Abstract
Skeletal muscle tissue undergoes adaptive changes in response to stress and the genes that control these processes are incompletely characterised. NDRG2 (N-myc downstream-regulated gene 2), a stress- and growth-related gene, was investigated in skeletal muscle growth and adaption. While NDRG2 expression levels were found to be up-regulated in both differentiated human and mouse myotubes compared with undifferentiated myoblasts, the suppression of NDRG2 in C2C12 myoblasts resulted in slowed myoblast proliferation. The increased expression levels of the cell cycle inhibitors, p21 Waf1/Cip1 and p27 Kip1, and of various muscle differentiation markers in NDRG2-deficient myoblasts indicate that a lack of NDRG2 promoted cell cycle exiting and the onset of myogenesis. Furthermore, the analysis of NDRG2 regulation in C2C12 myotubes treated with catabolic and anabolic agents and in skeletal muscle from human subjects following resistance exercise training revealed NDRG2 gene expression to be down-regulated during hypertrophic conditions, and conversely, up-regulated during muscle atrophy. Together, these data demonstrate that NDRG2 expression is highly responsive to different stress conditions in skeletal muscle and suggest that the level of NDRG2 expression may be critical to myoblast growth and differentiation.
Skeletal muscle development and mass are influenced by both hypertrophy- and atrophy-causing agents (Glass, 2005). Growth factor expression needs to be regulated for effective myoblast proliferation and differentiation, and in vitro, reductions in growth factor concentrations induce myoblasts to withdraw from the cell cycle, to commence differentiation and enter the post-mitotic state prior to the formation of multinucleated myotubes (Spizz et al. 1986; Frith-Terhune et al. 1998). Key regulators of these processes include both the muscle-specific basic helix–loop–helix group (bHLH) of transcription factors (Olson & Klein, 1994) and two families of the cyclin-dependent kinase (CDK) inhibitors, p21 Waf1/Cip1 and p16 INK4a (Maddika et al. 2007). The former CDK inhibitor family consists of p21 Waf1/Cip1, p27 Kip1 and p57 Kip2, which inhibit all CDKs regulating G1 gap phase to the DNA synthesis (S) phase of cell cycle progression, while the expression of the bHLH protein, myogenin, is induced upon myoblast differentiation and directly controls myotube formation (Olson & Klein, 1994). Both the bHLH proteins and the CDK inhibitors appear to modulate each other's function to control cell cycle termination and muscle differentiation. While skeletal muscle demonstrates plasticity to different stress conditions such as the physiological stress of resistance exercise and the pathological stress of cancer and sepsis, the consequences of physiological stress are adaption and growth. However, with pathological stress, key molecular targets become dysfunctional and the muscle becomes susceptible to the development of myopathies and dystrophies resulting in atrophy, myoblast apoptosis and reduced muscle function. Hence, the elucidation of novel genes that control the response of skeletal muscle to these stressors is essential in understanding the regulation of cellular proliferation and differentiation in the maintenance of muscle homeostasis.
The N-myc downstream-regulated gene (NDRG) family has been linked to stress responses and to cell proliferation and differentiation. There are four family members and expression analysis studies reveal each gene member to display distinct tissue localisation with NDRG1 being the most ubiquitously expressed (Qu et al. 2002). In contrast, NDRG2 is predominantly expressed in the brain, liver, heart and skeletal muscle in multiple species including human (Qu et al. 2002), rat (Boulkroun et al. 2002) and mouse (Murray et al. 2004). NDRG2 has been proposed to act as a tumour suppressor gene as decreased expression is evident in numerous cancer cell lines and tissues (Deng et al. 2003; Hu et al. 2004; Lusis et al. 2005; Liu et al. 2007; Lorentzen et al. 2007). NDRG2 overexpression studies result in reduced glioblastoma and breast cancer cell proliferation (Deng et al. 2003; Park et al. 2007) indicating a role for NDRG2 in cell proliferation control; however, the molecular mechanisms mediating this effect are unknown. In addition, NDRG2 is found to be up-regulated following the differentiation of dendritic cells (Choi et al. 2003) and PC12 neuronal cells (Takahashi et al. 2005), and is induced following hypoxia-induced stress (Wang et al. 2008).
In skeletal muscle, NDRG2 is a candidate substrate for key signalling serine–threonine kinases including Akt/PKB, p70 S6 kinase, p90 ribosomal S6 kinase, and SGK1 (serum- and glucocorticoid-induced kinase 1) (Burchfield et al. 2004; Murray et al. 2004). While the functional consequences of the phosphorylation of NDRG2 by these kinases are currently unknown, many of these kinases including Akt regulate skeletal muscle cell cycle progression, and hypertrophy and atrophy signalling (reviewed in Liang & Slingerland, 2003; Glass, 2005; Frost & Lang, 2007). Therefore, we hypothesise that NDRG2 plays a role in mediating the effects of these kinases in skeletal muscle signalling and thus may represent a new target for myopathies and dystrophies. Here, we sought to investigate the role of NDRG2 in skeletal muscle function. The aims of this study were to characterise NDRG2 expression during myoblast differentiation and to investigate the effect of reduced NDRG2 levels on myoblast proliferation and differentiation. The response of NDRG2 in C2C12 myotubes treated with anabolic and catabolic agents and in skeletal muscle from resistance exercise-trained individuals was also analysed. Our results identify for the first time that NDRG2 is a novel regulator of myoblast function and may play a role in skeletal muscle homeostasis.
Methods
Ethical approval
All human experimental procedures were approved by Deakin University and Barwon Health Human Research Ethics Committees and informed written consent was obtained from each participant prior to obtaining samples. This study conforms to the standards outlined by the Declaration of Helsinki.
Resistance exercise training
The description of the participants, details of the exercise training regime and skeletal muscle biopsies taken in this study are described previously (Carey et al. 2007). Briefly, 16 young (18–25 years old) and 15 older (60–75 years old) men (see Table 1 for participant characteristics) underwent a single bout of resistance exercise consisting of three sets of 12 repetitions of a maximal single-leg knee extension exercise on the Cybex NORM dynamometer (Cybex International Inc., Measham, UK) with a 2 min rest between sets. Biopsy samples from the vastus lateralis muscle in the fasted state were obtained both at rest and 2 h post-exercise. Following the acute training, all participants underwent a supervised 12 week progressive resistance exercise training program 3 days a week with a minimum of 48 h rest between each session. Training sessions involved two sets of exercise bouts consisting of 8 and 12 repetitions of leg press, bench press, seated row, leg extension, dumbbell shoulder press, and sit-up initially set at 50% of each individual's assessed repetition maximum (RM) strength with a specific rest period between sets. A progressive increase in the weights lifted each week was instigated until each individual reached 80% of their RM by week 6. The resistance exercise training was maintained at 80% RM thereafter for the remaining 6 weeks. At the end of the 12 week program, biopsy samples were taken from the vastus lateralis muscle of participants in the fasted state both at rest and 2 h following a single bout of resistance exercise as described above. All tissue was rapidly frozen in liquid nitrogen and stored at −80°C prior to RNA extraction with RNA-Bee reagent (Tel-Test, Friendswood, TX, USA).
Table 1.
Participant characteristics
| Characteristics | Younger men | Older men |
|---|---|---|
| Age (years) | 20 ± 0.5 | 68 ± 1* |
| Height (cm) | 181 ± 2 | 172 ± 1* |
| Weight (kg) | 72 ± 2 | 82 ± 2* |
| BMI | 22 ± 0.6 | 27 ± 0.7* |
| Concentric isokinetic peak torque (N m) | ||
| Pre-training | 207 ± 10 | 143 ± 12* |
| Post-training | 243 ± 9^ | 184 ± 11*^ |
| Eccentric isokinetic peak torque (N m) | ||
| Pre-training | 227 ± 11 | 179 ± 12* |
| Post-training | 286 ± 14^ | 215 ± 13*^ |
P < 0.01 compared with younger men;
P < 0.01 compared with pre-training values. BMI = body mass index. Data represented as mean ±s.e.m.
Cell culture
For the human primary cell cultures, cells were obtained from the vastus lateralis muscle of three men and three women (average age 69.1 ± 3 years; weight 79 ± 6 kg; height 168 ± 2 cm), who were screened for co-morbidities that compromise skeletal muscle health including type 2 diabetes, neuromuscular disorders, cardiovascular and renal disease. The harvesting and culturing of skeletal muscle satellite cells was performed as outlined previously with minor modifications (Gaster et al. 2001; Berggren et al. 2005). Briefly, biopsy muscle tissue was minced in ice-cold phosphate-buffered solution (PBS) prior to repeat digestions in 0.1% collagenase and 0.05% dispase (Gibco) for 20 min with agitation at 37°C. Released cells were grown in 5.5 mm glucose DMEM containing Glutamax, 2% fetal bovine serum (FBS), 0.5 mg ml−1 bovine serum albumin (BSA), 0.5 mg ml−1 fetuin (Bovogen, Melbourne, Australia), 20 ng ml−1 human epidermal growth factor (PeproTech, NJ, USA), 0.39 μg ml−1 dexamethasone (Sigma Aldridge, Melbourne, Australia), 0.6% penicillin–streptomycin and 0.6% amphostat B (ThermoTRACE, Melbourne, Australia). To remove contaminating fibroblasts, cells were pre-plated for 30 min in uncoated flasks prior to transferring the cell suspension to extra-cellular matix-coated flasks (Sigma Chemical Co., St Louis, MO, USA) and cultured in 5% O2 and 5% CO2 at 37°C. Myoblasts were expanded for three passages before using for experimentation. To differentiate, myoblasts were grown to 90% confluence prior to the addition of differentiation medium (5.5 mm glucose DMEM containing Glutamax, 2% horse serum, 0.5 mg ml−1 BSA, 0.5 mg ml−1 fetuin, 0.6% penicillin–streptomycin and 0.6% amphostat B). Cells were differentiated over 4 days and the medium was changed every 48 h.
