted to assess interaction effects between particular time points and treatment groups. Alpha was set to <0.05. All analyses were performed with SigmaStat software (version 3.5).
Results
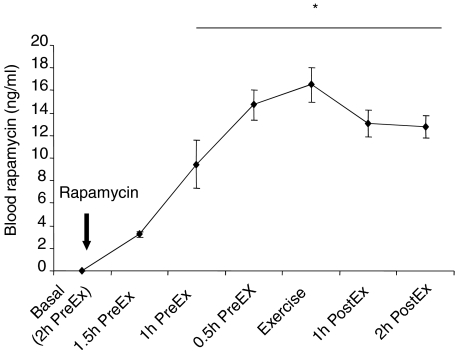
Subjects in the rapamycin group ingested 12 mg (∼0.15 mg (kg of body weight)−1) rapamycin 2 h before initiating exercise. At various time intervals, rapamycin was measured in the blood. Figure 1 illustrates the time course of rapamycin in the blood following rapamycin ingestion. Blood rapamycin concentration peaked immediately prior to initiation of the exercise protocol, and remained significantly elevated (P < 0.05) throughout the entire post-exercise period.
Figure 1. Data represent the time course of rapamycin in the blood measured immediately after rapamycin consumption.
PreEx is time before exercise while PostEx is time following exercise. *Significantly different from 1.5 h PreEx time period (P < 0.05).
Exercise data
Both groups had a similar 1RM (control: 96 ± 5 kg; rapamycin: 97 ± 9 kg) and worked at the same relative exercise intensity (∼70%). Although workload was occasionally adjusted in relation to the subjects’ ability to complete 10 repetions per set, the relative exercise intensity was consistent between groups. Blood lactate concentrations during the exercise bout were similar between groups (control: 9.6 ± 0.9 mg dl−1; rapamycin: 7.4 ± 0.6 mg dl−1; P > 0.05) indicating that exercise intensity was the same in both groups.
Blood hormone concentrations (Table 1)
Table 1.
Serum hormone concentrations before and after resistance exercise
| Basal | Exercise | 1 h PostEx | 2 h PostEx | |
|---|---|---|---|---|
| Cortisol (μmol l−1) | ||||
| Control | 0.20 ± 0.03 | 0.61 ± 0.23† | 0.70 ± 0.22† | 0.33 ± 0.09 |
| Rapamycin | 0.25 ± 0.02 | 0.42 ± 0.06† | 0.36 ± 0.06† | 0.28 ± 0.04 |
| Insulin (pmol l−1) | ||||
| Control | 51 ± 8 | 106 ± 20† | 71 ± 12 | 45 ± 5 |
| Rapamycin | 62 ± 11 | 93 ± 9† | 55 ± 10 | 30 ± 4 |
Values are means ±s.e.m. from control (n= 8) and rapamycin (n= 7) subjects.
Main effect for time (P < 0.05).
Cortisol concentrations were elevated to a similar extent in both groups immediately and 1 h post exercise (PostEx) in comparison to basal (P < 0.05) and returned to near-resting values at 2 h PostEx (P > 0.05). Insulin concentrations, in comparison to baseline, were increased in both groups immediately following exercise (P > 0.05) and returned to basal levels at 1 h and 2 h PostEx (P > 0.05) (see Table 1.
Blood amino acid concentrations (Table 2)
Table 2.
Branched-chain amino acid and phenylalanine venous concentrations (μmol l−1) before and after resistance exercise
| Basal | Exercise | 1 h PostEx | 2 h PostEx | |
|---|---|---|---|---|
| Leucine (μmol l−1) | ||||
| Control | 172 ± 6 | 168 ± 8 | 143 ± 10* | 150 ± 7* |
| Rapamycin | 187 ± 7 | 184 ± 7 | 167 ± 6*# | 158 ± 5* |
| Isoleucine (μmol l−1) | ||||
| Control | 58 ± 2 | 55 ± 3 | 47 ± 4* | 49 ± 3* |
| Rapamycin | 66 ± 5 | 62 ± 4 | 53 ± 3* | 50 ± 2* |
| Valine (μmol l−1) | ||||
| Control | 254 ± 8 | 235 ± 9* | 211 ± 12* | 207 ± 10* |
| Rapamycin | 260 ± 12 | 255 ± 13 | 232 ± 10* | 210 ± 7* |
| Phenylalanine (μmol l−1) | ||||
| Control | 60 ± 2 | 58 ± 2 | 59 ± 1 | 57 ± 2 |
| Rapamycin | 56 ± 6 | 56 ± 4 | 50 ± 3 | 50 ± 5 |
Values are means ±s.e.m. from control (n= 8) and rapamycin (n= 7) subjects.
Significantly different from basal (P < 0.05).
Significantly different from control (P < 0.05).
