Abstract
The catalytic and structural properties of the H67A and H349A altered dapE-encoded N-succinyl-l,l-diaminopimelic acid desuccinylase (DapE) from H. influenzae were investigated. Based on sequence alignment with CPG2 both H67 and H349 were predicted to be Zn(II) ligands. Catalytic activity was observed for the H67A altered DapE enzyme which exhibited kcat = 1.5 ± 0.5 sec−1 and Km = 1.4 ± 0.3 mM. No catalytic activity was observed for H349A under the experimental conditions used. The EPR and electronic absorption data indicate that the Co(II) ion bound to H349A-DapE is analogous to WT DapE after the addition of a single Co(II) ion. The addition of one equivalent of Co(II) to H67A altered DapE provides spectra that are very different from the first Co(II) binding site of the WT enzyme, but similar to the second binding site. The EPR and electronic absorption data, in conjunction with the kinetic data, are consistent with the assignment of H67 and H349 as active site metal ligands for the DapE from H. influenzae. Furthermore, the data suggest that H67 is a ligand in the first metal binding site while H349 resides in the second metal binding site. A three-dimensional homology structure of the DapE from H. influenzae was generated using the X-ray crystal structure of the DapE from N. meningitidis as a template and superimposed on the structure of AAP. This homology structure confirms the assignment of H67 and H349 as active site ligands. The superimposition of the homology model of DapE with the dizinc(II) structure of AAP indicates that within 4.0 Å of the Zn(II) binding sites of AAP, all of the amino acid residues of DapE are nearly identical.
Keywords: biomedicine, biosynthesis, electron paramagnetic resonance, enzyme kinetics, homology model, site-directed mutagenesis, structure-function relationship
Introduction
Bacterial infections are a significant and growing medical problem in both the United States and around the world (1). The CDC recently reported that there are now several strains of Staphylococcus aureus that are resistant to all known antibiotics including vancomycin (2). At least four other strains of common bacterial species capable of causing life-threatening illnesses (Enterococcus faecalis, Mycobacterium tuberculosis, Escherichia coli O157:H7, and Pseudomonas aeruginosa) have already shown resistant to many of the drugs in a clinicians arsenal (3, 4) and a fifth, the multi-drug resistant Acinetobacter baumannii (MDRAB), strain is posing a major health problem for returning wounded Iraq military personnel (5, 6). To overcome this problem, new enzymatic targets must be identified and small molecule inhibitors designed and synthesized to target these enzymes. The dapE-encoded N-succinyl-l,l-diaminopimelic acid desuccinylase (DapE; EC 3.5.1.18), a member of the mDAP/lysine biosynthetic pathway present in both Gram-positive and Gram-negative bacteria, is just such a target (7, 8).
DapE's catalyze the hydrolysis of N-succinyl-l,l-diaminopimelic acid to l,l-diaminopimelic acid (mDAP) and succinate and have been purified from Helicobacter pylori, E. coli, Haemophilus influenzae, and Neisseria meningitides (7, 8). The genes that encode for DapE's have been sequenced from nearly all pathogenic bacteria including the multi-drug resistant A. baumannii (MDRAB), E. coli (O157:H7), Vibrio cholerae, P. aeruginosa, Bordetella pertussis, Staphacocus aureus (strain MRSA252), Rickettsia prowazekii, and M. tuberculosis, to name a few. It has been shown that deletion of the gene encoding for DapE is lethal to H. pylori and M. smegmatis (9, 10). Even in the presence of lysine supplemented media H. pylori was unable to grow. Therefore, DapEs are essential for cell growth and proliferation and are apart of a biosynthetic pathway that is the only source for lysine in most bacteria. The fact that there are no similar pathways in mammals suggests that inhibitors of enzymes in the mDAP/lysine pathway may provide selective toxicity against bacteria and have little or no effect on humans (7).
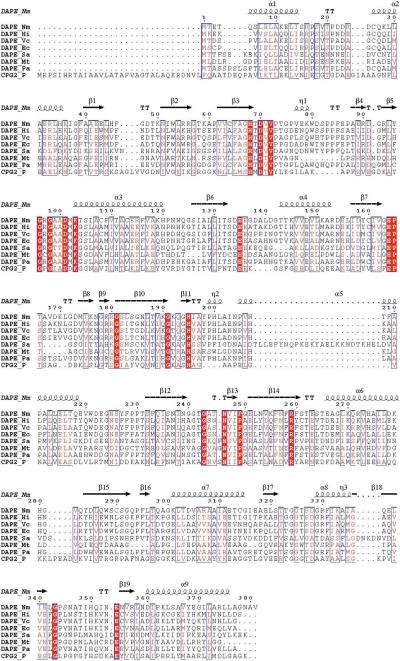
Biochemical studies have revealed that the DapE's from E. coli and H. influenzae are small (∼42 kDa) and dimeric in nature requiring two Zn(II) ions per mole of polypeptide for full enzymatic activity (8, 11, 12). The only X-ray crystallographic data available for a DapE enzyme is the 1.9 Å structure of the apo-form of the DapE from N. meningitidis (13). The absence of divalent metal ions makes it difficult to determine which residues constitute the catalytic active site. Based on sequence alignments with the aminopeptidase from Aeromonas proteolytica (AAP) (14), the carboxypeptidase from Pseudomonas sp strain-RS-16 (CPG2) (15), and several DapE gene sequences, all of the residues that function as ligands for the dinuclear active site in AAP and CPG2 are strictly conserved (Figure 1) (8, 14, 15). Since both AAP and CPG2 possess a (μ-aquo)(μ-carboxylato)dizinc(II) core with one terminal carboxylate and one histidine residue at each metal site, a similar active site was proposed for DapE based on EXAFS spectroscopic studies (16). Moreover, these sequence data suggest that the putative bridging water/(OH−) molecule likely forms a hydrogen bond to an active site carboxylate group, Glu134, which has been shown to function as the general acid/base in the catalytic process (17).
Figure 1.
