Abstract
In an effort to probe the role of the Zn(II) sites in metallo-β-lactamase L1, mononuclear metal ion containing and heterobimetallic analogs of the enzyme were generated and characterized using kinetic and spectroscopic studies. Mononuclear Zn(II)-containing L1, which binds Zn(II) in the consensus Zn1 site, was shown to be slightly active; however, this enzyme did not stabilize a nitrocefin-derived reaction intermediate that had been previously detected. Mononuclear Co(II)- and Fe(III)-containing L1 were essentially inactive, and NMR and EPR studies suggest that these metal ions bind to the consensus Zn2 site in L1. Heterobimetallic analogs (ZnCo and ZnFe) analogs of L1 were generated, and stopped-flow kinetic studies revealed that these enzymes rapidly hydrolyze nitrocefin and that there are large amounts of the reaction intermediate formed during the reaction. The heterobimetallic analogs were reacted with nitrocefin, and the reactions were rapidly freeze quenched. EPR studies on these samples demonstrate that Co(II) is five-coordinate in the resting state, proceeds through a four-coordinate species during the reaction, and is five-coordinate in the enzyme-product complex. These studies demonstrate that the metal ion in the Zn1 site is essential for catalysis in L1 and that the metal ion in the Zn2 site is crucial for stabilization of the nitrocefin-derived reaction intermediate.
INTRODUCTION
β-Lactam containing compounds are the most widely used antibiotics, and they exert their antimicrobial activity by inhibiting the crosslinking of the peptidoglycan building blocks of bacterial cell walls1. Ever since the introduction of these antibiotics in the clinic, there have been an increasing number of bacterial strains that are resistant to these drugs. The most common way that bacteria become resistant to β-lactams is through the production of β-lactamases, which cleave the β-lactam bond and inactivate the drug2. There are 500 known β-lactamases, and these enzymes have been classified into 4 groups3. Although groups A, C, and D exhibit different substrate specificities and susceptibilities to clinical inhibitors, they are similar in the fact that they utilize an active site serine as a nucleophile to attack the β-lactam carbonyl, generating a tetrahedral intermediate1. The group B enzymes, on the other hand, require 1−2 Zn(II) ions to hydrolyze β-lactams and thus are called metallo-β-lactamases (Mβls)4. Mβls have been further categorized into three subgroups according to amino acid homology, substrate preference, and the number of Zn(II) ions required for full activity. The B1 subgroup, represented by CcrA, BcII, and IMP-1, have two metal binding sites: Zn1, which consists of three histidines and a bridging hydroxide to coordinate Zn(II), and Zn2, which consists of one histidine, one aspartate, one cysteine, the bridging hydroxide, and a terminally-bound H2O to coordinate Zn(II). The B2 enzymes, represented by CphA and ImiS, bind Zn(II) at the consensus Zn2 site, which contains one histidine, one aspartate, one cysteine, and a solvent molecule to coordinate Zn(II). The B3 enzymes, represented by L1 and FEZ bind two Zn(II) ions, contain the same Zn1 site as the B1 enzymes, and utilize a Zn2 site, which consists of two histidines, one aspartate, one terminally-bound water, and the bridging hydroxide. Recently, a B2/B3 hybrid, metallo-β-lactamase GOB from E. meningoseptica, binds only 1 Zn(II) in the Zn2 site5.
There exists considerable controversy about the metal content of the nominally dinuclear Zn(II)-containing (B1 and B3) Mβl's. The initial crystal structure of BcII showed a single Zn(II) ion in the Zn1 site of the enzyme6; however, subsequent structures have shown a dinuclear Zn(II) site in BcII7,8. Similar conflicting data on the metal content of L1, IMP-1, and CcrA have not been reported; however, Wommer et al. used in vitro binding assays to predict that all Mβl's are metal-free in vivo and become mononuclear enzymes only in the presence of substrate9. Wommer et al. continued by concluding that dinuclear Zn(II)-containing Mβl's are isolation artifacts. Nonetheless, Page and coworkers have recently reported that BcII containing only one equivalent of Co(II) is inactive, and the loss of metal ion during catalysis is the reason for the burst kinetics exhibited by this enzyme10-12. In constrast, Vila and coworkers have recently published that the BcII containing only one equivalent of Zn(II) is catalytically-active13,14 and that B3 subgroup member, GOB, requires only a single metal ion in the Zn2 to be active, analogous to the B2 enzymes5.