Mouse C2C12 myoblasts (ATCC, Manassas, VA, USA) were cultured in 5% CO2 at 37°C in normal growth medium consisting of 25 mm glucose DMEM supplemented with 10% FBS. When visual evidence of spontaneous cell fusion was apparent, 2% horse serum and 25 mm glucose DMEM (differentiation medium) were added to enhance the differentiation process. Cells were differentiated over 6 days and medium was changed every 48 h. Anabolic and catabolic time-course treatments of myotubes were performed in C2C12 differentiation medium with 20 ng ml−1 of murine insulin-like growth factor (IGF)-1 (PeproTech), 100 nm insulin (Humulin, Eli Lilly, Indianapolis, IN, USA), 100 nm testosterone (Sigma), 40 ng ml−1 of murine tumour necrosis factor alpha (TNFα; PeproTech) or 10 μm of dexamethasone (Sigma). The medium for the 48 h treatment time-point was replenished after 24 h and all time-points were harvested concurrently.
Platinum E (Plat E) cells were a kind gift from D. James, Garvan Institute of Medical Research, Sydney, Australia. Plat E cells were cultured in 25 mm glucose DMEM with 10% heat-inactivated FBS and 1 μg ml−1 puromycin (Sigma) and 10 μg ml−1 blastocidin at 37°C in 5% CO2. All media components were obtained from Invitrogen (Melbourne, Australia) unless otherwise stated.
shRNA retroviral constructs and infection
The pSIREN RetroQ system kit (Clontech, Mountainview, CA, USA) was used to express the short hairpin RNAs (shRNAs). The shRNA used as a negative control for shRNA activity (shNegative) was provided with the kit. The shRNA targeting sequences used for NDRG2 were designed using an in-house algorithm. The sequences for shNDRG2a and shNDRG2b were 5′-TTGTTCTCATCAACATTGA-3′ and 5′-ATATGCAAGAGATCATACA-3′, respectively. Following cloning of each double-stranded hairpin oligonucleotide into pSIREN RetroQ vector, plasmid DNAs were transiently transfected into Plat E packaging cells (Morita et al. 2000) using Lipofectamine 2000 (Invitrogen) in the presence of OptiMem (Invitrogen) at a 1: 3.3 DNA: transfection reagent ratio as recommended by the manufacturer. Media containing each virus were harvested from the packaging cells 48 h post-transfection, filtered using a 0.45 μm filter (Millipore, North Ryde, NSW, Australia) and diluted 1: 1 with C2C12 growth medium before adding to 30% confluent C2C12 myoblasts. Polybrene (6 μg ml−1; Invitrogen) was added to the virus–medium mix prior to centrifugation for 45 min at 500 g. Infection continued for a further 3 h at 37°C, 20% O2 and 5% CO2, before removing the virus–medium mix and replacing with fresh C2C12 growth medium. The following day, 2.5 μg ml−1 of puromycin was added and cells were selected for 48 h prior to trypsinisation, counting and re-seeding at equal numbers for each experiment. Cells were harvested at the subconfluent myoblast, confluent myoblast or myotube stage. Imaging of the cells was performed using an IX81 inverted microscope (Olympus, Melbourne, Australia) and ImagePro software (Media Cybernetics, UK).
RNA extraction, quantitation and reverse transcription
RNA was extracted and purified using RNeasy mini kits (Qiagen, Clifton Hill, Victoria, Australia) and quantitated using the Agilent Bioanalyser and RNA 6000 Nano Assay kit (Agilent Technologies, USA). To prepare cDNA, reverse transcription was performed according to the manufacturer's instructions using the SuperScript III First-Strand Synthesis System (Invitrogen) in a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems, Foster City, CA, USA).
Quantitative PCR
Gene expression was analysed by quantitative PCR (qPCR) using Brilliant SYBR master mix (Stratagene, La Jolla, CA, USA) on the MX3005P QPCR system (Stratagene). The PCR conditions were: 95°C for 10 min (1 cycle); 95°C for 30 s and 60°C for 1 min (40 cycles). Relative gene expression was calculated at 2−ΔCt and normalised to the house-keeping gene, acidic ribosomal phosphoprotein, mRNA levels (Akamine et al. 2007) except for gene expression analyses of the human subjects and primary cell cultures where gene expression levels were normalised to cyclophilin. Primers were designed using Primer Express 2.0 software (Applied Biosystems) and were synthesised by Geneworks (Adelaide, Australia). See Table 2 for the list of primer sequences.
Table 2.
Primer sequences for qPCR analyses
| Gene | Accession no. | Forward primer (5′–3′) | Reverse primer (5′–3′) |
|---|---|---|---|
| Mouse NDRG2 | NM_013864 | gagttagctgcccgcatcc | gtgaccgagccataaggtgtc |
| Human NDRG2 | NM_201535 | tgccaagggttggatgga | tccggaatggaagaggtgag |
| Mouse myogenin | NM_031189 | tcggtcccaacccagga | gcagattgtgggcgtctg |
| Human myogenin | NM_002479 | ggtgcccagcgaatgc | tgatgctgtccacgatcga |
| Troponin I, fast twitch | NM_009405 | gaacactgcccgccactg | gctccttgctgctcttctgc |
| Myosin heavy chain | NM_010855 | tctccatctctgataacgcctacc | tgcctcctccttcttcttgtcc |
| Mouse MuRF1 | NM_001039048 | ccagtcggcccctgc | tgattttctcgtcttcgtgttcc |
| Human MuRF1 | NM_032588.2 | cctgagagccattgactttgg | cttcccttctgtggactcttcct |
| Mouse atrogin-1 | NM_026346 | agacctgcatgtgctcagtga | gcgaatctgcctctctgagaa |
| Human atrogin-1 | NM_058229.2 | gcagctgaacaacattcagatcac | cagcctctgcatgatgttcagt |
| p21 Waf/Cip1 | NM_007669 | gccttgtcgctgtcttgc | cgcttggagtgatagaaatctgtc |
| p27 Kip1 | NM_009875 | gagcagtgtccagggatgagg | tccacagtgccagcgttcg |
| cyclophilin | NM_021130.3 | catctgcactgccaagactga | ttcatgccttctttcactttgc |
| ARBP | NM_007475 | gctgcctcacatccggg | agctggcaccttattggcc |
NDRG2, N-myc downstream-regulated gene 2; MuRF1, muscle RING finger 1; ARBP; acidic ribosomal phosphoprotein.
Immunoblotting
To express NDRG2 ectopically, full-length human NDRG2 (amino acids 1–371) cDNA was ligated into the SalI and NotI sites of the pCMV-HA vector (BD Biosciences Clontech, Palo Alto, CA, USA) and transfected into C2C12 myoblasts using Lipofectamine 2000 (Invitrogen) at a 1: 4 DNA: transfection reagent ratio as recommended by the manufacturer. Cells were lysed in ice-cold buffer containing 50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% NP-40, 0.25% sodium deoxycholate, 1 mm phenylmethylsulfonyl fluoride, a cocktail of protease inhibitors, and phosphatase inhibitors (1 mm sodium pyrophosphate, 10 mm NaF, 1 mm Na3VO4) and left on ice for 20 min before centrifuging at 12 000 g for 10 min at 4°C to remove insoluble material. The protein concentration was determined using Bio-Rad protein assay reagent (Bio-Rad Laboratories, USA) and 20 μg of total protein were electrophoresed on 8 or 12.5% SDS-PAGE denaturing gels and transferred to polyvinylidene difluoride membranes by electroblotting. Immunoblotting was performed using 1 μg ml−1 of goat polyclonal anti-NDRG2 (E-20; cat. no. sc-19468), 1 μg ml−1 of mouse monoclonal anti-p21 Waf1/Cip1 (F-5; cat. no. sc-6246) or 0.5 μg ml−1 of mouse monoclonal anti-myogenin (FD5; cat. no. sc-12732) antibodies from Santa Cruz Biotechnology (Santa Cruz, CA, USA), or using 1: 1000 dilution of rabbit polyclonal anti-p27 Kip1 (cat. no. 2552) and anti-p15 INK4B (cat. no. 4822) from Cell Signaling Technology (MA, USA). Immunoblotting was performed overnight at 4°C in BLOTTO (5% skim milk and TBST (50 mm Tris-HCl, 750 mm NaCl, 0.25% Tween-20)). Following washing (3 × 5 min) in TBST, the detection of the primary antibodies was performed using a 1: 5000 dilution of rabbit anti-goat IgG, HRP-linked antibody (Sigma), or a 1: 2000 dilution of either goat anti-mouse or goat anti-rabbit IgG, HRP-linked antibodies (Cell Signaling Technology) for 1 h at room temperature in BLOTTO followed by washing in TBST (3 × 5 min). The immunoblots were developed in ECL chemiluminescence reagent (Amersham, GE Healthcare Biosciences, Rydalmere, NSW, Australia) and autoradiography was performed to visualise the immunoreactive bands. As a control for protein loading, membranes were re-probed with a 1: 10 000 dilution of a mouse monoclonal anti-β−actin antibody (AC-15; cat. no. 6276, Abcam, Taiwan). The ChemiGeniusII gel documentation system and Gene Snap software (Syngene, Cambridge UK) were used to visualise protein bands and densitometry was performed with Gene Tools software (Syngene).