Blood phenylalanine concentrations did not change throughout the study in either group (P > 0.05). Blood concentrations of the BCAAs (leucine, isoleucine and valine) were similar between groups at basal levels and following resistance exercise (P > 0.05). BCAA concentrations decreased slightly in both groups following exercise (see Table 2; P < 0.05).
Muscle protein synthesis
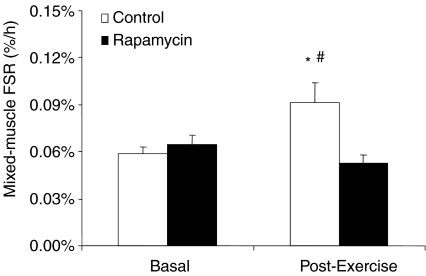
Mixed-muscle protein fractional synthetic rate (FSR) increased by ∼40% from baseline during the post-exercise recovery in the control group (P < 0.05; Fig. 2). Conversely, FSR did not change with exercise in the rapamycin group (P > 0.05 vs. basal) with a significant difference between groups (P < 0.05; Fig. 2).
Figure 2. Data represent mixed-muscle protein synthesis at basal and during the post-exercise period in control and rapamycin skeletal muscle.
*Significantly different from basal (P < 0.05). #Significantly different from rapamycin group.
Cell signalling
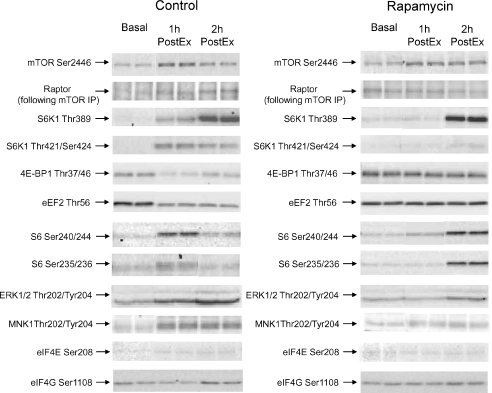
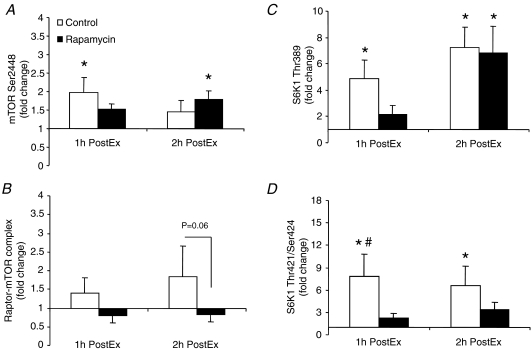
Representative blots for all proteins can be found in Fig. 3. In summary, as compared to basal, mTOR (Ser2448) phosphorylation was significantly elevated at 1 h PostEx in the control group and 2 h PostEx in the rapamycin group (Fig. 4A; P < 0.05). Raptor association with mTORC1 did not change statistically during post-exercise recovery in either group (P > 0.05); however, there was a trend for a group difference (i.e. raptor association with mTORC1 tended to be higher in the control group) at 2 h PostEx (Fig. 4B; P= 0.06). Ribosomal S6 kinase 1 (Thr389) phosphorylation increased from basal in the control group at 1 and 2 h PostEx, while it increased only at 2 h PostEx in the rapamycin group (Fig. 4C; P < 0.05). However, S6K1 phosphorylation at Thr421/Ser424 increased from basal only in the control group at 1 and 2 h PostEx (Fig. 4D; P < 0.05).
Figure 3.
Representative immunoblot protein images for control and rapamycin skeletal muscle samples at basal, 1 h PostEx and 2 h PostEx.
Figure 4. Data represent phosphorylation of mTOR at Ser2446 (A, control: n= 8; rapamycin: n= 7), raptor bound to mTOR (B, mTORC1; control: n= 6; rapamycin n= 7), S6K1 at Thr389 (C, control: n= 8; rapamycin: n= 7) and S6K1 at Thr421/Ser424 (D, control: n= 8; rapamycin: n= 7) in control and rapamycin skeletal muscle at basal, 1 h PostEx and 2 h PostEx.
Data are expressed as fold change from basal (mean ±s.e.m.). *Significantly different from basal (P < 0.05).
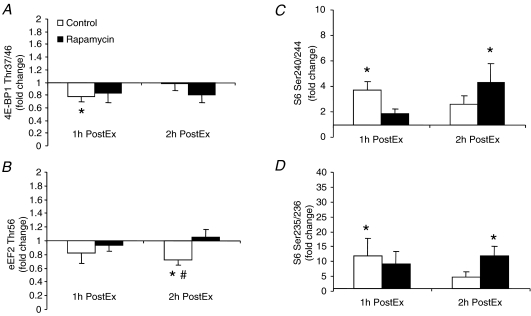
Phosphorylation of 4E-BP1 at Thr37/46 decreased significantly from basal at 1 h PostEx only in the control group (Fig. 5A). Eukaryotic elongation factor 2 (eEF2; Thr56) phosphorylation decreased significantly from basal at 2 h PostEx only in the control group and was significantly lower than the rapamycin group at this same time point (Fig. 5B; P < 0.05). Ribosomal protein S6 phosphorylation at Ser240/244 (Fig. 5C) and Ser235/236 (Fig. 5D) exhibited a differential behaviour: in the control group they were significantly increased from basal only at 1 h PostEx, while in the rapamycin group they were significantly increased from basal only at 2 h PostEx (P < 0.05).