The metal ion binding characteristics of L1, CcrA, and BcII have been investigated and characterized4,15. However, of much greater interest is the nature of the metal ion complement that is required for activity and the roles in catalysis, if any, of each of the metal ions that can be accommodated by these enzymes. The study of mononuclear or mixed-metal analogs of the enzymes provides one mechanism for the elucidation of the role of each metal and to indicate whether one or both are essential for activity. In addition, this information could be used to guide rational drug design efforts that use the Zn1, Zn2, or both sites as targets for inhibitors.
In this work, we describe the preparation and characterization of mononuclear metal ion and mixed-metal containing analogs of Mβl L1 from Stenotrophomonas maltophilia; kinetic, spectroscopic and spectrokinetic analyses of these species reveal roles for both metal ions in catalysis.
MATERIALS AND METHODS
Materials
E. coli strains DH5 and BL21(DE3)pLysS were purchased from Gibco BRL (Gaithesberg, MD) and Novagen (Madison, WI), respectively. Plasmids pET26b(+) and pUC19 were purchased from Novagen. Restriction enzymes, NcoI and HindIII, deoxynucleotides (dNTPs), thermopol buffer, MgSO4, and T4 DNA ligase were obtained from New England Biolabs (Beverly, MA), Promega Corporation (Madison, WI), and Gibco BRL. QuikChange site-directed mutagenesis kit was purchased from Stratagene. All mutagenic primers were purchased from Integrated DNA Technologies (IDT, Coralville, IA). Polymerase Chain Reaction (PCR) was performed using a Thermolyne Amplitron II from Barnstead (Dubuque, IA). DNA purification was performed by using a Qiagen Quick Gel Extraction kit (Velencia, CA). The QIAGEN-tip 100 kit and protocols were used for large-scale plasmid purifications. A Wizard Plus Miniprep kit from Promega was used for small-scale plasmid DNA preparations. Luria-Bertani (LB) media was purchased from Invitrogen (Carlsbad, CA). Isopropyl-β-D-thiogalactoside (IPTG) was purchased from Anatrace (Maumee, OH). All buffer solutions were prepared using chemicals purchased from Fisher Scientific (Pittsburgh, PA). All buffers and growth media were made with Barnstead NANOpure, ultrapure water. For metal-free solutions, Chelex 100 resin (Biorad Laboratories, Hercules, CA) was used, and the resulting solutions were filtered through a 0.45-micron filter membrane (Osmonic Inc.). Dialysis tubing was prepared as per Sambrook et al.16 from Spectro/Por regenerated cellulose, molecular porous membranes with a molecular weight cut off of 10,000 Da (Spectrum Corporation, Gardena, CA). A Fast Protein Liquid Chromatography (FPLC) system, chromatography columns, and resins were purchased from GE Healthcare. Nitrocefin was obtained from Becton Dickinson Microbiology System (Cockeysville, MD), and solutions of nitrocefin were prepared as previously described17.
Generation of histidine mutants of L1 by site-directed mutagenesis
The over-expression plasmids of four His→Cys L1 mutants, H116C, H118C, H121C and H196C were constructed using the L1 over-expression plasmid pET26b(+), which yields the full length form of the enzyme, and the QuikChange site-directed mutagenesis kit as per the instructions of the manufacturer. The following primers were used to generate the mutants:
H116Cfor CGGCTGATCCTGCTCAGCTGCGCACACGCCGACCATGCC
H116Crev GGCATGGTCGGCGTGTGCGCAGCTGAGCAGGATCAGCCG
H118C for ATCCTGCTCAGCCACGCATGCGCCGACCATGCCGGACCG
H118Crev CGGTCCGGCATGGTCGGCGCATGCGTGGCTGAGCAGGAT
H121Cfor CACGCACACGCCGACTGCGCCGGACCGGTGGCG
H121Crev CGCCACCGGTCCGGCGCAGTCGGCGTGTGCGTG
H196Cfor CACTTCATGGCGGGGTGCACCCCGGGCAGCACCGCG
H196Crev CGCGGTGCTGCCCGGGGTGCACCCCGCCATGAAGTG
H225Cfor GTGTTGCTGACACCGTGCCCGGGTGCCAGCAAC
H225Crev GTTGCTGGCACCCGGGCACGGTGTCAGCAACAC
Over-expression of His→Cys mutants
Large scale preparations of His→Cys L1 mutants were conducted by using the procedure of Crowder et al.17. L1 was quantitated by monitoring the absorbance at 280 nm and using an extinction coefficient of 54,600 M−1cm−117.