Immunohistochemistry and nuclei staining
Cells were cultured in glass chamber slides (BD Biosciences) and fixed in 2% paraformaldehyde–PBS for 20 min followed by a wash in 1× PBS. For immunohistochemical analysis, cells were blocked in 10% normal goat serum–PBS for 30 min, washed in 1× PBS and incubated overnight at 4°C in anti-myosin heavy chain antibody (cat. no. 18-0074; Zymed, Invitrogen) diluted 1: 25 in 1% BSA–PBS. Cells were washed well in 1× PBS and incubated with a pre-dilute mouse–rabbit dual link secondary antibody (cat. no. K4061; Dako Australia) for 1 h. Following a PBS wash, colour development was performed using the Dako Cytomation AEC + substrate chromagen and nuclei were counterstained with haematoxylin before coverslipping in Faramount aqueous mounting medium (Dako). For the fluorescent staining of myotube nuclei, fixed cells were incubated with 5 μg ml−1 DAPI (4′,6-diamidino-2-phenylindole, dihydrochloride; Molecular Probes, Invitrogen) in PBS for 15 min and then washed prior to mounting in fluorescence mounting medium (Dako) and counting of nuclei. Imaging was performed using a Zeiss Axioskop 2 upright microscope and Axiovision software (version 3.1.2.1) (Carl Zeiss, Australia).
Cell viability and proliferation assay
Mitochondrial dehydrogenase function as a measure of cell viability and proliferation was assayed by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT; Sigma) colourmetric method (Mosmann, 1983). shRNA-infected C2C12 myoblasts were seeded at 3 × 103 cells per well in a 96-well plate and assayed at days 1, 2 and 3 post-plating. Briefly, growth medium was removed and cells were incubated in serum-free DMEM containing 0.5 mg ml−1 MTT for 30–60 min at 37°C. Following the formation of the blue MTT formazan, cells were dissolved using 0.1 m HCl in isopropanol and the absorbance of each sample was determined at 570 nm (test wavelength) and 630 nm (reference wavelength) using an absorbance microplate reader (model 550; Bio-Rad). Data are expressed relative to the viability of shNegative-infected cells.
Cell cycle analysis
shRNA-infected and uninfected myoblasts were seeded at 1 × 106 cells and harvested 2 days later following two washes in 1 × PBS. Cells were centrifuged at 1000 g for 5 min and the pellet resuspended in 100 μl PBS before fixing in 900 μl 70% cold ethanol for 1 h on ice. Cells were recovered at 2000 g, washed twice more in cold PBS before centrifugation and resuspending the pellet in 0.2 mg ml−1 RNaseA (Sigma) in PBS and leaving at room temperature for 30 min. Cells were recovered once more and the pellets resuspended in 350 μl of 50 μg ml−1 propidium iodide (Invitrogen) in PBS before analysis by flow cytometry at 535 nm on a FACSCalibur machine (Becton Dickinson, North Ryde, NSW, Australia) using CELLQuest software. Ten thousand cell events were assessed and the percentage of cells in each phase of the cell cycle, G0/G1, S and G2/M, was determined by the ModFitLT V2.0 software (Becton Dickinson).
Statistical methods
All data are reported as the mean ± standard error of the mean (s.e.m.). Data normality were determined using the Kolmogorov–Smirnov test and statistical differences were assessed using an unpaired Student's t test for two-group comparisons or paired t tests for comparisons between the same groups at different time-points. For data involving three or more groups, data were subjected to one-way ANOVA using least significant differences or Games–Howell post hoc assessment for homogenous or non-homogeneous samples, respectively. Analyses were performed using Statistical Package for the Social Sciences software (SPSS version 14.1; Fullerton, CA, USA). Data were considered statistically significant at P < 0.05.
Results
Expression profile of NDRG2 during myoblast differentiation
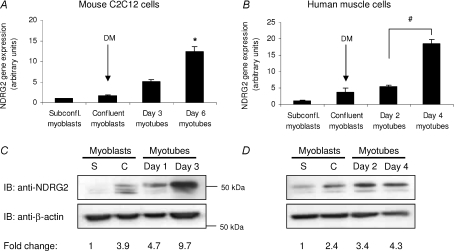
The expression profile of NDRG2 during both human and mouse muscle cell differentiation was analysed. Differentiation medium was added to both cell types when myoblasts were confluent and cells were then typically allowed to differentiate over a 4 day (human myotubes) or 6 day (C2C12 myotubes) period. Myogenin, a marker for commencement of myoblast fusion (Andres & Walsh, 1996), was analysed in differentiating C2C12 and human muscle cells and were found to increase 50- and 160-fold, respectively, indicating progressive myoblast differentiation (Fig. S1A and B). NDRG2 gene expression increased significantly in C2C12 and human myotubes compared with myoblasts following addition of differentiation media (P= 0.028 for C2C12 myotubes; Fig. 1A) (P < 0.009 for human myotubes; Fig. 1B). A nearly 10-fold increase in protein expression for NDRG2 was also measured in differentiating C2C12 myoblasts with at least three isoforms ranging from 45 to 50 kDa evident (Fig. 1C). The higher molecular weight band was more prominent in myotubes. In differentiating human muscle cells, NDRG2 protein levels also increased upon differentiation with two possible isoforms present (Fig. 1D). The different protein isoforms may represent variants that have been described previously (Boulkroun et al. 2002) or proteins that have undergone post-translational modification. Two isoforms of 45 and 50 kDa were also evident with exogenously expressed HA-tagged human NDRG2 (Fig. S1C).
Figure 1. NDRG2 expression in differentiating myoblasts.
Fold change in gene (A and B) and protein (C and D) expression of NDRG2 in a time-course representing differentiating mouse C2C12 myoblasts (A and C) and differentiating human primary muscle cell cultures (B and D). Differentiation media (DM) were added when myoblasts were confluent. *P= 0.028 and #P < 0.009 compared with subconfluent (subconfl.) myoblasts. Immunoblot (IB) analyses were performed with anti-NDRG2 and anti-β-actin antibodies. The fold change in NDRG2 protein levels is relative to NDRG2 expression in subconfluent myoblasts following normalisation to β-actin levels. Samples were analysed from both subconfluent (S) and confluent (C) myoblasts and from myotubes at various times (day 1, 2, 3 or 4) following the addition of differentiation media to confluent myoblasts.
Phenotypic effects on C2C12 myoblast differentiation following suppression of endogenous NDRG2
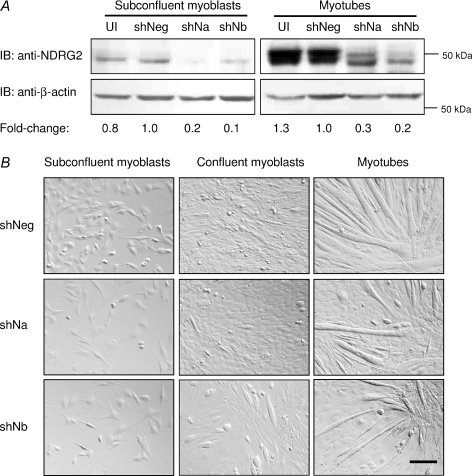
NDRG2 levels in C2C12 myoblasts were suppressed using two separate shRNA retroviral constructs targeting mouse NDRG2 mRNA (shNDRG2a and shNDRG2b). Following 48 h antibiotic selection to remove uninfected cells, the infected subconfluent myoblasts were counted and re-plated at equal numbers for each experiment. Gene expression analysis revealed that NDRG2 mRNA levels were suppressed by 80–90% in the infected C2C12 myoblasts and myotubes (data not shown). NDRG2 protein levels were decreased by 80–90% in C2C12 subconfluent myoblasts and by 70–80% in C2C12 myotubes (Fig. 2A). The shNDRG2b construct consistently gave a slightly greater level of suppression than shNDRG2a. Phenotypically, the shNDRG2-infected subconfluent myoblasts, particularly in the shNDRG2b cells, did not appear to proliferate as rapidly in comparison with the shNegative-infected cells (Fig. 2B; left panels). The control myoblasts reached confluence 4 days post-plating and commenced spontaneous myoblast fusion (Fig. 2B; top middle panel), at which time, low serum differentiation medium was added. At 4 days post-plating, spontaneous myoblast fusion was also evident in the shNDRG2-infected myoblasts despite the shNDRG2b-infected myoblasts remaining subconfluent (Fig. 2B; bottom middle panel). Following the addition of differentiation medium, the shNDRG2-infected C2C12 cells, particularly the shNDRG2b-infected samples, did not uniformly differentiate compared with the shNegative-infected myotubes (Fig. 2B; right panels).
Figure 2. Expression and phenotypic analyses of NDRG2-deficient C2C12 cells.