Figure 5. Data represent phosphorylation of 4E-BP1 at Thr37/46 (A, control: n= 8; rapamycin: n= 7), eEF2 at Thr56 (B, control: n= 8; rapamycin: n= 7), S6 at Ser240/244 (C, control: n= 8; rapamycin: n= 7) and S6 at Ser235/236 (D, control: n= 8; rapamycin: n= 7) in control and rapamycin skeletal muscle at basal, 1 h PostEx and 2 h PostEx.
Data are expressed as fold change from basal (mean ±s.e.m.). *Significantly different from basal (P < 0.05). #Significantly different from rapamycin group (P < 0.05).
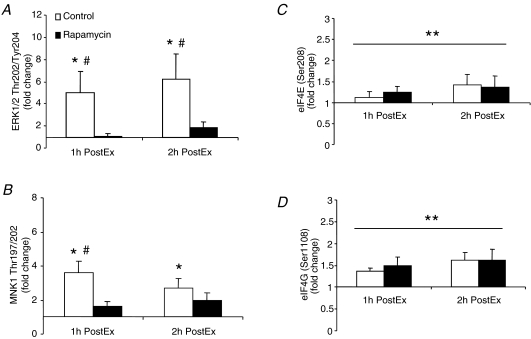
Extracellular signal-regulated kinase (ERK1/2) phosphorylation at Thr202/Tyr204 significantly increased from basal only in the control group at 1 and 2 h PostEx with a significant difference between groups at these same time points (Fig. 6A; P < 0.05). MAP kinase-interacting kinase 1 (MNK1) phosphorylation at Thr197/202, was significantly increased from basal only in the control group 1 and 2 h PostEx with a significant difference between groups at the 1 h PostEx (Fig. 6B; P < 0.05). There was a main effect for time in which the phosphorylation of eIF4E (Ser208; Fig. 6C) and eIF4G (Ser1108; Fig. 6D) were significantly increased to a similar extent, as compared to basal, at 1 and 2 h PostEx in both groups (P < 0.05).
Figure 6. Data represent phosphorylation of ERK1/2 at Thr202/Tyr204 (A, control: n= 8; rapamycin: n= 7), MNK1 at Thr197/202 (B, control: n= 8; rapamycin: n= 7), eIF4E at Ser208 (C, control: n= 5; rapamycin: n= 6) and eIF4G at Ser1108 (D, control: n= 4; rapamycin: n= 7) in control and rapamycin skeletal muscle at basal, 1 h PostEx and 2 h PostEx.
Data are expressed as fold change from basal (mean ±s.e.m.). *Significantly different from basal (P < 0.05). #Significantly different from rapamycin group (P < 0.05). **Main effect for time (P < 0.05).
Additional signalling data
Phosphorylation of Akt (Thr473) was significantly increased to a similar extent at 1 h and 2 h PostEx in both the control (1 h PostEx: 2.2 ± 0.3-fold; 2 h PostEx: 1.7 ± 0.2-fold) and rapamycin (1 h PostEx: 1.9 ± 0.2-fold; 2 h PostEx: 1.5 ± 0.2-fold) groups in comparison to basal (P < 0.05). Phosphorylation of AMP-activated protein kinase (AMPKα) at Thr172 was significantly increased to a similar extent for both the control (1.7 ± 0.4-fold) and rapamycin (1.9 ± 0.4-fold) groups at 1 h PostEx in comparison to basal (P < 0.05). Protein expression of eIF2Bɛ did not change in either group during the duration of the experiment (P > 0.05). In addition, there were no significant changes in eIF2Bɛ (Ser539) phosphorylation as compared to basal for control (1 h PostEx: 1.3 ± 0.2-fold; 2 h PostEx: 1.2 ± 0.1-fold) or rapamycin (1 h PostEx: 1.2 ± 0.3-fold; 2 h PostEx: 0.9 ± 0.2-fold) groups following exercise (P > 0.05).
Discussion
The current study was undertaken to mechanistically determine if mTORC1 activation is a fundamental regulator of the contraction-induced increase in skeletal muscle protein synthesis in humans. The novel findings from this study are the following: (1) the increase in human muscle protein synthesis during the early (1–2 h) post-exercise recovery period is abolished by rapamycin administration prior to exercise; (2) the exercise-induced change in the phosphorylation of a majority of proteins in the mTORC1 signalling pathway (i.e. mTOR, S6K1, rpS6) were delayed with rapamycin treatment; and (3) rapamycin completely blocked S6K1 (Thr421/Ser424), eEF2 (Thr56), ERK1/2 (Thr202/Tyr204), and blunted MNK1 (Thr197/202) phosphorylation during post-exercise recovery. These findings indicate that the stimulation of muscle protein synthesis following a bout of resistance exercise is, in part, rapamycin sensitive, and suggest that the mTORC1 signalling pathway is mechanistically important in regulating the contraction-induced increase in muscle protein synthesis. However, it appears that additional signalling pathways (e.g. ERK1/2) may also contribute to acute alterations in muscle protein synthesis in humans and may be indirectly influenced by rapamycin treatment.