Preparation of 1Zn-, 1Co-, 1Fe-L1
Mature L1 (M-L1) was over-expressed as previously described by adding 100 μM ZnCl2, CoCl2, or Fe(NH4)2(SO4)2 to the minimal medium18. After protein over-expression and centrifugation to collect the E. coli cells, the pellet was resuspended in 300 mL of 50 mM Hepes, pH 6.0, and the suspension was centrifuged for 15 minutes (8200xg). The resulting pellet was resuspended in 50 mM Hepes, pH 6.0, and the cells were lysed by using a French press as previously described17. The cleared supernatant (centrifugation for 25 minutes at 23,400g) was loaded onto a 25 mL SP-Sepharose column that was equilibrated with 50 mM Hepes, pH 6.0, and bound proteins were eluted from the column using a linear 0 − 500 mM NaCl gradient in the same buffer. L1 typically eluted at 80−120 mM NaCl, and the fractions were analyzed for the presence of L1 by using SDS-PAGE, as previously described17.
Preparation of ZnCoL1, ZnFeL1, FeFeL1, and CoCoL1 samples
The ZnFe analog of L1 was prepared by adding 3 equivalents of Zn(II) to as-isolated 1FeL1 or 3 equivalents of Fe(II) to as-isolated 1ZnL1, followed by dialysis against 4 × 1L of Chelex-treated 50 mM Hepes, containing 50 mM NaCl, to remove unbound metal. The ZnCo analog was prepared by adding 3 equivalents of Zn(II) to 1CoL1. The FeFe- and CoCo- analogs of L1 were prepared by refolding apo-L1 in the presence of 100 μM Fe(II) or Co(II), as recently described18.
Metal analyses
The metal content of the protein samples was determined by using a Varian Liberty 150 Inductively Coupled Plasma spectrometer with atomic emission spectroscopy detection (ICP-AES). All the proteins were diluted to 10 μM with 50 mM Hepes, pH 7.0. A calibration curve with 4 standards and a correlation coefficient of greater than 0.999 was generated using Zn(II), Fe, and Co(II) reference solutions from Fisher Scientific. The following emission wavelengths were chosen to ensure the lowest detection limits possible: Zn(II), 213.856 nm, Fe, 259.940 nm, and Co(II), 238.892 nm.
1H NMR spectroscopy
1H NMR spectra were collected on a Bruker Avance 500 spectrometer operating at 500.13 MHz, 298 K, magnetic field of 11.7 T, recycle delay (AQ) of 41 ms, and sweep width of 400 ppm. Proton chemical shifts were calibrated by assigning the H2O signal the value of 4.70 ppm. A modified presaturation pulse sequence (zgpr) was used to suppress the proton signals originating from solvent. The presaturation pulse was as short as possible (500 ms) to avoid saturation of solvent-exchangeable proton signals. The concentration of NMR samples was generally in the range of 1.0 − 1.2 mM. Samples in D2O were prepared by performing three or more dilution/concentration cycles in a Centricon-10.
Rapid-freeze-quench (RFQ) and EPR spectroscopy
L1 (0.5 mM) was reacted with 1.5 mM nitrocefin in 50 mM cacodylate buffer, pH 7.0, and at 3 ± 1 °C, and the reaction mixture was freeze-quenched for EPR spectroscopy using a system described in earlier work 19, 20; the calibrated reaction time was 10.4 ± 0.5 ms. Following EPR data collection, some samples were thawed by agitation of the sample tubes in water at 25 °C for 2 min and refrozen in liquid nitrogen. Low temperature EPR spectroscopy was carried out using a Bruker EleXsys E600 spectrometer equipped with an Oxford Instruments ITC503 liquid helium flow system. EPR was recorded at 9.63 GHz (B0⊥B1) or 9.37 GHz (B0∥B1) using an ER4116DM dual-mode cavity, with 100 kHz magnetic field modulation. Other EPR recording parameters are given in the legends to figures.