Sequence alignment between the structurally characterized DapE from N. meningitidis (Nm) with DapE's from H. influenzae (Hi), V. cholerae (Vc), E. coli strain O157:H7 (Ec), Bordetella pertussis, S. aureus strain MRSA252 (Sa), M. tuberculosis (Mt), P. aeruginosa (Pa) and the structurally characterized carboxypeptidase from Pseudomonas sp strain-RS-16 (CPG2).
In an effort to clearly define the active site residues and provide insight into the structural properties of each divalent metal ion in DapE enzymes, we have prepared the H67A and H349A altered DapE enzymes from H. influenzae. Based on sequence alignment with CPG2 (Figure 1), both H67 and H349 were predicted to be Zn(II) ligands (8, 13-15, 18). These two altered enzymes were investigated via kinetic, electronic absorption, and EPR spectroscopy and these data verify that H67A and H349A are in fact active site Zn(II) ligands.
Materials and Methods
Reagents
d,l-α,ε-DAP (98% pure), succinic anhydride and ion exchange resin (Dowex 50WX8−200, H+ form) were purchased from Sigma. Technically pure 2-naphthalenesulfonic acid (70%) was purchased from Aldrich and crystallized from a 5% aqueous solution of HCl, dried in a vacuum desiccator over CaCl2 to give a white solid of 2-Naphthalenesulfonic acid 1-hydrate (confirmed by NMR, Mp 124−125°C). Microcrystalline cellulose was purchased from EM Science. All chemicals used in this study were purchased from commercial sources and were of the highest quality available.
Site directed mutagenesis
The Pet-23(a)+ vector containing the DapE gene was isolated from E. coli JM109 cells using a Promega Wizard Miniprep DNA isolation kit. Altered forms of DapE were obtained by PCR mutagenesis using the following primers: (H67A) 5'- GCT TTT GCA GGT GCG ACG GAT GTT GTG CC -3' and 5'- GGC ACA ACA TCC GTC GCA CCT GCA AAA GC -3' and (H349A) 5'- GTT AAA TTC AAC CAT TGC GAA AGT CAA TGA ATG -3' and 5'- CAT TCA TTG ACT TTC GCA ATG GTT GAA TTT AAC -3'. Site directed mutants were obtained using the Quick Change™ Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA) following Stratagene's procedure. Reaction products were transformed into E. coli BL21 competent cells and grown on LB-agarose plates containing carbenicillin at a concentration of 50 μg/ml. A single colony of each mutant was grown in 50 mL LB containing 50 μg/ml of carbenicillin. Plasmids were isolated using Wizard Plus Minipreps DNA purification kit (Promega, Madison, WI, USA) or Qiaprep® Spin Miniprep kit (Qiagene, Valencia, CA). Each mutation was confirmed by DNA sequencing (USU Biotechnology Center).
Protein expression and purification
The recombinant WT DapE from H. influenzae was expressed and purified, as previously described with minor modifications, from a stock culture kindly provided by Professor John Blanchard (8, 12). Site-directed mutant forms were purified using the same methodology. Each purified altered form of DapE exhibited a single band on SDS-PAGE indicating Mr = ∼41,500 Da. Protein concentrations were determined from the absorbance at 280 nm using a molar absorptivity calculated using the method developed by Gill and Hippel (ε280 = 36,040 M−1 cm−1). The protein concentration determined using this molar absorptivity was in close agreement to that obtained using a Bradford assay. Purified enzyme samples submitted for metal contact analyses were typically at a concentration of 30 μM. Metal analyses were performed using inductively coupled plasma-atomic emission spectrometry (ICP-AES). Individual aliquots of purified altered forms of DapE were typically stored in liquid nitrogen until needed.
Metal removal from altered DapE enzymes
Apo-DapE enzymes were prepared by extensive dialysis for 3 to 4 days against 10 mM EDTA in 50 mM HEPES buffer, pH 7.5. Each DapE enzyme was then exhaustively dialyzed against metal-free (Chelex-100 treated) 50 mM HEPES buffer, pH 7.5. The presence of any remaining metal ions in solution or bound by DapE were estimated by comparing the activity of the apo-enzyme with a sample that had been reconstituted with Zn(II). DapE enzyme samples incubated with EDTA typically had less than 5% residual activity after dialysis.
Synthesis of N-succinyl-diaminopimelic acid (l,l-SDAP)
The d,d- and l,l-isoforms of DAP were separated from the d,l- isoform using the method described by Bergmann and Stein (19). SDAP was synthesized using the procedure described by Lin et al. (20) providing an overall yield of 41% (1.84g; 5.7mmol). NMR (D2O, 270MHz), δ (ppm) 1.25−1.45 (m, 2H), 1.55−1.90 (m, 4H), 2.29−2.52 (m, 4H), 3.64 (dd, 1H), 4.05 (dd, 1H), 4.76 (bs, H2O + ammonium ion). The l,l- and d,d- SDAP isoforms were separated using an HPLC (Shimadzu SCL-10A VP) with a Chirobiotic T column (250 × 10 mm; Alltec). The isocratic mixture of 20% methanol in water (adjusted to pH 4) was used as the eluting solvent. The two isoforms were present in an approximate 1:1 ratio.
Enzymatic assay of altered DapE enzymes
The specific activity, Vmax (velocity) and Km (Michaelis constant) of each DapE enzyme was determined in triplicate by monitoring amide bond cleavage of l,l-SDAP at 225 nm in Chelex-100 treated 50 mM HEPES buffer, pH 7.5. Enzyme activities are expressed as units/mg where one unit is defined as the amount of enzyme that cleaves 1 μmol of l,l-SDAP at 30 °C in 1 min. Catalytic activities were determined with an error of ± 10 %. Initial rates were fit directly to the Michaelis-Menten equation to obtain the catalytic constants Km and kcat.