The mTORC1 signalling pathway has been widely studied for its role in regulating cellular size in animals and humans (i.e. resistance exercise, cancer, ageing, etc.). Previous work from our research group has consistently identified changes in the mTOR signalling pathway immediately following potent anabolic stimuli, such as resistance exercise and essential amino acid ingestion, that correlated with changes in skeletal muscle protein synthesis (Dreyer et al. 2006, 2008; Fujita et al. 2007; Drummond et al. 2008; Fujita et al. 2008). Therefore, in the current study, we sought to evaluate whether changes in protein synthesis during the early recovery period following resistance exercise was mechanistically regulated by mTORC1 in human skeletal muscle. To our knowledge, this is the first study in humans specifically designed to determine the role of mTORC1 signalling in exercise-induced muscle protein accretion. We found that protein synthesis (as measured by labelled phenylalanine incorporation into the muscle) was increased in the control group (∼40%) but unchanged in the rapamycin group following resistance exercise (Fig. 2). Rapamycin has been used previously to block changes in protein synthesis following insulin administration in L6 myoblasts (Kimball et al. 1998) and in rodents following resistance exercise (Kubica et al. 2005). For example, rapamycin injection through the tail vein of rodents 2 h prior to an acute bout of resistance exercise, prevented the increase in muscle protein synthesis 16 h following exercise (Kubica et al. 2005). Although we measured post-exercise protein synthesis at much earlier time-points (1–2 h post-exercise), the findings in the current study are similar to the previous work done by Kubica and colleagues in rodents.
To determine whether rapamycin altered cell signalling associated with the regulation of muscle protein synthesis, we evaluated both the mTORC1 and ERK1/2 signalling pathways: Each pathway participates in translation initiation and elongation following resistance exercise in skeletal muscle of rodents and humans (Bolster et al. 2003; Dreyer et al. 2006; Williamson et al. 2006b). Our data indicate that Akt (Ser473), a positive upstream regulator of mTORC1, was increased (∼2-fold) equally in both treatment groups during post-exercise recovery. This is in line with previous work showing that Akt can be independently increased by muscle contraction (Sakamoto et al. 2002) and is consistent with the increase in blood insulin concentrations detected immediately following exercise (see Table 1). Incidentally, we also found that blood cortisol concentrations post-exercise were similar between groups and cannot explain why muscle protein synthesis was blocked in the rapamycin group. However, since rapamycin is a specific inhibitor of mTORC1 we thoroughly examined several downstream components of the mTORC1 signalling pathway.
First, we found that the phosphorylation of mTOR (Ser2448; Fig. 4A), S6K1 (Thr389; Fig. 4C) and rpS6 (Ser240/244 and 235/236; Fig. 5C and D) was not completely blocked with rapamycin administration prior to exercise, but instead delayed as compared to the control group. A possible reason for this finding could be that, for safety reasons, the rapamycin dose we used was lower than that utilized in animal studies. For instance, our subjects ingested approximately 0.15 mg (kg body weight)−1 as compared to a typical dose of 0.75 mg (kg body weight)−1 in rodents. This is consistent with a previous study in which a lower dose of rapamycin (0.08 mg (kg body weight)−1) did not completely block insulin-induced mTORC1 signalling in humans, resulting only in a partial inhibition of S6K1 and insulin receptor substrate 1 serine phosphorylation, which increased glucose uptake (Krebs et al. 2007). Another protein that has been found to be rapamycin sensitive is eIF4G (Vary et al. 2007). However, in the current study we show that eIF4G phosphorylation increases to a similar extent in both groups during post-exercise recovery (Fig. 6D). This may indicate that the dosage of rapamycin used in the current study may have been insufficient for complete inhibition of the mTORC1 pathway. However, it is important to underscore that in our study the amount of raptor protein associated with mTORC1 tended to be lower at 2 h post-exercise in the rapamycin group (Fig. 4B), which implies that mTORC1 signalling in these subjects was at least partially inhibited. Interestingly, rapamycin reduced the binding of raptor to mTOR after rapamycin treatment in HEK293 cells (Oshiro et al. 2004) while, in another study, the raptor–mTOR association was increased following exercise in mice (Williamson et al. 2006b). Taken together, these findings show that although post-exercise protein synthesis was inhibited by rapamycin administration in humans, this was only partially supported by differences in raptor–mTORC1 association, as mTORC1 signalling was not completely blocked possibly due to the rapamycin dose used.