A, Immunoblot (IB) analyses of NDRG2 (top panel) and β-actin (bottom panel) protein levels in uninfected cells (UI) and in cells infected with the negative control (shNeg) or with NDRG2-specific shRNAs (shNa and shNb). The fold change in NDRG2 protein expression compared with shNeg control is indicated following normalisation to β-actin levels. B, brightfield microscopy of subconfluent myoblasts, confluent myoblasts and myotubes following shRNA infections. Scale bar, 50 μm.
Cell viability and cell cycle analyses of shNDRG2-infected C2C12 myoblasts
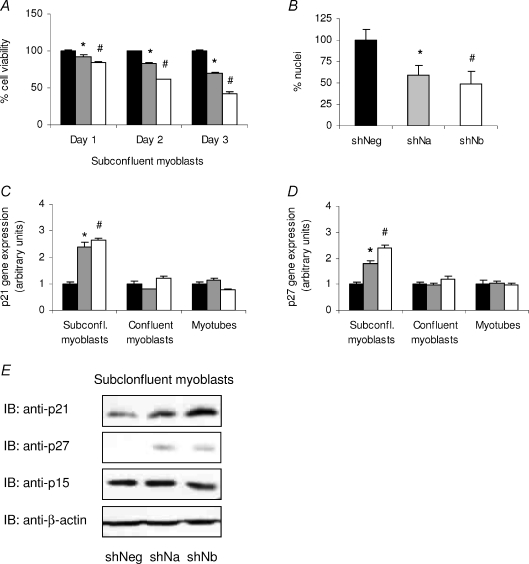
To ascertain whether a reduction in C2C12 myoblast proliferation was occurring in NDRG2-deficient C2C12 myoblasts, C2C12 subconfluent myoblasts infected with the negative control or with NDRG2-specific shRNAs were assessed by MTT assay at days 1, 2 and 3 post-plating. The shNDRG2a-infected cells revealed a significant reduction in cell number by 7, 17 and 30% on days 1, 2 and 3, respectively (P < 0.015), when compared with the shNegative cells on each day (Fig. 3A). The shNDRG2b-infected cells demonstrated an even greater reduction with a 15, 38 and 57% decrease measured on days 1, 2 and 3, respectively (P < 0.001). A significantly lower number of nuclei remained evident in the shNDRG2-infected myotubes compared with the negative control myotubes (Fig. 3B). Increased cell death was not visually apparent in the shNDRG2-infected cells nor was the activation of caspase 3, a key effector of apoptosis. No change in total caspase 3 levels or evidence of its 17/19 kDa cleavage products were measured by immunoblotting (data not shown). A reduced rate of cell proliferation was further evident by FACS analyses of shRNA-infected subconfluent myoblasts where 16% more shNDRG2b-infected myoblasts were found retained in the stationary G0/G1 phase with a corresponding 14% decrease in cell number occurring in the S phase of the cell cycle compared with uninfected and shNegative myoblasts (see Supplementary Fig. S2).
Figure 3. Effect of reduced NDRG2 levels on C2C12 myoblast proliferation.
A, MTT assay of C2C12 subconfluent myoblasts infected with shNeg (black bars), shNa (grey bars) or shNb (white bars) at day 1, 2 and 3 post-plating. Data are representative of three independent experiments (n= 4 per sample). *P < 0.014 and #P < 0.001 compared with the shNeg value on each day. B, number of nuclei per field of view (n= 3) in shNeg-, shNa- and shNb-infected myotubes. Data are expressed as a % change to shNeg. *P= 0.015 and #P= 0.010 compared with shNeg value. C and D, fold change gene expression in control (shNeg, black bars) and shNDRG2-infected (shNa, grey bars; and shNb, white bars) subconfluent (subconfl.) myoblasts, confluent myoblasts and myotubes for p21 Waf/Cip1 (C) and p27 Kip1 (D). Data are normalised to shNegative-infected cell values in each group and are representative of three independent experiments (n= 3 per sample). *P < 0.003 and #P < 0.002 compared with shNeg value in each cell type. E, immunoblot analyses (IB) of p21, p27, p15 INK4B and β-actin protein levels in shRNA-infected subconfluent myoblasts.
We next investigated whether the expression of specific regulators of the cell cycle were altered in shNDRG2-deficient subconfluent myoblasts. The gene expression of p21 Waf1/Cip1 and p27 Kip1 was analysed and found to be induced 2- to 2.5-fold when compared with the control myoblasts (P < 0.004 for p21 and P < 0.002 for p27) (Fig. 3C and D). There were no differences measured between both genes in the confluent myoblasts or in the myotubes. In agreement with the mRNA levels, the protein levels for p21 and p27 increased in both types of shNDRG2-infected subconfluent myoblasts in contrast to the shNegative-infected cells while no change in expression of an alternate CDK inhibitor, p15 INK4B, was evident (Fig. 3E).
Expression analyses of muscle cell differentiation genes in shNDRG2-infected C2C12 cells
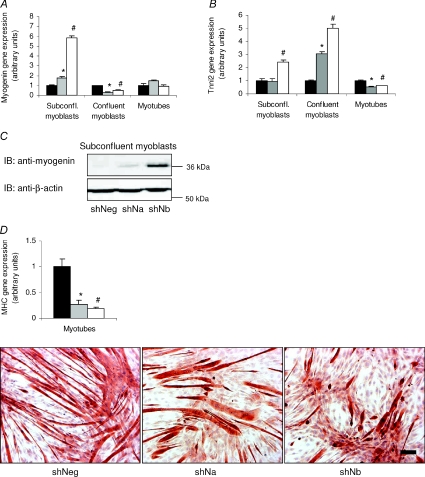
Markers of muscle cell differentiation, myogenin and troponin I type 2 (Tnni2), the fast twitch skeletal muscle isoform that is activated upon myoblast fusion (Koppe et al. 1989; Lin et al. 1994), were both analysed in shRNA-infected C2C12 cells. Myogenin gene expression was significantly up-regulated in both shNDRG2a- and shNDRG2b-deficient subconfluent myoblasts (P < 0.004; Fig. 4A) where it was increased 6-fold in the latter compared with the negative control myoblasts. Myogenin protein levels reflected the gene expression changes in the subconfluent myoblasts (Fig. 4C). In contrast to the subconfluent myoblasts, a 2- to 3-fold decrease in myogenin gene expression (P < 0.002) in the shNDRG2a- and shNDRG2b-infected confluent myoblasts was measured while no difference was found in the myotubes (Fig. 4A). Elevated Tnni2 mRNA levels were also measured, with a 2.5-fold increase in the shNDRG2b-infected subconfluent myoblasts and both a 3-fold and a 5-fold increase in the shNDRG2a- and shNDRG2b-infected confluent myoblasts, respectively, were found in relation to the shNegative cells (P= 0.005 in the subconfluent myoblasts and P < 0.004 in the confluent myoblasts) (Fig. 4B). Tnni2 mRNA levels, however, were reduced 2-fold in both the shNDRG2a- and shNDRG2b-infected myotubes (P < 0.004) (Fig. 4B). Moreover, myosin heavy chain (MHC) gene expression, a marker of skeletal muscle fibres, also decreased significantly in the shNDRG2-infected myotubes (P < 0.015) (Fig. 4D) while immunostaining for myosin protein expression revealed a more sporadic presence of myotubes in the NDRG2-deficient C2C12 cells suggesting reduced MHC expression (Fig. 4D).
Figure 4. Effect of reduced NDRG2 levels on C2C12 cell differentiation.
Fold change gene expression in control (shNeg, black bars) and shNDRG2-infected (shNa, grey bars; and shNb, white bars) subconfluent (subconfl.) myoblasts,confluent myoblasts and myotubes for myogenin (A) and Tnni2 (B). Data are normalised to shNegative-infected cell values in each group and are representative of three independent experiments (n= 3 per sample). *P < 0.004 and #P < 0.003 compared with shNeg value in each cell type. C, immunoblot analyses (IB) of myogenin and β-actin protein levels in shRNA-infected myoblasts. D, gene expression (top panel) and immunostaining of myosin heavy chain protein expression (bottom panels) in shRNA-infected myotubes. *P < 0.03 and #P < 0.004 compared with the shNeg value. Scale bar, 100 μm.