Spectroscopic measurements
Electronic absorption spectra were recorded on a Shimadzu UV-3101PC spectrophotometer. All apo-DapE samples used in spectroscopic measurements were made rigorously anaerobic prior to incubation with Co(II) (CoCl2≥99.999% Strem Chemicals, Newburyport, MA) for ∼30 min at 25°C. All Co(II)-containing protein samples were handled in an anaerobic glove box (Ar/5 % H2, ≤ 1 ppm O2; Coy Laboratories) until frozen. Electronic absorption spectra were normalized for protein concentration and the absorption due to uncomplexed Co(II) (ε512 nm = 6.0 M−1 cm−1 (21). Low-temperature EPR spectroscopy was performed at 9.64 GHz using a Bruker ESP-300E spectrometer equipped with an ER 4116 DM dual mode X-band cavity and an Oxford Instruments ESR-900 helium flow cryostat. Spectra were recorded using 12 G magnetic field modulation at 100 kHz; other parameters are given in the figure legends. EPR simulations were carried out using matrix diagonalization (XSophe, Bruker Biospin), assuming a spin Hamiltonian H = βg.H.S + S.D.S. Enzyme concentrations for EPR were typically 1 to 2 mM. Altered DapE enzymes were incubated with the appropriate amounts of Co(II) for 30 min at 25 °C then frozen in an EPR tube for analysis.
Results
Kinetic parameters of H349A and H67A altered DapE enzymes
The kinetic parameters for the His67A and H349A altered DapE enzymes were determined in the presence of three equivalents of Zn(II), after a one-hour incubation period, by monitoring the hydrolysis of l,l-SDAP at pH 7.5 (Table 1). The H67A altered DapE enzyme catalyzed the hydrolysis of l,l-SDAP with a kcat = 1.5 ± 0.5 sec−1 and Km = 1.4 ± 0.2 mM. No enzymatic activity was detected for H349A under the experimental conditions used. For comparison, the WT DapE enzyme from H. influenzae exhibits a kcat = 140 ± 1.0 sec−1 and Km = 0.73 ± 0.05 mM (Table 1).
Table 1.
Kinetic constants and metal ion content for the H349A, H65A, and WT DapE enzymes from H. influenzae.
| Kinetic Constants | H67A | H349A | WT |
|---|---|---|---|
| Km (mM) | 1.4 ± 0.2 | --- | 0.73 ± 0.05 |
| kcat (s−1) | 1.5 ± 0.5 | NDB | 149 ± 1.0 |
|
kcat/Km (M−1s−1) |
1,070 |
--- |
204,110 |
| Metal ContentA | 0.5 ± 0.1 | 0 | 1 ± 0.1 |
As purified.
None Detected.
Zn(II) binding to DapE altered enzymes
The number of tightly bound divalent metal ions per monomer for the “as purified” WT and altered DapE enzymes was determined by ICP-AES analysis (Table 1) (8). For H67A DapE, 0.5 ± 0.1 Zn(II) ions were found to be bound to the enzyme whereas no detectable Zn(II) ions were found in samples of the H349A enzyme. For comparison, WT DapE was found to contain 1.1 ± 0.1 tightly bound Zn(II) ions (8, 12).
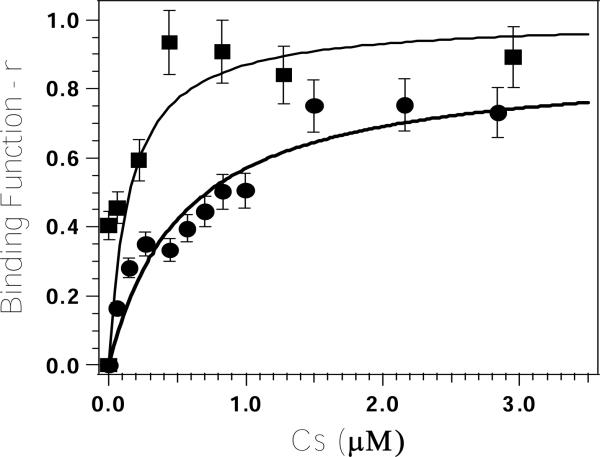
Titration of Zn(II) into apo-H67A DapE (at a concentration of 9 μM) revealed that ∼70% of the observed catalytic activity was recovered after the addition of only one equivalent of metal ion (8, 12). The dissociation constant (Kd) for this divalent metal binding site was obtained by fitting these titration data to equation 1:(22)
| (1) |
where p is the number of sites for which interaction with Zn(II) is governed by the intrinsic dissociation constant, Kd, Cs is the free metal concentration, and r is the binding function calculated by conversion of the fractional saturation (fa) (22). Values for Kd and p were obtained by fitting these data via an iterative process that allowed Kd and p to vary (Figure 2). The best fits obtained for Zn(II) binding to the H67A DapE from H. influenzae provided a p value of 0.9 and a Kd value of 0.54 ± 0.15 μM. A similar titration performed on the WT DapE from H. influenzae provided a p value of 1.0 and a Kd value of 0.14 ± 0.05 μM (12).
Figure 2.
Plot of Binding function-r, vs. Cs (the concentration of free metal ions in solution) for Zn(II) titration into apo-H67A altered DapE (circles) and WT DapE (squares) in 50 mM HEPES buffer, pH 7.5.
Electronic absorption spectra of Co(II)-loaded H67A and H349A DapE
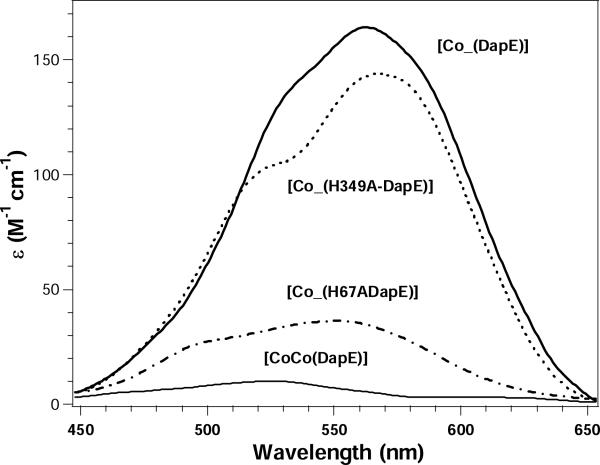
The binding of Co(II) to the H67A and H349A altered DapE enzymes from H. influenzae was investigated by electronic absorption spectroscopy (Figure 3). The addition of one equivalent of Co(II) to apo-H67A DapE (in Chelex-100 treated 50 mM HEPES buffer, pH 7.5) provided a visible absorption spectrum with a maximum at 550 nm (ε550 ∼35 M−1cm−1) along with a shoulder at 490 nm (ε490 ∼25 M−cm−). For the H349A altered DapE enzyme a maximum at 565 nm (ε565 ∼145 M−cm−) was observed along with a shoulder at 525 nm (ε525 ∼100 M−cm−).