It is also important to highlight that although a majority of mTORC1 signalling proteins were not completely blocked following rapamycin treatment, eEF2 phosphorylation (Thr56; Fig. 5B) was reduced only in the control group following resistance exercise, a major component of peptide elongation. The decrease in eEF2 phosphorylation, and subsequent increase in muscle protein synthesis, has been consistently demonstrated in our laboratory following acute resistance exercise and essential amino acid ingestion in humans (Dreyer et al. 2006, 2008; Fujita et al. 2007; Drummond et al. 2008). A decrease in eEF2 phosphorylation, by relieved inhibition of eEF2 kinase, activates eEF2 to assist in the relocation of the ribosome to the mRNA transcript (Browne & Proud, 2002). It has been previously shown that eEF2 is rapamycin sensitive via S6K1 but rapamycin insensitive via p90 ribosomal protein S6 kinase polypeptide 1 (RSK1) (Wang et al. 2001; Wang & Proud, 2002). In our study, we found that eEF2 and S6K1 (Thr421/Ser424; Fig. 4D) phosphorylation was unchanged in the rapamycin group while the phosphorylation of both of these molecules was significantly altered at 2 h post-exercise in the control group. An extensive review of S6K1 phosphorylation events has suggested that full activation of S6K1 requires phosphorylation of multiple sites (Pullen & Thomas, 1997). Therefore, although the phosphorylation of several mTORC1 pathway proteins were delayed post-exercise in the rapamycin-treated subjects, the reduced activation of S6K1 probably allowed eEF2 kinase to remain active which resulted in an unaltered eEF2 phosphorylation. We propose that partial inhibition of S6K1 and hence a reduced rate of translation elongation is a plausible mechanism for the lack of increase in muscle protein synthesis in the rapamycin group.
To further address the role of translation elongation, we evaluated the ERK signalling pathway since ERK1/2 can activate eEF2 activity, independent of mTOR, through RSK1 (Wang et al. 2001; Wang & Proud, 2002). ERK1/2 and its downstream substrates are rapidly phosphorylated and activated following anabolic stimuli (Williamson et al. 2003, 2006b; Drummond et al. 2008) and this is an important regulator of muscle protein synthesis (Fluckey et al. 2004, 2006). An intriguing finding of our study was that ERK1/2 phosphorylation was nearly abolished and MNK1 phosphorylation was blunted in the rapamycin group while being dramatically increased in the control group (Fig. 6A and B). We were puzzled as to how rapamycin treatment is capable of blocking the ERK1/2 signalling pathway since rapamycin does not affect ERK1/2 or MNK1 directly (Bain et al. 2007). In order to determine whether the MAPK pathway is completely inhibited in the rapamycin group, we measured eIF4E (Ser209) phosphorylation, a downstream target of MNK1. We found that eIF4E (Ser209) phosphorylation was significantly increased to a similar extent following exercise in both groups (Fig. 6C) which indicates that reduced ERK/MNK signalling is not directly due to rapamycin and is probably a result of a yet to be identified indirect mechanism. One possibility may be that rapamycin could have indirect effects on other signalling pathways via altered Ca2+ release (Su et al. 2003; MacMillan et al. 2005; Schoffstall et al. 2005).
We previously mentioned that partial inhibition of S6K1 phosphorylation and activity may contribute to the blocked muscle protein synthesis following rapamycin treatment by inhibiting translation elongation. However, since rpS6 phosphorylation was blunted in the rapamycin group at 1 h post-exercise we cannot rule out partial inhibition of translation initiation as well (Fig. 5C and D). It should be noted that ERK1/2 signalling (via RSK1) can also phosphorylate rpS6 Ser235/236 (Pende et al. 2004; Roux et al. 2007), leading to enhanced translation initiation. Although our data strongly support the hypothesis that a reduced signalling through mTORC1 appears to be important in blocking the contraction-induced increase in muscle protein synthesis, our data also support the notion that reduced ERK1/2 signalling may also be regulating both translation initiation and elongation following rapamycin administration. Therefore, we hypothesize that dual activation of the mTORC1 and ERK1/2 signalling pathways is critical for full stimulation of human skeletal muscle protein synthesis following an acute bout of resistance exercise.
The delay in phosphorylation for several key proteins, in the subjects receiving rapamycin, may suggest that other regulators of translation are involved in controlling the rate of muscle protein synthesis following resistance exercise. One example is the activation of eIF2B, particularly the phosphorylation of the ɛ subunit (human Ser539; rat Ser535). Since eIF2B activation is dependent upon ERK signalling (Kleijn & Proud, 2000), is rapamycin sensitive (Kubica et al. 2004), and is dephosphorylated following resistance exercise in humans (Glover et al. 2008) and rodents (Kubica et al. 2004), we also examined whether eIF2Bɛ expression and phosphorylation were altered in the rapamycin group. We found that eIF2Bɛ expression and phosphorylation (Ser539) was unchanged in both of our treatment groups following resistance exercise but this may have been due to the shorter time course of the current study since eIF2Bɛ appears to change at <6 h post-exercise (Kubica et al. 2004; Glover et al. 2008). In any event future work should examine multiple signalling pathways in order to better explain the effect of rapamycin on human muscle protein synthesis. In addition, extending the time course of future experiments will be valuable since muscle protein synthesis rates remain elevated for 24–48 h following an acute bout of resistance exercise.