Steady-state kinetics
All kinetic studies were conducted on a Agilent 8453 UV-Vis diode array spectrophotometer at 25 °C. Steady-state kinetic parameters, the Michaelis constant Km and the turnover number kcat, were determined by monitoring product formation at 485 nm using nitrocefin as substrate in 50 mM Chelex-treated, cacodylate, pH 7.0. The rate of change in the absorbance at 485 nm was converted into the rate of change in the concentration of the product by dividing the absorbance (path length = 1 cm) by the extinction coefficient of the product 17,420 M−1cm−1 17.
Stopped-flow kinetic studies
Stopped-flow kinetic experiments were performed on an Applied Photophysics SX18MV spectrophotometer equipped with a constant temperature circulating water bath as previously described21-23. All experiments were performed in 50 mM Chelex-treated, cacodylate buffer, pH 7.0, at 10 °C. All the proteins were diluted with 50 mM Chelex-treated, cacodylate buffer to 100 μM, and the substrate was prepared and diluted to 100 μM in the same buffer.
RESULTS
Mutations of metal binding histidines
Our previous attempts to prepare a mixed-metal analog of L1 by adding Co(II) to apo- or 1Zn-L1 were unsuccessful primarily due to the oxidation of Co(II) to Co(III)24. Another potential problem with generating a mixed-metal analog is the reported dissociation constants for metal binding to the Zn1 and Zn2 sites in L1. Wommer et al. reported that the Zn(II) binding constants to the two sites in L1 are 2.6 and 6 nM9. This result suggests that the addition of different metal ions to apo-L1 would result in sample with mixtures of possible metal centers. Therefore, we attempted to prepare a mixed-metal analog of L1 by weakening one of the metal binding sites through mutation of one of the histidine groups in each metal binding site. For example, we reasoned that the mutation of His116 to Cys in the Zn1 site would result in a mutant that binds the first added metal ion tightly to the Zn2 site and the second metal ion much less tightly to the Zn1 site.
Five metal binding mutants of L1 (H116C, H118C, H121C, H160C, and H263C) were successfully prepared using nondegenerate oligonucleotides, the QuikChange Site Directed Mutagenesis kit, and polymerase chain reaction. DNA sequencing of the resulting L1 genes in both directions was used to confirm that only the desired mutations were present. Small-scale growth cultures showed that all five mutants were over-expressed at levels comparable to that of wild-type L1. However, large-scale (4 L) over-expression and purification of these mutants showed that only the H116C and H121C mutants were soluble and could be purified.
The purified mutants were analyzed for metal binding. After purification, the H116C mutant was shown to bind 0.33 equivalents of Zn(II), while the H121C mutant bound 0.11 equivalents of Zn(II) (Table 1). The mutants were incubated with a 10 molar excess of Zn(II), and the resulting enzymes were then exhaustively dialyzed versus Chelex-treated buffer. Zn(II)-loaded H116C and H121C mutants were shown to bind 0.85 and 0.98 equivalents of Zn(II), respectively (Table 1), which is one-half of the metal bound by recombinant wild-type L1. The as-isolated and Zn(II)-loaded mutants were characterized by using steady state kinetic studies. As-isolated H116C and H121C mutants exhibited kcat values of < 0.01 s−1 when using nitrocefin as substrate; however, the Zn(II)-loaded H116C and H121C mutants exhibited kcat values of 0.38 and 2.3 s−1 and Km values of 20 and 72 μM, respectively. The inclusion of 100 μM Zn(II) in the steady-state kinetics assay buffer resulted in no change in the steady-state kinetic constants for the H116C mutant, and a kcat = 33 s−1 and a Km = 99 μM for the H121C mutant, when using nitrocefin as the substrate. While we were successful in preparing analogs of L1 with differential metal binding affinities for the Zn1 and Zn2 sites, one of the mutants exhibited very little activity (H116C) and the other exhibited a Km value that suggested a large change in the active site of the enzyme (H121C).
Table 1.