Figure 3.
UV-Vis spectra of Co(II)-substituted WT, H67A, and H349A DapE enzymes at pH 7.5. The spectrum shown for [CoCo(DapE)] is the difference spectrum of the observed UV-Vis spectra of [Co_(DapE)] and [CoCo(DapE)]. All UV-Vis spectra were recorded at 25 °C with an enzyme concentration of 1 mM. All spectra were recorded under strict anaerobic conditions and the absorbance due to apo-DapE was subtracted.
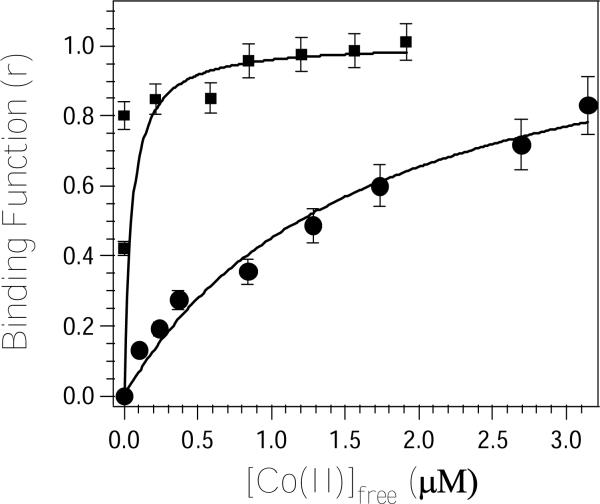
The Co(II)-binding properties of H349A and H67A DapE were also investigated using electronic absorption spectroscopy by titrating Co(II) into apo-H349A and apo-H67A DapE (1 mM) and monitoring the absorption at 565 and 550 nm, respectively. These data were fit via an iterative process to equation 1 allowing Kd and p to vary (Figure 4). The best fits obtained for H67A and H349A DapE provided p values of 1.2 and 1.0 and Kd values of 1,700 ± 100 and 50 ± 15 μM, respectively.
Figure 4.
Plot of Binding function-r, vs. Cs (the concentration of free metal ions in solution) of the molar absorptivities at 565 nm (circles) and 525 nm (squares) for a 65 μM DapE sample in 50 mM HEPES buffer, pH 7.5 titrated with Co(II).
EPR analysis of the H67A and H349A DapE enzymes
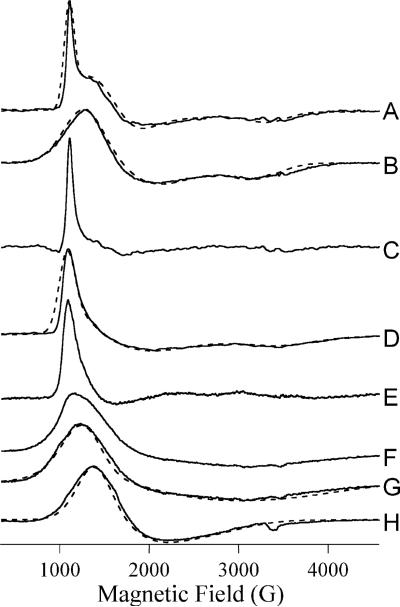
EPR spectra of both the mono- and dicobalt(II)-loaded H67A and H349A altered DapE enzymes from H. influenzae were recorded (Figure 5). Spectra recorded at multiple temperatures and microwave powers indicated that the spectrum of [Co_(H349A-DapE)] was complex and two distinct signals were obtained, a low temperature signal at 4 K (Figure 5D) and a high temperature signal at 15 K (Figure 5F). The low temperature signal was similar, but not identical, to that (Figure 5A) of [Co_(WT-DapE)]. The high temperature signal was broad and exhibited only one well-defined resonance position at 1,215 G. Simulation of the dominant component of the high temperature signal (Figure 5G) suggested uniformly low greal values if the signal is indeed due to a single chemical species. The EPR signal from [Co_(H67A-DapE)], in contrast, was essentially invariant from 4 − 15 K, consistent with a single chemical species (Figure 5H). Computer simulation was also consistent with a single EPR species. This signal was clearly distinct from WT DapE signals and was similar to the previously described signal from [Co_(E134D-DapE)] (17).
Figure 5.
EPR spectra of WT and altered DapE enzymes. Solid lines are experimental or experimentally-derived spectra and dashed lines are calculated spectra. Experimental spectra are from (A) [Co_(DapE)]; (B) [CoCo(DapE)]; (D) and (F) [Co_(H349A-DapE)]; and (H) [Co_(H67ADapE)]. (A), (B) and (H) were recorded at 10 K, 0.2 mW; (D) was recorded at 4 K, 0.2 mW and (F) was recorded at 15 K, 0.2 mW. (C) is a difference spectrum generated by subtraction of an arbitrary amount of (B) from (A); similarly (E) = (D) – (F) and (G) = (F) – (D). Simulations were calculated assuming , and D >> gβHS (50 cm−1). Other spin Hamiltonian parameters employed were (A), g⊥ = 2.60, g∥ = 2.18, E/D = 0.14, σ(E/D) = 0.06; (B) g⊥ = 2.32, g∥ = 2.22, E/D = 0.10; (D) g⊥ = 2.60, g∥ = 2.10, E/D = 0.16, σ(E/D) = 0.10; (G) g⊥ = 2.14, g∥ = 2.05, E/D = 0.15 and (H) g⊥ = 2.15, g∥ = 2.55, E/D = 0.08.