In summary, the contraction-induced increase in muscle protein synthesis is blocked while mTORC1 and ERK1/2 signalling are inhibited or delayed in healthy human subjects following rapamycin treatment. We conclude that the stimulation of the skeletal muscle protein synthesis during the early post-exercise (1–2 h) recovery period is rapamycin dependent, implying that mTORC1 plays a key role (at least in part) in post-exercise muscle protein anabolism. We further propose that a full activation of muscle protein synthesis probably requires input from other signalling pathways, such as ERK1/2, which appear at least indirectly affected by rapamycin.
Acknowledgments
We would like to thank the nurses and staff at the GCRC for their assistance in screening, admitting, and assisting with the subjects during data collection and Ming Zheng and Shelley Medina for technical assistance. Furthermore, we would also like to thank Dr Scot Kimball for his technical advice with the immunoprecipitation methodology. This study was supported by grants from the National Institute of Arthritis, Musculoskeletal and Skin Diseases AR049877 and the National Institute on Aging P30 AG024832. Additional support came from grant no. M01 RR00073 from the General Clinical Research Branch, National Center for Research Resources.
References
- Anthony JC, Yoshizawa F, Anthony TG, Vary TC, Jefferson LS, Kimball SR. Leucine stimulates translation initiation in skeletal muscle of postabsorptive rats via a rapamycin-sensitive pathway. J Nutr. 2000;130:2413–2419. doi: 10.1093/jn/130.10.2413. [DOI] [PubMed] [Google Scholar]
- Bain J, Plater L, Elliott M, Shpiro N, Hastie CJ, McLauchlan H, Klevernic I, Arthur JS, Alessi DR, Cohen P. The selectivity of protein kinase inhibitors: a further update. Biochem J. 2007;408:297–315. doi: 10.1042/BJ20070797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Bolster DR, Kubica N, Crozier SJ, Williamson DL, Farrell PA, Kimball SR, Jefferson LS. Immediate response of mammalian target of rapamycin (mTOR)-mediated signalling following acute resistance exercise in rat skeletal muscle. J Physiol. 2003;553:213–220. doi: 10.1113/jphysiol.2003.047019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne GJ, Proud CG. Regulation of peptide-chain elongation in mammalian cells. Eur J Biochem. 2002;269:5360–5368. doi: 10.1046/j.1432-1033.2002.03290.x. [DOI] [PubMed] [Google Scholar]
- Calder AG, Anderson SE, Grant I, McNurlan MA, Garlick PJ. The determination of low d5-phenylalanine enrichment (0.002–0.09 atom percent excess), after conversion to phenylethylamine, in relation to protein turnover studies by gas chromatography/electron ionization mass spectrometry. Rapid Commun Mass Spectrom. 1992;6:421–424. doi: 10.1002/rcm.1290060704. [DOI] [PubMed] [Google Scholar]
- Chung J, Kuo CJ, Crabtree GR, Blenis J. Rapamycin-FKBP specifically blocks growth-dependent activation of and signaling by the 70 kd S6 protein kinases. Cell. 1992;69:1227–1236. doi: 10.1016/0092-8674(92)90643-q. [DOI] [PubMed] [Google Scholar]
- Cuthbertson DJ, Babraj J, Smith K, Wilkes E, Fedele MJ, Esser K, Rennie M. Anabolic signaling and protein synthesis in human skeletal muscle after dynamic shortening or lengthening exercise. Am J Physiol Endocrinol Metab. 2006;290:E731–738. doi: 10.1152/ajpendo.00415.2005. [DOI] [PubMed] [Google Scholar]
- Dreyer HC, Drummond MJ, Pennings B, Fujita S, Glynn EL, Chinkes DL, Dhanani S, Volpi E, Rasmussen BB. Leucine-enriched essential amino acid and carbohydrate ingestion following resistance exercise enhances mTOR signaling and protein synthesis in human muscle. Am J Physiol Endocrinol Metab. 2008;294:E392–E400. doi: 10.1152/ajpendo.00582.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyer HC, Fujita S, Cadenas JG, Chinkes DL, Volpi E, Rasmussen BB. Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J Physiol. 2006;576:613–624. doi: 10.1113/jphysiol.2006.113175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MJ, Dreyer HC, Pennings B, Fry CS, Dhanani S, Dillon EL, Sheffield-Moore M, Volpi E, Rasmussen BB. Skeletal muscle protein anabolic response to resistance exercise and essential amino acids is delayed with aging. J Appl Physiol. 2008;104:1452–1461. doi: 10.1152/japplphysiol.00021.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckey JD, Dupont-Versteegden EE, Knox M, Gaddy D, Tesch PA, Peterson CA. Insulin facilitation of muscle protein synthesis following resistance exercise in hindlimb-suspended rats is independent of a rapamycin- sensitive pathway. Am J Physiol Endocrinol Metab. 2004;287:E1070–E1075. doi: 10.1152/ajpendo.00329.2004. [DOI] [PubMed] [Google Scholar]