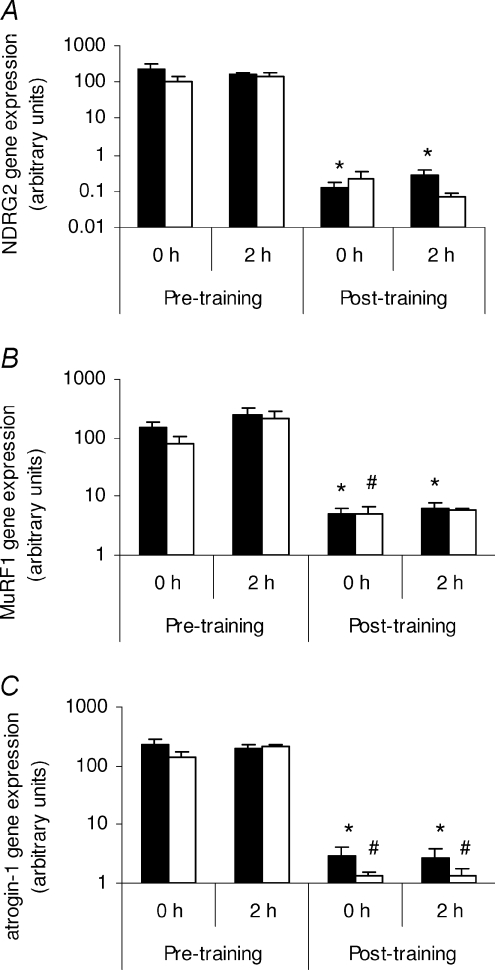
NDRG2, MuRF1 and atrogin-1 gene expression in resistance exercise trained skeletal muscle from young and older individuals
Having established a role for NDRG2 in myoblast growth and differentiation, NDRG2 expression was next investigated in different metabolically stressed states. Long-term resistance exercise training is a model of physiological stress and muscle hypertrophy (reviewed by Fluck & Hoppeler, 2003) and we sought to determine whether NDRG2 was regulated under this condition. We also analysed the gene expression of two markers of atrophy, the E3 ubiquitin ligases atrogin-1/MAFbx and MuRF1 (muscle RING finger 1), to help molecularly characterise the state of the muscle tissue. Induction of atrogin-1 and MuRF1 gene expression indicates increased proteolysis via the ubiquitin proteosome pathway while their decreased expression indicates hypertrophic conditions (Glass, 2005). The gene expression of NDRG2, MuRF1 and atrogin-1 was each found to decrease significantly in the younger individuals following a 12 week resistance exercise training program compared with the pre-resistance trained, rested (0 h) state (P < 0.038 for NDRG2, Fig. 5A; P < 0.023 for MuRF1, Fig. 5B; P < 0.019 for atrogin-1, Fig. 5C). A similar finding was measured in the chronically resistance exercise trained older individuals, where NDRG2 gene expression also tended to decrease (P= 0.078, Fig. 5A) and MuRF1 (P= 0.047, Fig. 5B) and atrogin-1 (P < 0.020, Fig. 5C) mRNA levels were significantly reduced in older individuals when compared with the pre-resistance trained state. No difference was found in any of the gene expression levels measured between the two different age groups at any time point nor was any difference found between each group following a single acute bout of resistance exercise.
Figure 5. The effects of resistance exercise training on NDRG2, MuRF1 and atrogin-1 gene expression in skeletal muscle.
NDRG2 (A), MuRF1 (B) and atrogin-1 (C) gene expression in the vastus lateralis muscle from young (black bars; n= 16) and older (white bars; n= 15) males at 0 h and 2 h following completion of a single bout of extension exercise before (pre-training) and after (post-training) 12 weeks of resistance exercise training. A, *P < 0.04 compared with 0 h pre-training in younger males. B, *P < 0.032 compared with 0 h pre-training in younger males; #P= 0.047 compared with 0 h pre-training in older males. C, *P < 0.02 compared with 0 h pre-training in younger males; #P < 0.012 compared with 0 h pre-training in older males.
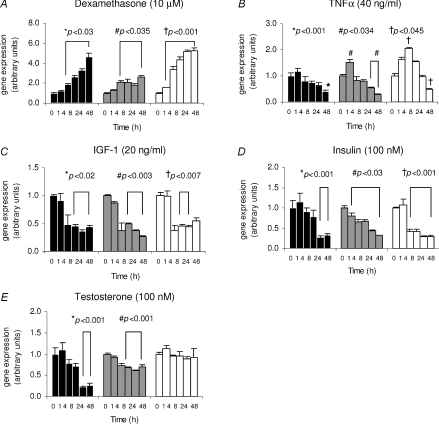
NDRG2 gene expression in atrophic and hypertrophic C2C12 myotubes
To follow on from the resistance training results, the effects of anabolic and catabolic stimuli on NDRG2, MuRF1 and atrogin-1 gene expression were assessed in C2C12 myotubes. C2C12 myotubes were treated with the atrophy-causing catabolic (dexamethasone and TNFα) and hypertrophy-causing anabolic (IGF-1, insulin and testosterone) agents for up to 48 h. Treatment with dexamethasone caused a significant increase in NDRG2 gene expression in nearly all times measured with a 4.7-fold induction found after 48 h (P < 0.03) (Fig. 6A). Similar profiles for MuRF1 (P < 0.035) and atrogin-1 (P < 0.001) mRNAs were measured with up to 2.6- and 5.3-fold increases, respectively, by 48 h post-treatment. These data are in agreement with previous studies in dexamethasone-treated C2C12 myotubes (Dehoux et al. 2004; Stitt et al. 2004). Treatment with TNFα produced a peak induction of mRNA expression at 1 and 4 h post-TNFα treatment for MuRF1 (P= 0.033) and atrogin-1 (P= 0.044) (Fig. 6B), respectively; the latter result agreeing with prior studies (Li et al. 2005). Following initial induction, a progressive decline in mRNA levels for both genes then occurred, reaching significance by 24 h or 48 h (P < 0.004 for MuRF1 and P= 0.038 for atrogin-1). TNFα did not significantly induce NDRG2 mRNA, although at 24 h and 48 h post-treatment expression levels were reduced (P < 0.001) (Fig. 6B). The effects of the anabolic agents, insulin, IGF-1 and testosterone, were also assessed. As a result, IGF-1 and insulin caused a 2- and 3-fold decrease, respectively, in mRNA levels for each gene compared with the 0 h samples (Fig. 6C and D). The IGF-1-mediated effects were evident by 4 h or 8 h post-treatment and sustained during later time-points for all genes (P < 0.02 for NDRG2, P < 0.003 for MuRF1 and P < 0.007 for atrogin-1) (Fig. 6C), which again is in agreement with prior studies relating to MuRF1 and atrogin-1 regulation in C2C12 myotubes (Dehoux et al. 2004; Sacheck et al. 2004). Following insulin treatment, a significant decrease in MuRF1 gene expression was observed at all times (P < 0.03) while reductions in atrogin-1 and NDRG2 mRNA levels were evident by 4 h (P < 0.001) and 24 h (P < 0.001), respectively, compared with the 0 h samples (Fig. 6D). Testosterone treatment decreased NDRG2 mRNA up to 4.5-fold (P < 0.001) while MuRF1 expression was reduced by 1.6-fold (P < 0.002) with no effect on atrogin-1 mRNA levels at the times measured (Fig. 6E).
Figure 6. NDRG2, MuRF1 and atrogin-1 gene expression in C2C12 myotubes following time-course treatments.
Fold change in gene expression of NDRG2 (black bars), MuRF1 (grey bars) and atrogin-1 (white bars) following dexamethasone (A; 10 μm), TNFα (B; 40 ng ml−1), IGF-1 (C; 20 ng ml−1), insulin (D; 100 nm) and testosterone (E; 100 nm) treatments. Time-course (h) is on the x-axis. *, # or †P < 0.05 compared with the 0 h time-point for NDRG2, MuRF1 or atrogin-1, respectively. Each experiment is a representative of 2–3 independent experiments (n= 3 per sample).
Discussion
Here, we describe the first functional characterisation of NDRG2 in skeletal muscle cells where NDRG2 displays high expression and is a potential substrate for growth factor-regulated kinases. NDRG2 expression levels were found to increase during muscle cell differentiation while a deficiency of NDRG2 resulted in slowed C2C12 myoblast proliferation and induced cell cycle exit and myogenesis. In response to stimuli which modulate myofibre growth or atrophy, NDRG2 expression was found to be regulated both in vitro and in vivo. Resistance exercise training effects in individuals led to a decrease in NDRG2 gene expression in skeletal muscle. Furthermore, atrophy and hypertrophy agents significantly regulated NDRG2 mRNA levels in C2C12 myotubes. Specifically, the growth factors IGF-1, insulin and testosterone decreased NDRG2 expression while the atrophy agent dexamethasone increased NDRG2 mRNA levels. Together, these data indicate a novel role for NDRG2 in contributing to the control of muscle mass and homeostasis.
A role for NDRG2 as a putative tumour suppressor in cancer cell growth control has been suggested. Decreased NDRG2 gene expression has been measured in multiple cancer cell types (Deng et al. 2003; Hu et al. 2004; Lusis et al. 2005; Liu et al. 2007; Lorentzen et al. 2007) and gene-silencing studies have also resulted in increased cell proliferation in cancer cell lines (Choi et al. 2007b). Conversely, the overexpression of NDRG2 has led to decreased cancer cell growth (Deng et al. 2003; Choi et al. 2007a; Park et al. 2007). Based on these reports, we had predicted that by reducing endogenous NDRG2 levels, increased myoblast growth would occur. Contrary to our hypothesis, the knockdown of NDRG2 with the two individual NDRG2-specific shRNA constructs resulted in reduced myoblast proliferation and an earlier onset of myogenesis, with the shNDRG2b construct displaying a more enhanced effect. This outcome, however, is not unprecedented as a similar result has been reported for NDRG1. NDRG1 shares 53–65% amino acid homology with the other NDRG family members (Qu et al. 2002), and is also considered a tumour suppressor agent as it exhibits reduced expression in cancer and metastatic tissues and its expression is induced in differentiating cells (reviewed in Ellen et al. 2007). Additional studies have identified that a lack of NDRG1 results in the inhibition of liver cancer cell proliferation in vitro and in vivo (Yan et al. 2008) and decreased trophoblast cell viability/proliferation (Chen et al. 2006). The latter study also found that during stress conditions (hypoxia) decreased levels of NDRG1 resulted in the induction of apoptosis (Chen et al. 2006). During basal conditions in NDRG2-deficient myoblasts, we did not observe increased cell death nor detect cleavage of caspase 3. However, it is possible that under specific stress conditions, lack of NDRG2 may affect cell apoptosis. The apparent differing roles for both NDRG2 and NDRG1 as either anti- or pro-proliferative factors could be dictated by different cell types and requires further investigation.