Modeling of the DapE structure
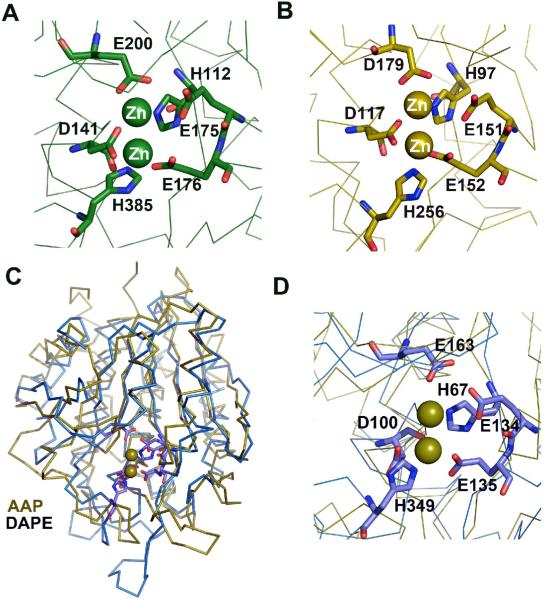
Homology modeling techniques were used to construct a three-dimensional model of the DapE from H. influenzae using the X-ray crystal structure of the DapE from N. meningitidis (1vgy) as a template (23, 24). Sequence analysis of the target and template show that these two proteins are of similar length, share broad sequence similarity (55% sequence identity and 72 % similarity), and exhibit no sequence gaps. The ModWeb-based homology building server was used to construct a structural homology model of the DapE from H. influenzae. Since the X-ray crystal structure of the DapE from N. meningitidis contains no metal ions, the modeled structure of the DapE from H. influenzae was superimposed on the structure of AAP using the SSM server (14). Both structures superimpose well with a Z-score of 5.45 rmsd with 2.46 Å resolution for 216 Cα atoms. The homology model of DapE reveals that within 4.0 Å of the Zn(II) binding sites of DapE, all of the amino acid residues are nearly identical to that observed in CPG2 and AAP (Figure 6) (14, 15).
Figure 6.
A) Close-up view of the catalytic center of CPG2 (15). B) Close-up view of the catalytic center of AAP (14). C) Superimposition of the structures of the catalytic domain of DapE (blue) and the structure of AAP (green). D) Close-up view of the catalytic center of homology modeled DapE from H. influenzae (blue) superimposed on the structure of AAP (olive). The proposed catalytic residues are shown as sticks whereas metal (olive color) positions are approximated based on the AAP structure and are shown in corresponding olive color.
Discussion
The meso-diaminopimelate (mDAP)/lysine biosynthetic pathway offers several potential anti-bacterial targets, such as DapE, that have yet to be explored (25-27). One of the products of this pathway, lysine, is required for protein biosynthesis and is also used in the peptidoglycan layer of Gram-positive bacterial cell walls. A second product, the amino acid meso-diaminopimelate (mDAP), is an essential component of the peptidoglycan layer for Gram-negative bacteria, providing a link between polysaccharide strands. However, a major limitation in developing a novel class of antibiotics that target DapE enzymes is the lack of knowledge about their active site structure (7, 8). The only X-ray crystal structure reported for a DapE enzyme is the apo-form (13) making it difficult to not only determine which residues constitute the catalytic active site but whether one or two metal ions are present and what their coordination geometry looks like. In order to shed some light on the active site structure of DapE enzymes and to verify that two histidine residues act as Zn(II) ligands, we have prepared the H67A and H349A altered DapE enzymes from H. influenzae. Both H67 and H349 are strictly conserved in DapE gene sequences and have been suggested to function as Zn(II) ligands (Figure 1) (8).
The specific activity of the H67A and H349A altered DapE enzymes from H. influenzae was determined in the presence of l,l-SDAP and three equivalents of Zn(II). Comparison of the kinetic parameters for the Zn(II)-loaded WT and H67A enzymes indicate that the decrease in activity of the H67A mutant is due to a decrease in kcat. The kcat value for the H67A altered DapE decreased by ∼95-fold. Interestingly, the Km value determined for the H67A altered enzyme only decreased by ∼2-fold compared to the WT DapE enzyme. Calculation of the catalytic efficiency (kcat/ Km) for the H67A altered enzyme results in a value ∼180-fold less than the wild type enzyme. The histidine to alanine mutation at this position, while having some impact on substrate binding, has primarily affected a key aspect of the mechanism of the protein.
Based on the ICP-AES data, the H67A mutated DapE still contains ∼0.5 molar equivalents of Zn(II) after extensive dialysis. With a Kd of ∼0.54 μM derived from Zn(II) activity titrations, some Zn(II) may have been lost to equilibrium effects during dialysis. Under saturating metal concentrations, this variant likely still binds one full equivalent of Zn(II), similar to the 1.1 equivalents of Zn(II) determined to be present in the WT DapE. The H349A variant, on the other hand, contained no significant quantities of Zn(II). This data implies that the remaining metal binding site in H349A DapE has a much larger Kd, and is unable to bind metal in the absence of saturating concentrations of Zn(II).
The hydrolysis of L,L-SDAP by the H349A altered enzyme could not be detected under the assay conditions employed. The lack of measurable activity for the H349A mutant enzyme may be the result of the reaction conditions required for detecting enzymatic activity. Since the amide bond cleavage of l,l-SDAP is monitored at 225 nm, enzyme concentrations must be < 10 μM in order to not overwhelm the amide bond absorption with background protein absorption. The impaired ability of this variant to bind Zn(II) is also consistent with a severely reduced/eliminated enzymatic activity. This further substantiates the hypothesis that this point mutation has knocked out a key metal ligand. If a reduction in substrate affinity and/or the ability to bind Zn(II) is affected by alteration of H349, catalytic activity will also be difficult to detect due to the limited solubility of l,l-SDAP. Taken together, these kinetic data indicate that important residues, consistent with the proposal that they function as active site Zn(II) ligands.