Steady state kinetic parameters and metal content of HXXC mutants of L1.
| Mutant | kcat (s−1) | Km (μM) | Zn(II) Content |
|---|---|---|---|
| H116Ca | < 0.01 | ND | 0.33 ± 0.01 |
| H121Ca | < 0.01 | ND | 0.11 ± 0.01 |
| H116Cb | 0.38 ± 0.01 | 20 ± 1 | 0.85 ± 0.05 |
| H121Cb | 2.3 ± 0.2 | 72 ± 25 | 0.98 ± 0.05 |
| H116Cc | 0.35 ± 0.01 | 18 ± 1 | NA |
| H121Cc | 33 ± 4 | 99 ± 15 | NA |
| As-isolated/wild-type L1c | 41±1 | 4±1 | 1.90 ± 0.01 |
as isolated
after adding 2 equivalents of Zn(II) and then dialysis
reaction with 100 μM Zn(II) in buffer
ND – not determined
NA – not applicable
Preparation and characterization of the ZnCo-analog of L1
Our initial attempts to prepare Co(II)-substituted L1 by biological incorporation were unsuccessful because of the oxidation of Co(II) to Co(III) presumably during protein purification24. In these studies, the gene for L1 contained a leader sequence that directed the export of over-expressed L1 into the periplasm of E. coli, and our recent studies strongly suggest that folding and metallation of L1 occurs in the periplasm18. In this same study, we demonstrated that the removal of the leader sequence from the L1 gene resulted in the enzyme being folded and metallated in the cytoplasm of E. coli. Significantly, the metal content of the resulting enzyme could be affected greatly by the addition of metal ions in the growth medium.
In an effort to prepare a Co(II)-substituted form of L1, we over-expressed L1 in minimal medium containing 100 μM CoCl2 using the L1 gene without the leader sequence. The resulting, purified enzyme (called 1Co-L1) was pink, and the color did not change up to two months in 4 °C. Metal analyses revealed that the protein bound 0.9 equivalents of cobalt and 0.1 equivalents of Zn(II) (Table 2). Steady-state kinetic studies revealed that the enzyme exhibited a kcat of 11 ± 1 s−1 and a Km of 4.3 ± 0.1 μM, when using nitrocefin as a substrate (Table 2). These steady-state kinetic constants are different than those of ZnZn-L1 (kcat of 39 s−1; Km of 5.9 μM); 1Zn-L1 (kcat of 30 s−1; Km of 5.5 μM), and CoCo-L1 (kcat of 63 s−1; Km of 20 μM) (Table 2). In addition, the kcat values exhibited by the mononuclear metal ion containing analogs are not one-half of those exhibited by the dinuclear metal ion containing analogs, suggesting that the samples of mononuclear metal ion containing analogs are not made up of one-half dinuclear metal ion containing analogs and one-half apo-enzymes.
Table 2.
Steady state kinetics of different metal bound analogs of L1
| Species | kcat (s−1) | Km (μM) | Metal Content |
|---|---|---|---|
| ZnZn-L1 | 39 ± 1 | 5.9 ± 0.5 | 2.0 ± 0.1 Zn(II) added |
| 1Zn-L1 | 30 ± 1 | 5.5 ± 0.7 | 1.0 ± 0.1 Zn(II) |
| 1Co-L1 | 11 ± 1 | 4.3 ± 0.1 | 0.9 ± 0.1 Co, 0.10 ± 0.01 Zn(II) |
| 1Fe-L1 | 2.6 ± 1 | 53 ± 25 | 0.9 ± 0.1 Fe, 0.20 ± 0.01 Zn(II) |
| CoCo-L1a | 63 ± 3 | 20 ± 1 | 1.80 ± 0.20 Co |
| FeFe-L1a | 0 | N/A | 1.90 ± 0.01 Fe |
| ZnCo-L1 | 26 ± 0.3 | 2.3 ± 0.1 | 1.0 ± 0.1 Zn(II); 1.0 ± 0.1 Co |
| ZnFe-L1b | 20 ± 2 | 3.6 ± 1 | 1.0 ± 0.1 Zn(II); 1.0 ± 0.1 Fe |
| ZnFe-L1c | 24 ± 3 | 4.0 ± 1 | 1.2 ± 0.1 Zn(II); 0.9 ± 0.1 Fe |
data from Hu et al. 26
Fe added to Zn-L1 and then dialysis
Zn(II) added to Fe-L1 and then dialysis
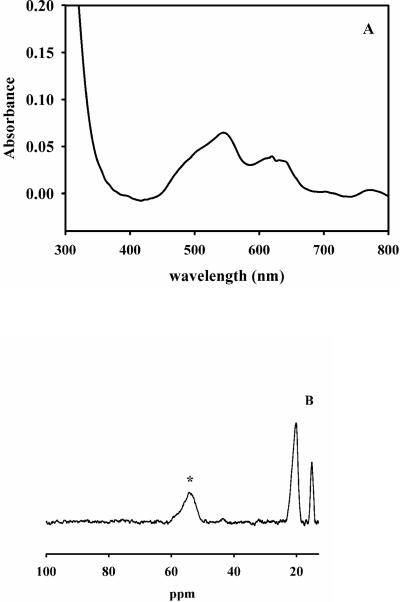
The UV-Vis difference spectrum of 1Co-L1 revealed a broad, weak peak between 500 − 650 nm (Figure 1A), which was assigned to ligand field transitions of high-spin Co(II), and the extinction coefficient at 550 nm was 130 M−1cm−1, which suggests that the Co(II) is 5-coordinate25. This spectrum is different than that of CoCo-L1, which was prepared by adding Co(II) to TCEP (tris(2-carboxyethyl)phosphine)-treated apo-L1, in that there is no broad absorbance peak between 330−360 nm corresponding to a S to Co(II) ligand to metal charge transfer band26. The addition of 1 eq. of Zn(II) to 1Co-L1 did not change the UV-Vis spectrum (Figure 1A).