- Fluckey JD, Knox M, Smith L, Dupont-Versteegden EE, Gaddy D, Tesch PA, Peterson CA. Insulin-facilitated increase of muscle protein synthesis after resistance exercise involves a MAP kinase pathway. Am J Physiol Endocrinol Metab. 2006;290:E1205–E1211. doi: 10.1152/ajpendo.00593.2005. [DOI] [PubMed] [Google Scholar]
- Fujita S, Dreyer HC, Drummond MJ, Glynn EL, Cadenas JG, Yoshizawa F, Volpi E, Rasmussen BB. Nutrient signalling in the regulation of human muscle protein synthesis. J Physiol. 2007;582:813–823. doi: 10.1113/jphysiol.2007.134593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita S, Dreyer HC, Drummond MJ, Glynn EL, Volpi E, Rasmussen BB. Essential amino acid and carbohydrate ingestion prior to resistance exercise does not enhance post-exercise muscle protein synthesis. J Appl Physiol. 2008 doi: 10.1152/japplphysiol.90395.2008. June 5 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover EI, Oates BR, Tang JE, Moore DR, Tarnopolsky MA, Phillips SM. Resistance exercise decreases eIF2Bɛ phosphorylation and potentiates the feeding-induced stimulation of p70S6K1 and rpS6 in young men. Am J Physiol Regul Integr Comp Physiol. 2008;295:R604–R610. doi: 10.1152/ajpregu.00097.2008. [DOI] [PubMed] [Google Scholar]
- Karlsson HK, Nilsson PA, Nilsson J, Chibalin AV, Zierath JR, Blomstrand E. Branched-chain amino acids increase p70S6k phosphorylation in human skeletal muscle after resistance exercise. Am J Physiol Endocrinol Metab. 2004;287:E1–E7. doi: 10.1152/ajpendo.00430.2003. [DOI] [PubMed] [Google Scholar]
- Kimball SR, Horetsky RL, Jefferson LS. Signal transduction pathways involved in the regulation of protein synthesis by insulin in L6 myoblasts. Am J Physiol Cell Physiol. 1998;274:C221–C228. doi: 10.1152/ajpcell.1998.274.1.C221. [DOI] [PubMed] [Google Scholar]
- Kleijn M, Proud CG. The activation of eukaryotic initiation factor (eIF)2B by growth factors in PC12 cells requires MEK/ERK signalling. FEBS Lett. 2000;476:262–265. doi: 10.1016/s0014-5793(00)01743-9. [DOI] [PubMed] [Google Scholar]
- Krebs M, Brunmair B, Brehm A, Artwohl M, Szendroedi J, Nowotny P, Roth E, Furnsinn C, Promintzer M, Anderwald C, Bischof M, Roden M. The Mammalian target of rapamycin pathway regulates nutrient-sensitive glucose uptake in man. Diabetes. 2007;56:1600–1607. doi: 10.2337/db06-1016. [DOI] [PubMed] [Google Scholar]
- Kubica N, Bolster DR, Farrell PA, Kimball SR, Jefferson LS. Resistance exercise increases muscle protein synthesis and translation of eukaryotic initiation factor 2Bɛ mRNA in a mammalian target of rapamycin-dependent manner. J Biol Chem. 2005;280:7570–7580. doi: 10.1074/jbc.M413732200. [DOI] [PubMed] [Google Scholar]
- Kubica N, Kimball SR, Jefferson LS, Farrell PA. Alterations in the expression of mRNAs and proteins that code for species relevant to eIF2B activity after an acute bout of resistance exercise. J Appl Physiol. 2004;96:679–687. doi: 10.1152/japplphysiol.00962.2003. [DOI] [PubMed] [Google Scholar]
- MacMillan D, Currie S, Bradley KN, Muir TC, McCarron JG. In smooth muscle, FK506-binding protein modulates IP3 receptor-evoked Ca2+ release by mTOR and calcineurin. J Cell Sci. 2005;118:5443–5451. doi: 10.1242/jcs.02657. [DOI] [PubMed] [Google Scholar]
- Oshiro N, Yoshino K, Hidayat S, Tokunaga C, Hara K, Eguchi S, Avruch J, Yonezawa K. Dissociation of raptor from mTOR is a mechanism of rapamycin-induced inhibition of mTOR function. Genes Cells. 2004;9:359–366. doi: 10.1111/j.1356-9597.2004.00727.x. [DOI] [PubMed] [Google Scholar]