In order to determine the effects of altering H67 and H349 on the metal binding properties of the DapE from H. influenzae, the number of tightly bound metal ions and their corresponding Kd values were determined and compared to WT DapE. Activity titration of apo-H67A DapE with Zn(II) provided a Kd value of 0.54 μM. Since only one metal ion is bound, this Kd value corresponds to the microscopic binding constant for the binding of a single metal ion to H67A DapE. This Kd value is ∼4-fold weaker than the Kd value obtained for WT DapE (12) but is similar to Zn(II) Kd values reported for other Zn(II) enzymes that contain mixed histidine-carboxylate active sites (28-30). The ability of the H67 and H349 altered DapE enzymes from H. influenzae to bind Co(II) was also investigated by titrating Co(II) into apo-DapE samples and monitoring the molar absorptivity of the λmax for the Co(II) d-d bands. The Kd value determined for Co(II) binding to H67A DapE is 1,700 μM while the value determined for H349A is 50 μM. Since alteration of either H67 or H349 will greatly perturb and likely inhibit the ability of their respective metal binding sites to bind metal ions, these Kd values likely correspond to a single metal binding event. Interestingly, the Kd value determined for Co(II) binding to the H67A altered DapE enzyme is ∼3,150 times weaker than the Kd value obtained for Zn(II) binding. Moreover, the Kd value obtained for Co(II) binding to H349A is 6 times stronger than that obtained for WT DapE. These data suggest that Co(II) and Zn(II) bind significantly differently to the histidine altered DapE enzymes. This difference is likely due to Zn(II)'s preference for four-coordinate geometries vs. Co(II)'s preference for five or six-coordinate structures (31). Alternatively, these data may suggest that Zn(II) and Co(II) bind preferentially to different sides of the dinuclear active site.
Structural and electronic properties of the H67A and H349A altered DapE enzymes from H. influenzae were gleaned from electronic absorption and EPR spectroscopy. Upon the addition of one equivalent of Co(II) to H67A altered DapE ([Co_(H67A-DapE)]), two resolvable d-d transitions at 490 and 550 nm (ε = 25 and 35 M−cm−1, respectively) are observed. Ligand-field theory predicts that optical transitions of four-coordinate Co(II) complexes give rise to intense absorptions (ε > 300 M−cm−) in the higher wavelength region (625 ± 50 nm) due to a comparatively small ligand-field stabilization energy, while transitions of octahedral Co(II) complexes have very weak absorptions (ε < 30 M−cm−) at lower wavelengths (525 +/−50 nm). Five-coordinate Co(II) centers show intermediate features: i.e. moderate absorption intensities (50 < ε < 250 M−cm−) with several maxima between 525 and 625 nm (32, 33). Based on the molar absorptivities and d-d absorption maxima of [Co_(H67A-DapE)] the resulting metal binding site is likely a distorted five- or six-coordinate geometry. On the other hand, upon the addition of one equivalent of Co(II) to H349A altered DapE, two resolvable d-d transitions at 525 and 565 nm (ε = 100 and 145 M−cm−, respectively) are observed suggesting the metal binding site is likely a distorted five-coordinate geometry. For comparison, the WT DapE from H. influenzae in the presence of one equivalent of Co(II) exhibits a maximum at 562 nm (ε562 ∼ 165 M−1cm−1) with a shoulder at 535 nm (ε535 ∼135 M−1cm−1) while the addition of a second equivalent of Co(II) to WT DapE increases the absorption intensities at 562 and 535 nm by only ∼10 M−1cm−1 suggesting that the second metal binding site is octahedral (12).
EPR analysis is consistent with the electronic absorption data. The EPR spectrum of [Co_(H349A)] clearly indicated two components, one component observed at low temperature and a second at higher temperature. The low temperature component was simulated as a single species with parameters consistent with a five-coordinate Co(II) center, although significant strain in E/D was required to obtain the unusual line shape. The persistence of this signal at 4K was suggestive, however, of an signal (34, 35) and subtraction of the high temperature signal yielded a difference spectrum (Figure 5E) characterized by a spike at geff. ∼6.3, consistent with and a distorted tetrahedral geometry. The difference spectrum was not sufficiently devoid of other features to be conclusive but prompted a reanalysis of the hitherto incompletely described data from WT DapE. The spectrum of [Co_(WT-DapE)] could also be simulated as a single, likely five-coordinate, species (Figure 2A). However, the coincidence of some resonance positions with those from [CoCo(WT-DapE)] suggested that in [Co_(WT-DapE)] a nominally small proportion of the molecules are actually [CoCo(WT-DapE)]. Subtraction of the spectra yielded a difference spectrum with a sharp spike at g ∼ 6.3 and little other EPR absorption. The overall similar EPR behavior of [Co_(WT-DapE)] and [Co_(H349A-DapE)], and the clearer data for [Co_(WT-DapE)], suggest that both [Co_(WT-DapE)] and [Co_(H349A-DapE)] contain some tetrahedral Co(II) and some Co(II) of higher coordination. The EPR spectrum from [Co_(H67A-DapE)], in contrast, was very different from that of [Co_(WT-DapE)] and resembled that from [Co_(E134D-DapE)] and, to a lesser extent, from [CoCo(WT-DapE)]. The broad lines and low E/D suggest relatively relaxed six-coordinate geometry with water molecules occupying non-protein coordination sites.
The EPR and electronic absorption data indicate very clearly that the Co(II) in H349A-DapE is analogous to the first-binding Co(II) in WT DapE whereas the Co(II) in H67-DapE is very different from the first Co(II) but is similar to the second-binding Co(II) in WT DapE. These data, along with the kinetic data, are consistent with the assignment of H67 and H349 as active site ligands for the DapE from H. influenzae and that H67 is a ligand in the first metal binding site while H349 resides in the second metal binding site. While the current lack of structural data preclude definitive assignment of the active site residues, a three-dimensional homological structure of the DapE from H. influenzae, generated using the X-ray crystal structure of the DapE from N. meningitidis as a template and superimposed on the structure of AAP, confirms the assignment of H67 and H349 as active site ligands. The superimposition of the homology model of DapE with the dizinc(II) structure of AAP indicates that within 4.0 Å of the Zn(II) binding sites of AAP, all of the amino acid residues of DapE are nearly identical. Based on this model, the H67 residue in the DapE from H. influenzae corresponds to H97 in AAP while H349 corresponds to H256 in AAP. However, the X-ray crystal structure of the butane boronic acid inhibited form of AAP provided evidence that H97 functions as a ligand in the second metal binding site (36). This apparent reversal of the metal ion roles for DapE vs. AAP may be the result of the bulky methyl group of alanine altering divalent metal ion binding or, more likely, indicate that the catalytic roles of the active site metal ions in DapE's are reversed from that proposed for AAP. The latter suggestion is not necessarily surprising since AAP cleaves from the N-terminus while DapE cleaves the equivalent of a C-terminal carboxylate group.