Figure 1. UV-Vis and NMR spectra of 1Co-L1.
A: UV-Vis difference spectrum of 1Co-L1 prepared using biological incorporation method. The enzyme concentration was 550 μM, and the buffer was 50 mM Hepes, pH 7.0. B: 1H NMR spectrum of 550 μM 1Co-L1. The asterisk signifies the peak that is solvent-exchangeable.
The 1H NMR spectrum of 1Co-L1 showed one broad peak, which integrated to 2 protons, at 50 ppm, and the peak was solvent-exchangeable (Figure 1C). Since there are two histidines in the Zn2 site and three histidines in the Zn1 site27, we assign these peaks to the NH protons on Co(II)-bound His121 and His263, which indicates the Co(II) is bound to the Zn2 site in L1. The addition of 1 eq. of Zn(II) to 1Co-L1 did not change the NMR spectrum (Figure 1C).
Previously, we reported that 1Zn-L1 could be prepared by addition of 1 equivalent of Zn(II) to apo-L1, and this sample was characterized with steady-state kinetics and EXAFS spectroscopy28. The enzyme exhibited a kcat of 30 s−1 and a Km of 5.5 μM when using nitrocefin as the substrate (Table 2), but we were uncertain whether these constants reflected an enzyme sample that contained significant amount of ZnZn-L1 due to the amounts of adventitious Zn(II) found in buffers9,29. The addition of Co(II) to 1Zn-L1 resulted in a pink coloration that immediately turned orange in less than 10 seconds, indicating oxidation of Co(II) to Co(III). On the other hand, the addition of Zn(II) to 1Co-L1, which was prepared by the biological incorporation method described above, resulted in a protein that remained pink in color. The ZnCo-L1 (this notation indicates Zn(II) in the Zn1 site and Co(II) in the Zn2 site) analog of L1 exhibited a kcat of 26 s−1 and a Km of 2.3 μM, when using nitrocefin as substrate. These values are similar to those of 1Zn-L1 and ZnZn-L1, and it is not possible with steady-state kinetics alone to determine if the ZnCo-L1 analog is responsible for the observed activity.