As NDRG2 is a substrate for several serine–threonine protein kinases including Akt and SGK1 in skeletal muscle (Burchfield et al. 2004; Murray et al. 2004), it is possible that the functional effects we observed in NDRG2-deficient myoblasts are due to a reduction in NDRG2 phosphoprotein levels that may be impacting on muscle function. While SGK1 is considered a physiological regulator of NDRG2 in skeletal muscle (Murray et al. 2004), its role in this tissue is not so clearly defined as Akt. Both Akt1 and Akt2 are key components of skeletal muscle cell proliferation and differentiation in vitro (Bouzakri et al. 2006; Heron-Milhavet et al. 2006; Wilson & Rotwein, 2007) and in vivo (Cho et al. 2001a,b). Akt either directly or indirectly regulates p21 and p27 activity or transcription during cell proliferation and cell cycle exiting in cancer cells (Maddika et al. 2007). IGF-1 also regulates p21 and p27 in proliferating muscle cells (Chakravarthy et al. 2000; Lawlor & Rotwein, 2000; Machida et al. 2003; Heron-Milhavet et al. 2006). The increased expression of p21 and p27 measured in NDRG2-deficient myoblasts, in addition to the proliferation data and cell cycle analysis presented here, suggest that myoblasts lacking NDRG2 are slowing in their ability to proliferate and entering a post-mitotic state earlier than the negative control-infected cells despite the NDRG2-deficient cells being subconfluent and existing in normal growth (high serum) culture conditions. While NDRG2 may be targeting the G1–S cell cycle progression via p21 and p27, its effect on CDK inhibitors appears specific as no change in the expression of p15 INK4B, a member of the p16 INK4a protein family, was observed. It is perhaps unexpected that the knockdown of NDRG2 reduced myoblast proliferation given myoblasts display significantly lower NDRG2 expression levels compared with the differentiated myotubes. This outcome, however, is not unusual if compared with Akt2. Akt2 expression increases during myoblast differentiation (Fujio et al. 2001) as well as functioning to control cell cycle exiting and differentiation of myoblasts (Bouzakri et al. 2006; Heron-Milhavet et al. 2006) and possessing a multitude of activities as a key signalling molecule in myotubes (Cho et al. 2001a; Bouzakri et al. 2006; Frost & Lang, 2007). As NDRG2 is a potential effector of Akt (Burchfield et al. 2004; Murray et al. 2004), perhaps NDRG2 also affects similar processes as Akt in both myoblasts and myotubes.
With the increase in NDRG2 expression during muscle differentiation, a possible outcome associated with the knockdown of NDRG2 would be a reduced ability of the myoblasts to differentiate. The data from this study appear to indicate that the onset of differentiation in NDRG2-deficient myoblasts was able to commence in conjunction with cell cycle exiting. However, and perhaps due to insufficient cell numbers, incorrect myoblast alignment and fusion may have occurred, which lead to reduced formation of myotubes as indicated by the level of Tnni2 and MHC expressed and the phenotypic appearance of the cells. The role for NDRG2 in cell differentiation is still unclear as its knockdown did not appear to affect the ability of monocytes to differentiate into dendritic cells (Choi et al. 2007a) although enhanced differentiation of trophoblasts was observed following NDRG2 overexpression (Chen et al. 2006). With our current data, it is difficult to assess whether NDRG2 affects differentiation or cell cycle exiting specifically as the role of cell cycle inhibitors and myogenin in muscle cell differentiation are intertwined. While p21 (and p57) are required for muscle differentiation in vivo (Zhang et al. 1999), in C2C12 myoblasts, myogenin expression is induced independently and prior to both p21 induction and cell cycle arrest, and hence, prior to phenotypic changes involving cell fusion in response to low serum conditions (Andres & Walsh, 1996; Zhang et al. 1999). In turn, p27 is considered a ‘trigger’ CDK inhibitor where its increased expression has been mapped to occur prior to myogenin and p21 induction (Zabludoff et al. 1998). It may be that altering the level of NDRG2 affects the normal crosstalk and ratios between the proliferation and differentiation protein markers, which leads to premature differentiation. Further studies would be required to determine whether NDRG2 directly or indirectly regulates the molecules involved in the onset of myoblast differentiation or cell cycle exit.
Another role for Akt is to mediate growth factor and cytokine signalling during muscle hypertrophy and atrophy (Guttridge, 2004; Frost & Lang, 2007). Long-term resistance exercise can cause skeletal muscle growth involving increased satellite cell activation and myoblast proliferation and fusion, and contractile protein synthesis (Kadi & Thornell, 2000; Seynnes et al. 2007). In addition, resistance training can be associated with increased production of anabolic factors, and conversely, a reduction in cortisol levels in both young and older individuals (Kraemer et al. 1999). The down-regulation of NDRG2 mRNA levels following resistance training accordingly may be due to the effects of myoblast proliferation and/or increased anabolic and decreased catabolic circulating agents. Our data demonstrating similarly decreased MuRF1 and atrogin-1 gene expression levels support this hypothesis as Akt indirectly down-regulates these genes during muscle hypertrophy in cell and animal models (Stitt et al. 2004; Marino et al. 2008). To further add to the resistance training findings, we have shown novel down-regulation of NDRG2 by insulin, IGF-1 and testosterone in myofibres. The latter finding is consistent with a previous study describing decreased NDRG2 mRNA expression by androgens in rat kidney (Boulkroun et al. 2005). Glucocorticoid regulation of NDRG2 has also been reported previously although with varying outcomes. The induction of NDRG2 gene expression by dexamethasone and glucocorticoids occurs in the brain and heart of rats (Nichols, 2003). In dexamethasone-induced de-differentiation of dendritic cells, however, decreased NDRG2 gene expression was observed (Choi et al. 2007a) while dexamethasone exhibited no effect on gene expression in kidney cells (Boulkroun et al. 2002). These differences in dexamethasone regulation of NDRG2 gene expression may reflect variable experimental conditions or different cell types and possibly alternate signalling pathways. As TNFα did not significantly increase NDRG2 gene expression, it suggests that not all atrophic agents regulate NDRG2 similarly in muscle cells. Interestingly, we did not observe a difference in NDRG2 gene expression between the young versus older individuals in the basal state or following resistance training. A reduction in both muscle mass (sarcopenia) and circulating anabolic hormones with age are well described (reviewed in Solomon & Bouloux, 2006) so why was there no age-related differences with NDRG2 mRNA levels in skeletal muscle? Both MuRF1 and atrogin-1 gene expression levels were also not found to increase with age in this study, which is consistent with other reports where their levels of expression did not change in individuals with age-related sarcopenia (Whitman et al. 2005; Leger et al. 2008). These results suggest that different regulatory molecules are potentially responsible for alternate forms of muscle atrophy, and furthermore, indicate a potential co-regulation of NDRG2 with the ubiquitin ligases MuRF1 and atrogin-1 under specific conditions.
We have provided in this study novel insight into the role of NDRG2 in skeletal muscle function. We have identified that a lack of NDRG2 affects myoblast proliferation and ultimately myotube differentiation. The mechanism of this regulation requires further investigation and it will be of interest to determine whether NDRG2 regulates other aspects of muscle adaption such as satellite cell activity and myoblast fusion. As myotubes have undergone terminal division and NDRG2 is up-regulated upon muscle cell differentiation, it is likely that increased levels of NDRG2 may be required for myotube function and homeostasis. Moreover, the in vitro regulation of NDRG2 by atrophy and hypertrophy agents indicates that NDRG2 may contribute to muscle mass control. Hence, the identification of novel interacting muscle proteins, and tissue- and temporal-specific gene targeting studies in the future, could help further elucidate the role for NDRG2 in skeletal muscle.
Acknowledgments
We thank Dr JC Molero for provision of the myogenin antibody and for helpful discussions in conjunction with Professor D James and Dr A Russell. We are grateful to Dr F Collier for generous assistance and use of the FACSCalibur. This work was supported by Verva Pharmaceuticals Ltd and by the Molecular Medicine and Nutrition Cluster, Deakin University.