- Pende M, Um SH, Mieulet V, Sticker M, Goss VL, Mestan J, Mueller M, Fumagalli S, Kozma SC, Thomas G. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5'-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol Cell Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proud CG. Cell signaling. mTOR, unleashed. Science. 2007;318:926–927. doi: 10.1126/science.1150653. [DOI] [PubMed] [Google Scholar]
- Pullen N, Thomas G. The modular phosphorylation and activation of p70s6k. FEBS Lett. 1997;410:78–82. doi: 10.1016/s0014-5793(97)00323-2. [DOI] [PubMed] [Google Scholar]
- Roux PP, Shahbazian D, Vu H, Holz MK, Cohen MS, Taunton J, Sonenberg N, Blenis J. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282:14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvinsky I, Meyuhas O. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci. 2006;31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev. 2005;19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Hirshman MF, Aschenbach WG, Goodyear LJ. Contraction regulation of Akt in rat skeletal muscle. J Biol Chem. 2002;277:11910–11917. doi: 10.1074/jbc.M112410200. [DOI] [PubMed] [Google Scholar]
- Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159–168. doi: 10.1016/j.molcel.2006.03.029. [DOI] [PubMed] [Google Scholar]
- Schoffstall B, Kataoka A, Clark A, Chase PB. Effects of rapamycin on cardiac and skeletal muscle contraction and crossbridge cycling. J Pharmacol Exp Ther. 2005;312:12–18. doi: 10.1124/jpet.104.073445. [DOI] [PubMed] [Google Scholar]
- Su Z, Sugishita K, Li F, Ritter M, Barry WH. Effects of FK506 on [Ca2+]i differ in mouse and rabbit ventricular myocytes. J Pharmacol Exp Ther. 2003;304:334–341. doi: 10.1124/jpet.102.041210. [DOI] [PubMed] [Google Scholar]
- Vary TC, Anthony JC, Jefferson LS, Kimball SR, Lynch CJ. Rapamycin blunts nutrient stimulation of eIF4G, but not PKCɛ phosphorylation, in skeletal muscle. Am J Physiol Endocrinol Metab. 2007;293:E188–E196. doi: 10.1152/ajpendo.00037.2007. [DOI] [PubMed] [Google Scholar]
- von Manteuffel SR, Gingras AC, Ming XF, Sonenberg N, Thomas G. 4E-BP1 phosphorylation is mediated by the FRAP-p70s6k pathway and is independent of mitogen- activated protein kinase. Proc Natl Acad Sci U S A. 1996;93:4076–4080. doi: 10.1073/pnas.93.9.4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Proud CG. Regulation of the phosphorylation of elongation factor 2 by MEK-dependent signalling in adult rat cardiomyocytes. FEBS Lett. 2002;531:285–289. doi: 10.1016/s0014-5793(02)03536-6. [DOI] [PubMed] [Google Scholar]
- Wang X, Li W, Williams M, Terada N, Alessi DR, Proud CG. Regulation of elongation factor 2 kinase by p90(RSK1) and p70 S6 kinase. Embo J. 2001;20:4370–4379. doi: 10.1093/emboj/20.16.4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Proud CG. The mTOR pathway in the control of protein synthesis. Physiology (Bethesda) 2006;21:362–369. doi: 10.1152/physiol.00024.2006. [DOI] [PubMed] [Google Scholar]
- Widrick JJ, Stelzer JE, Shoepe TC, Garner DP. Functional properties of human muscle fibers after short-term resistance exercise training. Am J Physiol Regul Integr Comp Physiol. 2002;283:R408–R416. doi: 10.1152/ajpregu.00120.2002. [DOI] [PubMed] [Google Scholar]
- Williamson D, Gallagher P, Harber M, Hollon C, Trappe S. Mitogen-activated protein kinase (MAPK) pathway activation: effects of age and acute exercise on human skeletal muscle. J Physiol. 2003;547:977–987. doi: 10.1113/jphysiol.2002.036673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson DL, Bolster DR, Kimball SR, Jefferson LS. Time course changes in signaling pathways and protein synthesis in C2C12 myotubes following AMPK activation by AICAR. Am J Physiol Endocrinol Metab. 2006a;291:E80–E89. doi: 10.1152/ajpendo.00566.2005. [DOI] [PubMed] [Google Scholar]
- Williamson DL, Kubica N, Kimball SR, Jefferson LS. Exercise-induced alterations in extracellular signal-regulated kinase 1/2 and mammalian target of rapamycin (mTOR) signalling to regulatory mechanisms of mRNA translation in mouse muscle. J Physiol. 2006b;573:497–510. doi: 10.1113/jphysiol.2005.103481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe RR, Chinkes DL. Isotope Tracers in Metabolic Research. Hobokon, NJ: Wiley-Liss; 2005. Calculation of concentration by the internal standard technique; p. 127. [Google Scholar]