Acknowledgments
†This work was supported by the National Science Foundation (CHE- 0652981, RCH) and the National Institutes of Health (AI056321, RR001980 BB). The Bruker ESP-300E EPR spectrometer was purchased with funds provided by the National Science Foundation (BIR-9413530).
Abbreviations
- DapE
dapE-encoded N-succinyl-l,l-diaminopimelic acid desuccinylase
- AAP
aminopeptidase from Aeromonas proteolytica
- CPG2
carboxypeptidase from Pseudomonas sp strain-RS-16
- MetAPs
methionyl aminopeptidases
- SDAP
N-succinyl-diaminopimelic acid
- EDTA
Ethylenediaminetetraacetic acid
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- Tris
tris(hydroxymethyl)aminomethane
- SDS-PAGE
Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis
- HPLC
High Performance Liquid Chromatography
- ICP-AES
Inductively Coupled Plasma Atomic Emission Spectrometry
- EPR
Electron Paramagnetic Resonance
References
- 1.Henery CM. Antibiotic resistance. C&E News. 2000:41–58. [Google Scholar]
- 2.Prevention C. f. D. C. a. Hospital infection control practices advisory committee's recommendations for preventing the spead of vancomycin resistance. MMWR Morb. Mortal. Wkly Rep. 1995;44:1–13. [PubMed] [Google Scholar]
- 3.Levy SB. The challenge of antibiotic resistance. Sci. Am. 1998;278:46–53. doi: 10.1038/scientificamerican0398-46. [DOI] [PubMed] [Google Scholar]
- 4.Chin J. Resistance is useless. New Scientist. 1996;152:32–35. [Google Scholar]
- 5.Nemec A, Krízová L, Maixnerová M, Diancourt L, van der Reijden TJ, Brisse S, van den Broek P, Dijkshoorn L. Emergence of carbapenem resistance in Acinetobacter baumannii in the Czech Republic is associated with the spread of multidrug-resistant strains of European clone II. J. Antimicrob. Chemother. 2008 May 13; doi: 10.1093/jac/dkn205. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 6.Shih MJ, Lee NY, Lee HC, Chang CM, Wu CJ, Chen PL, Ko NY, Ko WC. Risk factors of multidrug resistance in nosocomial bacteremia due to Acinetobacter baumannii: a case-control study. J. Microbiol. Immunol. Infect. 2008;41:118–123. [PubMed] [Google Scholar]
- 7.Velasco AM, Leguina JI, Lazcano A. Molecular evolution of the lysine biosynthetic pathways. J. Mol. Evol. 2002;55:445–459. doi: 10.1007/s00239-002-2340-2. [DOI] [PubMed] [Google Scholar]
- 8.Born TL, Zheng R, Blanchard JS. Hydrolysis of N-succinyl-L,-Ldiaminopimelic acid by the Haemophilus influenzae dapE-encoded desuccinylase: metal activation, solvent isotope effects, and kinetic mechanism. Biochemistry. 1998;37:10478–10487. doi: 10.1021/bi9806807. [DOI] [PubMed] [Google Scholar]
- 9.Karita M, Etterbeek ML, Forsyth MH, Tummuru MR, Blaser MJ. Characterization of Helicobacter pylori dapE and construction of a conditionally lethal dapE mutant. Infect. Immun. 1997;65:4158–4164. doi: 10.1128/iai.65.10.4158-4164.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pavelka MS, Jacobs WR. Biosynthesis of diaminopimelate, the precursor of lysine and a component of peptidoglycan, is an essential function of Mycobacterium smegmatis. J. Bacteriol. 1996;178:6496–6507. doi: 10.1128/jb.178.22.6496-6507.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouvier J, Richaud C, Higgins W, Bögler O, Stragier P. Cloning, characterization, and expression of the dapE gene of Escherichia coli. J. Bacteriol. 1992;174:5265–5271. doi: 10.1128/jb.174.16.5265-5271.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bienvenue DL, Gilner DM, Davis RS, Bennett B, Holz RC. Substrate Specificity, Metal Binding Properties, and Spectroscopic Characterization of the dapE-encoded-N-succinyl-L,L-Diaminopimelic Acid Desuccinylase from Haemophilus influenzae. Biochemistry. 2003;42:10756–10763. doi: 10.1021/bi034845+. [DOI] [PubMed] [Google Scholar]
- 13.Badger J, Sauder JM, Adams JM, Antonysamy S, Bain K, Bergseid MG, Buchanan SG, Buchanan MD, Batiyenko Y, Christopher JA, Emtage S, Eroshkina A, Feil I, Furlong EB, Gajiwala KS, Gao X, He D, Hendle J, Huber A, Hoda K, Kearins P, Kissinger C, Laubert B, Lewis HA, Lin J, Loomis K, Lorimer D, Louie G, Maletic M, Marsh CD, Miller I, Molinari J, Muller-Dieckmann HJ, Newman JM, Noland BW, Pagarigan B, Park F, Peat TS, Post KW, Radojicic S, Ramos A, Romero R, Rutter ME, Sanderson WE, Schwinn KD, Tresser J, Winhoven J, Wright TA, Wu L, Xu J, Harris TJ. Structural analysis of a set of proteins resulting from a bacterial genomics project. Proteins. 2005;60:787–796. doi: 10.1002/prot.20541. [DOI] [PubMed] [Google Scholar]
- 14.Chevrier B, Schalk C, D'Orchymont H, Rondeau J-M, Moras D, Tarnus C. Crystal structure of Aeromonas proteolytica aminopeptidase: a prototypical member of the co-catalytic zinc enzyme family. Structure. 1994;2:283–291. doi: 10.1016/s0969-2126(00)00030-7. [DOI] [PubMed] [Google Scholar]