Glossary
Abbreviations
- bHLH
basic helix–loop–helix
- CDK
cyclin-dependent kinase
- IGF-1
insulin-like growth factor-1
- MHC
myosin heavy chain
- MuRF1
muscle RING finger 1
- NDRG2
N-myc downstream-regulated gene 2
- p21
p21 Waf1/Cip1
- p27
p27 Kip1
- SGK1
serum- and glucocorticoid-induced kinase 1
- TNFα
tumour necrosis factor alpha
- Tnni2
troponin I type 2
Supplemental material
Online supplemental material for this paper can be accessed at:
http://jp.physoc.org/cgi/content/full/jphysiol.2008.167882/DC1
References
- Akamine R, Yamamoto T, Watanabe M, Yamazaki N, Kataoka M, Ishikawa M, Ooie T, Baba Y, Shinohara Y. Usefulness of the 5′ region of the cDNA encoding acidic ribosomal phosphoprotein P0 conserved among rats, mice, and humans as a standard probe for gene expression analysis in different tissues and animal species. J Biochem Biophys Methods. 2007;70:481–486. doi: 10.1016/j.jbbm.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Andres V, Walsh K. Myogenin expression, cell cycle withdrawal, and phenotypic differentiation are temporally separable events that precede cell fusion upon myogenesis. J Cell Biol. 1996;132:657–666. doi: 10.1083/jcb.132.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berggren JR, Tanner CJ, Koves TR, Muoio DM, Houmard JA. Glucose uptake in muscle cell cultures from endurance-trained men. Med Sci Sports Exerc. 2005;37:579–584. doi: 10.1249/01.mss.0000158180.11224.e8. [DOI] [PubMed] [Google Scholar]
- Boulkroun S, Fay M, Zennaro MC, Escoubet B, Jaisser F, Blot-Chabaud M, Farman N, Courtois-Coutry N. Characterization of rat NDRG2 (N-Myc downstream regulated gene 2), a novel early mineralocorticoid-specific induced gene. J Biol Chem. 2002;277:31506–31515. doi: 10.1074/jbc.M200272200. [DOI] [PubMed] [Google Scholar]
- Boulkroun S, Le Moellic C, Blot-Chabaud M, Farman N, Courtois-Coutry N. Expression of androgen receptor and androgen regulation of NDRG2 in the rat renal collecting duct. Pflugers Arch. 2005;451:388–394. doi: 10.1007/s00424-005-1410-x. [DOI] [PubMed] [Google Scholar]
- Bouzakri K, Zachrisson A, Al-Khalili L, Zhang BB, Koistinen HA, Krook A, Zierath JR. siRNA-based gene silencing reveals specialized roles of IRS-1/Akt2 and IRS-2/Akt1 in glucose and lipid metabolism in human skeletal muscle. Cell Metab. 2006;4:89–96. doi: 10.1016/j.cmet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Burchfield JG, Lennard AJ, Narasimhan S, Hughes WE, Wasinger VC, Corthals GL, Okuda T, Kondoh H, Biden TJ, Schmitz-Peiffer C. Akt mediates insulin-stimulated phosphorylation of Ndrg2: evidence for cross-talk with protein kinase C theta. J Biol Chem. 2004;279:18623–18632. doi: 10.1074/jbc.M401504200. [DOI] [PubMed] [Google Scholar]
- Carey KA, Farnfield MM, Tarquinio SD, Cameron-Smith D. Impaired expression of Notch signaling genes in aged human skeletal muscle. J Gerontol A Biol Sci Med Sci. 2007;62:9–17. doi: 10.1093/gerona/62.1.9. [DOI] [PubMed] [Google Scholar]
- Chakravarthy MV, Abraha TW, Schwartz RJ, Fiorotto ML, Booth FW. Insulin-like growth factor-I extends in vitro replicative life span of skeletal muscle satellite cells by enhancing G1/S cell cycle progression via the activation of phosphatidylinositol 3′-kinase/Akt signaling pathway. J Biol Chem. 2000;275:35942–35952. doi: 10.1074/jbc.M005832200. [DOI] [PubMed] [Google Scholar]
- Chen B, Nelson DM, Sadovsky Y. N-myc down-regulated gene 1 modulates the response of term human trophoblasts to hypoxic injury. J Biol Chem. 2006;281:2764–2772. doi: 10.1074/jbc.M507330200. [DOI] [PubMed] [Google Scholar]
- Cho H, Mu J, Kim JK, Thorvaldsen JL, Chu Q, Crenshaw EB, 3rd, Kaestner KH, Bartolomei MS, Shulman GI, Birnbaum MJ. Insulin resistance and a diabetes mellitus-like syndrome in mice lacking the protein kinase Akt2 (PKBβ) Science. 2001a;292:1728–1731. doi: 10.1126/science.292.5522.1728. [DOI] [PubMed] [Google Scholar]
- Cho H, Thorvaldsen JL, Chu Q, Feng F, Birnbaum MJ. Akt1/PKBα is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem. 2001b;276:38349–38352. doi: 10.1074/jbc.C100462200. [DOI] [PubMed] [Google Scholar]
- Choi SC, Kim KD, Kim JT, Kim JW, Lee HG, Kim JM, Jang YS, Yoon DY, Kim KI, Yang Y, Cho DH, Lim JS. Expression of human NDRG2 by myeloid dendritic cells inhibits down-regulation of activated leukocyte cell adhesion molecule (ALCAM) and contributes to maintenance of T cell stimulatory activity. J Leukoc Biol. 2007a;83:89–98. doi: 10.1189/jlb.0507300. [DOI] [PubMed] [Google Scholar]
- Choi SC, Kim KD, Kim JT, Kim JW, Yoon DY, Choe YK, Chang YS, Paik SG, Lim JS. Expression and regulation of NDRG2 (N-myc downstream regulated gene 2) during the differentiation of dendritic cells. FEBS Lett. 2003;553:413–418. doi: 10.1016/s0014-5793(03)01030-5. [DOI] [PubMed] [Google Scholar]
- Choi SC, Yoon SR, Park YP, Song EY, Kim JW, Kim WH, Yang Y, Lim JS, Lee HG. Expression of NDRG2 is related to tumor progression and survival of gastric cancer patients through Fas-mediated cell death. Exp Mol Med. 2007b;39:705–714. doi: 10.1038/emm.2007.77. [DOI] [PubMed] [Google Scholar]
- Dehoux M, Van Beneden R, Pasko N, Lause P, Verniers J, Underwood L, Ketelslegers JM, Thissen JP. Role of the insulin-like growth factor I decline in the induction of atrogin-1/MAFbx during fasting and diabetes. Endocrinology. 2004;145:4806–4812. doi: 10.1210/en.2004-0406. [DOI] [PubMed] [Google Scholar]
- Deng Y, Yao L, Chau L, Ng SS, Peng Y, Liu X, Au WS, Wang J, Li F, Ji S, Han H, Nie X, Li Q, Kung HF, Leung SY, Lin MC. N-Myc downstream-regulated gene 2 (NDRG2) inhibits glioblastoma cell proliferation. Int J Cancer. 2003;106:342–347. doi: 10.1002/ijc.11228. [DOI] [PubMed] [Google Scholar]
- Ellen T, Ke Q, Zhang P, Costa M. NDRG1, a growth and cancer related gene: regulation of gene expression and function in normal and disease states. Carcinogenesis. 2007;29:2–8. doi: 10.1093/carcin/bgm200. [DOI] [PubMed] [Google Scholar]
- Fluck M, Hoppeler H. Molecular basis of skeletal muscle plasticity–from gene to form and function. Rev Physiol Biochem Pharmacol. 2003;146:159–216. doi: 10.1007/s10254-002-0004-7. [DOI] [PubMed] [Google Scholar]
- Frith-Terhune A, Koh KN, Jin WJ, Chung KB, Park SK, Koh GY. Programmed changes of cell cycle regulators by serum deprivation regardless of skeletal myocyte differentiation. Mol Cells. 1998;8:637–646. [PubMed] [Google Scholar]
- Frost RA, Lang CH. Protein kinase B/Akt: a nexus of growth factor and cytokine signaling in determining muscle mass. J Appl Physiol. 2007;103:378–387. doi: 10.1152/japplphysiol.00089.2007. [DOI] [PubMed] [Google Scholar]
- Fujio Y, Mitsuuchi Y, Testa JR, Walsh K. Activation of Akt2 inhibits anoikis and apoptosis induced by myogenic differentiation. Cell Death Differ. 2001;8:1207–1212. doi: 10.1038/sj.cdd.4400919. [DOI] [PubMed] [Google Scholar]
- Gaster M, Kristensen SR, Beck-Nielsen H, Schroder HD. A cellular model system of differentiated human myotubes. APMIS. 2001;109:735–744. doi: 10.1034/j.1600-0463.2001.d01-140.x. [DOI] [PubMed] [Google Scholar]
- Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Guttridge DC. Signaling pathways weigh in on decisions to make or break skeletal muscle. Curr Opin Clin Nutr Metab Care. 2004;7:443–450. doi: 10.1097/01.mco.0000134364.61406.26. [DOI] [PubMed] [Google Scholar]
- Heron-Milhavet L, Franckhauser C, Rana V, Berthenet C, Fisher D, Hemmings BA, Fernandez A, Lamb NJ. Only Akt1 is required for proliferation, while Akt2 promotes cell cycle exit through p21 binding. Mol Cell Biol. 2006;26:8267–8280. doi: 10.1128/MCB.00201-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XL, Liu XP, Lin SX, Deng YC, Liu N, Li X, Yao LB. NDRG2 expression and mutation in human liver and pancreatic cancers. World J Gastroenterol. 2004;10:3518–3521. doi: 10.3748/wjg.v10.i23.3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadi F, Thornell LE. Concomitant increases in myonuclear and satellite cell content in female trapezius muscle following strength training. Histochem Cell Biol. 2000;113:99–103. doi: 10.1007/s004180050012. [DOI] [PubMed] [Google Scholar]
- Koppe RI, Hallauer PL, Karpati G, Hastings KE. cDNA clone and expression analysis of rodent fast and slow skeletal muscle troponin I mRNAs. J Biol Chem. 1989;264:14327–14333. [PubMed] [Google Scholar]
- Kraemer WJ, Hakkinen K, Newton RU, Nindl BC, Volek JS, McCormick M, Gotshalk LA, Gordon SE, Fleck SJ, Campbell WW, Putukian M, Evans WJ. Effects of heavy-resistance training on hormonal response patterns in younger vs. older men. J Appl Physiol. 1999;87:982–992. doi: 10.1152/jappl.1999.87.3.982. [DOI] [PubMed] [Google Scholar]
- Lawlor MA, Rotwein P. Insulin-like growth factor-mediated muscle cell survival: central roles for Akt and cyclin-dependent kinase inhibitor p21. Mol Cell Biol. 2000;20:8983–8995. doi: 10.1128/mcb.20.23.8983-8995.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leger B, Derave W, De Bock K, Hespel P, Russell AP. Human sarcopenia reveals an increase in SOCS-3 and myostatin and a reduced efficiency of Akt phosphorylation. Rejuvenation Res. 2008;11:163–175B. doi: 10.1089/rej.2007.0588. [DOI] [PubMed] [Google Scholar]
- Li YP, Chen Y, John J, Moylan J, Jin B, Mann DL, Reid MB. TNF-α acts via p38 MAPK to stimulate expression of the ubiquitin ligase atrogin1/MAFbx in skeletal muscle. FASEB J. 2005;19:362–370. doi: 10.1096/fj.04-2364com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang J, Slingerland JM. Multiple roles of the PI3K/PKB (Akt) pathway in cell cycle progression. Cell Cycle. 2003;2:339–345. [PubMed] [Google Scholar]
- Lin Z, Lu MH, Schultheiss T, Choi J, Holtzer S, DiLullo C, Fischman DA, Holtzer H. Sequential appearance of muscle-specific proteins in myoblasts as a function of time after cell division: evidence for a conserved myoblast differentiation program in skeletal muscle. Cell Motil Cytoskeleton. 1994;29:1–19. doi: 10.1002/cm.970290102. [DOI] [PubMed] [Google Scholar]
- Liu N, Wang L, Liu X, Yang Q, Zhang J, Zhang W, Wu Y, Shen L, Zhang Y, Yang A, Han H, Yao L. Promoter methylation, mutation, and genomic deletion are involved in the decreased NDRG2 expression levels in several cancer cell lines. Biochem Biophys Res Commun. 2007;358:164–169. doi: 10.1016/j.bbrc.2007.04.089. [DOI] [PubMed] [Google Scholar]
- Lorentzen A, Vogel LK, Lewinsky RH, Saebo M, Skjelbred CF, Godiksen S, Hoff G, Tveit KM, Lothe IM, Ikdahl T, Kure EH, Mitchelmore C. Expression of NDRG2 is down-regulated in high-risk adenomas and colorectal carcinoma. BMC Cancer. 2007;7:192. doi: 10.1186/1471-2407-7-192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis EA, Watson MA, Chicoine MR, Lyman M, Roerig P, Reifenberger G, Gutmann DH, Perry A. Integrative genomic analysis identifies NDRG2 as a candidate tumor suppressor gene frequently inactivated in clinically aggressive meningioma. Cancer Res. 2005;65:7121–7126. doi: 10.1158/0008-5472.CAN-05-0043. [DOI] [PubMed] [Google Scholar]
- Machida S, Spangenburg EE, Booth FW. Forkhead transcription factor FoxO1 transduces insulin-like growth factor's signal to p27Kip1 in primary skeletal muscle satellite cells. J Cell Physiol. 2003;196:523–531. doi: 10.1002/jcp.10339. [DOI] [PubMed] [Google Scholar]
- Maddika S, Ande SR, Panigrahi S, Paranjothy T, Weglarczyk K, Zuse A, Eshraghi M, Manda KD, Wiechec E, Los M. Cell survival, cell death and cell cycle pathways are interconnected: implications for cancer therapy. Drug Resist Updat. 2007;10:13–29. doi: 10.1016/j.drup.2007.01.003. [DOI] [PubMed] [Google Scholar]
- Marino JS, Tausch BJ, Dearth CL, Manacci MV, McLoughlin TJ, Rakyta SJ, Linsenmayer MP, Pizza FX. β2-integrins contribute to skeletal muscle hypertrophy in mice. Am J Physiol Cell Physiol. 2008;295:C1026–C1036. doi: 10.1152/ajpcell.212.2008. [DOI] [PubMed] [Google Scholar]
- Morita S, Kojima T, Kitamura T. Plat−E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 2000;7:1063–1066. doi: 10.1038/sj.gt.3301206. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Murray JT, Campbell DG, Morrice N, Auld GC, Shpiro N, Marquez R, Peggie M, Bain J, Bloomberg GB, Grahammer F, Lang F, Wulff P, Kuhl D, Cohen P. Exploitation of KESTREL to identify NDRG family members as physiological substrates for SGK1 and GSK3. Biochem J. 2004;384:477–488. doi: 10.1042/BJ20041057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols NR. Ndrg2, a novel gene regulated by adrenal steroids and antidepressants, is highly expressed in astrocytes. Ann N Y Acad Sci. 2003;1007:349–356. doi: 10.1196/annals.1286.034. [DOI] [PubMed] [Google Scholar]
- Olson EN, Klein WH. bHLH factors in muscle development: dead lines and commitments, what to leave in and what to leave out. Genes Dev. 1994;8:1–8. doi: 10.1101/gad.8.1.1. [DOI] [PubMed] [Google Scholar]
- Park Y, Shon SK, Kim A, Kim KI, Yang Y, Cho DH, Lee MS, Lim JS. SOCS1 induced by NDRG2 expression negatively regulates STAT3 activation in breast cancer cells. Biochem Biophys Res Commun. 2007;363:361–367. doi: 10.1016/j.bbrc.2007.08.195. [DOI] [PubMed] [Google Scholar]
- Qu X, Zhai Y, Wei H, Zhang C, Xing G, Yu Y, He F. Characterization and expression of three novel differentiation-related genes belong to the human NDRG gene family. Mol Cell Biochem. 2002;229:35–44. doi: 10.1023/a:1017934810825. [DOI] [PubMed] [Google Scholar]
- Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am J Physiol Endocrinol Metab. 2004;287:E591–E601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- Seynnes OR, de Boer M, Narici MV. Early skeletal muscle hypertrophy and architectural changes in response to high-intensity resistance training. J Appl Physiol. 2007;102:368–373. doi: 10.1152/japplphysiol.00789.2006. [DOI] [PubMed] [Google Scholar]
- Solomon AM, Bouloux PM. Modifying muscle mass – the endocrine perspective. J Endocrinol. 2006;191:349–360. doi: 10.1677/joe.1.06837. [DOI] [PubMed] [Google Scholar]
- Spizz G, Roman D, Strauss A, Olson EN. Serum and fibroblast growth factor inhibit myogenic differentiation through a mechanism dependent on protein synthesis and independent of cell proliferation. J Biol Chem. 1986;261:9483–9488. [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy-induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamada M, Ohata H, Honda K. Ndrg2 promotes neurite outgrowth of NGF-differentiated PC12 cells. Neurosci Lett. 2005;388:157–162. doi: 10.1016/j.neulet.2005.06.055. [DOI] [PubMed] [Google Scholar]
- Wang L, Liu N, Yao L, Li F, Zhang J, Deng Y, Liu J, Ji S, Yang A, Han H, Zhang Y, Zhang J, Han W, Liu X. NDRG2 is a new HIF-1 target gene necessary for hypoxia-induced apoptosis in A549 cells. Cell Physiol Biochem. 2008;21:239–250. doi: 10.1159/000113765. [DOI] [PubMed] [Google Scholar]
- Whitman SA, Wacker MJ, Richmond SR, Godard MP. Contributions of the ubiquitin-proteasome pathway and apoptosis to human skeletal muscle wasting with age. Pflugers Arch. 2005;450:437–446. doi: 10.1007/s00424-005-1473-8. [DOI] [PubMed] [Google Scholar]
- Wilson EM, Rotwein P. Selective control of skeletal muscle differentiation by Akt1. J Biol Chem. 2007;282:5106–5110. doi: 10.1074/jbc.C600315200. [DOI] [PubMed] [Google Scholar]
- Yan X, Chua MS, Sun H, So S. N-Myc down-regulated gene 1 mediates proliferation, invasion, and apoptosis of hepatocellular carcinoma cells. Cancer Lett. 2008;262:133–142. doi: 10.1016/j.canlet.2007.12.010. [DOI] [PubMed] [Google Scholar]
- Zabludoff SD, Csete M, Wagner R, Yu X, Wold BJ. p27Kip1 is expressed transiently in developing myotomes and enhances myogenesis. Cell Growth Differ. 1998;9:1–11. [PubMed] [Google Scholar]
- Zhang P, Wong C, Liu D, Finegold M, Harper JW, Elledge SJ. p21(CIP1) and p57(KIP2) control muscle differentiation at the myogenin step. Genes Dev. 1999;13:213–224. doi: 10.1101/gad.13.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.