- 15.Rowsell S, Pauptit RA, Tucker AD, Melton RG, Blow DM, Brick P. Crystal structure of carboxypeptidase G2, a bacterial enzyme with applications in cancer therapy. Structure. 1997;5:337–347. doi: 10.1016/s0969-2126(97)00191-3. [DOI] [PubMed] [Google Scholar]
- 16.Cosper NJ, Bienvenue DL, Shokes J, Gilner DM, Tsukamoto T, Scott* R, Holz* RC. The dapE-encoded N-succinyl-L,L-Diaminopimelic Acid Desuccinylase from Haemophilus influenzae is a Dinuclear Metallohydrolase. J. Am. Chem. Soc. 2004;125:14654–14655. doi: 10.1021/ja036650v. [DOI] [PubMed] [Google Scholar]
- 17.Davis R, Bienvenue D, Swierczek SI, Gilner DM, Rajagopal L, Bennett B, Holz RC. Kinetic and spectroscopic characterization of the E134A- and E134D-altered dapE-encoded N-succinyl-L,L-diaminopimelic acid desuccinylase from Haemophilus influenzae. J Biol Inorg Chem. 2006;11:206–216. doi: 10.1007/s00775-005-0071-8. [DOI] [PubMed] [Google Scholar]
- 18.Makarova KS, Grishin NV. The Zn-peptidase superfamily: functional convergence after evolutionary divergence. J. Mol. Biol. 1999;292:11–17. doi: 10.1006/jmbi.1999.3059. [DOI] [PubMed] [Google Scholar]
- 19.Bergmann M, Stein WH. J. Biol. Chem. 1939;129:609–618. [Google Scholar]
- 20.Lin Y, Myhrman R, Schrag ML, Gelb MH. Bacterial N-succinyl-L-diaminopimelic acid desuccinylase. Purification, partial characterization, and substrate specificity. J. Biol. Chem. 1988;263:1622–1627. [PubMed] [Google Scholar]
- 21.D'souza VM, Bennett B, Copik AJ, Holz RC. Characterization of the Divalent Metal Binding Properties of the Methionyl Aminopeptidase from Escherichia coli. Biochemistry. 2000;39:3817–3826. doi: 10.1021/bi9925827. [DOI] [PubMed] [Google Scholar]
- 22.Winzor DJ, Sawyer WH. Quantitative Characterization of Ligand Binding. Wiley-Liss; New York: 1995. [Google Scholar]
- 23.Eswar N, John B, Mirkovic N, Fiser A, Ilyin VA, Pieper U, Stuart AC, Marti-Renom MA, Madhusudhan MS, Yerkovich B, Sali A. Tools for comparative protein structure modeling and analysis. Nucleic Acids Res. 2003;31:3375–3380. doi: 10.1093/nar/gkg543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr. 2004;60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 25.Scapin G, Blanchard JS. Enzymology of bacterial lysine biosyntesis. Adv. Enzymol. 1998;72:279–325. doi: 10.1002/9780470123188.ch8. [DOI] [PubMed] [Google Scholar]
- 26.Born TL, Blanchard JS. Structure/function studies on enzymes in the diaminopimelate pathway of bacterial cell wall synthesis. Cur. Opin. Chem. Biol. 1999;3:607–613. doi: 10.1016/s1367-5931(99)00016-2. [DOI] [PubMed] [Google Scholar]
- 27.Girodeau J-M, Agouridas C, Masson M, R., P., LeGoffic F. The lysine pathway as a target for a new genera of synthetic antibacterial antibiotics? J. Med. Chem. 1986;29:1023–1030. doi: 10.1021/jm00156a021. [DOI] [PubMed] [Google Scholar]
- 28.Prescott JM, Wilkes SH. Aeromonas Aminopeptidase. Methods Enzymol. 1976;45:530–543. doi: 10.1016/s0076-6879(76)45047-4. [DOI] [PubMed] [Google Scholar]
- 29.Fleminger G, Yaron A. Soluble and immobilized clostridial aminopeptidase and aminopeptidase P as metal-requireing enzymes. Biochim. Biophys. Acta. 1984;789:245–256. doi: 10.1016/0167-4838(84)90180-8. [DOI] [PubMed] [Google Scholar]
- 30.de Seny D, Heinz U, Wommer S, Kiefer M, Meyer-Klaucke, Galleni M, Frere J-M, Bauer R, Adolph H-W. Metal ion binding and coordination geometry for wild-type and mutants of the metallo-β-lactamase from Bacillus cereus 569/H/9 (BcII) J. Biol. Chem. 2001;276:45065–45078. doi: 10.1074/jbc.M106447200. [DOI] [PubMed] [Google Scholar]
- 31.Holz RC. The Aminopeptidase from Aeromonas proteolytica: Structure and Mechanism of Co-Catalytic Metal Centers Involved in Peptide Hydrolysis. Coord. Chem. Rev. 2002;232:5–26. [Google Scholar]
- 32.Bertini I, Luchinat C. High-spin cobalt(II) as a probe for the investigation of metalloproteins. Adv. Inorg. Biochem. 1984;6:71–111. [PubMed] [Google Scholar]
- 33.Horrocks W, DeW I, Jr., Holmqust B, Thompson JS. J. Inorg. Chem. 1980;12:131–141. doi: 10.1016/s0162-0134(00)80124-5. [DOI] [PubMed] [Google Scholar]
- 34.Huntington KM, Bienvenue D, Wei Y, Bennett B, Holz RC, Pei D. Slow-Binding Inhibition of the Aminopeptidase from Aeromonas proteolytica by Peptide Thiols: Synthesis and Spectral Characterization. Biochemistry. 1999;38:15587–15596. doi: 10.1021/bi991283e. [DOI] [PubMed] [Google Scholar]
- 35.Crawford PA, Yang K-W, Sharma N, Bennett B, Crowder MW. Spectroscopic Studies on Cobalt(II)-Substituted Metallo-β-lactamase ImiS from Aeromonas veronii bv. sobria. Biochemistry. 2005;44:5168–5176. doi: 10.1021/bi047463s. [DOI] [PubMed] [Google Scholar]
- 36.DePaola C, Bennett B, Holz RC, Ringe D, Petsko G. 1-Butaneboronic Acid Binding to Aeromonas proteolytica Aminopeptidase: A Case of Arrested Development. Biochemistry. 1999;38:9048–9053. doi: 10.1021/bi9900572. [DOI] [PubMed] [Google Scholar]