EPR spectroscopy of metal-ion-substituted forms of L1
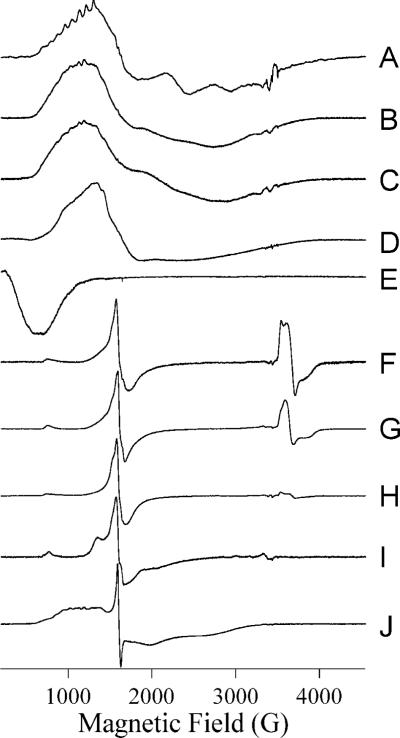
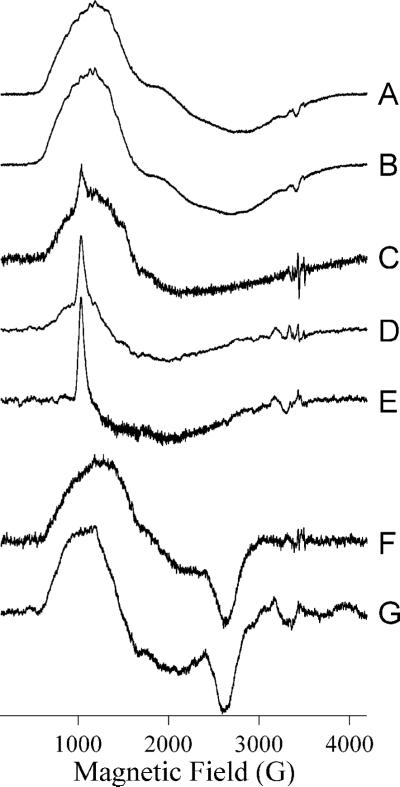
EPR spectra of L1 with increasing Co(II) complement show a complex but sequential pattern of Co(II) binding (Figure 2A – E). A sample of nominally 1Co-L1 that was found to contain only 0.8 eq Co(II) exhibited an EPR spectrum (Figure 2A) that contained two reasonably well-resolved components. A 59Co hyperfine pattern with A = 9.8 × 10−3 cm−1, centered at 996 G (geff. = 6.89), and a derivative feature at 2320 G (geff. = 2.97) were assigned to a rhombic species with greal(⊥) = 2.55 and E/D = 0.27. The second species exhibited no sharp resonances and was due to an axial species similar to that observed from Co(II) in L1 in earlier work20. Based on previous reports, these signals are consistent with an equilibrium of Co(II)-H2O and Co(II)-OH30-32. More typically, 1Co-L1 contained 0.9 − 1.0 eq Co(II), and the spectrum (Figure 2B) became less well-resolved, with only inflection points to suggest the presence of the distinct species observed at lower Co(II) complement. The spectrum of ZnCo-L1 (Figure 2C) was very similar to that of 1Co-L1 (Figure 2B), suggesting that the presence of Zn(II) in either of the binding sites did not significantly perturb the electronic structure of Co(II) in the remaining sites. In contrast, the spectrum of 2Co-L1 (Figure 2D) was markedly different from those of 1Co-L1 and ZnCo-L1; the spectrum could not be simulated assuming even two distinct species, and spin-Hamiltonian parameters could not be assigned. EPR absorption at very low field, 0 − 500 G, suggested the presence of a spin-coupled component in the spectrum, and this was confirmed by parallel mode EPR (Figure 2E), which revealed a resonance at geff. ∼ 10, consistent with S’ = 2 and/or S’ = 3 resonances in an S’ = 0, 1, 2, 3 spin ladder due to coupling of two S = 3/2 Co(II) ions.
Figure 2. EPR spectra from metal-containing species of L1.
Spectra are of the following species of L1: (A) L1 containing 0.8 eq Co(II) at 12 K, 25 mW; (B) 1Co-L1 at 12 K, 2 mW; (C) ZnCo-L1 at 10 K, 2 mW; (D) CoCo-L1 at 12 K, 10 mW; (E) CoCo-L1 at 7 K, 20 mW, B0∥B1; (F) 1Fe-L1 at 10 K, 2 mW; (G) ZnFe-L1 at 12 K, 10 mW; (H) FeFe-L1 at 10 K, 2 mW; (I) FeNi-L1 at 7 K, 50 mW; (J) FeCo-L1 at 10 K, 2 mW. Spectra are shown with arbitrary intensities.
Stopped-flow kinetic studies on ZnCo-L1
The accurate interpretation of steady-state kinetic studies on mixed-metal and mononuclear metal-containing analogs can be complicated by the presence of adventitious Zn(II) in the assay buffers. For example, typical steady-state kinetic studies contain 1−10 nM L1, and the amount of adventitious Zn(II) in buffers, even those that have been Chelex-treated, can be between 10−100 nM9,29. Therefore, it is probable that steady-state kinetic assays were conducted with enzymes containing a mixture of possible metal centers. Therefore, we characterized the mixed-metal analogs and proteins containing only one metal ion with presteady-state kinetic studies at or near single turnover conditions (∼50 μM enzyme and ∼50 μM nitrocefin). The advantage of this approach is that the enzyme concentrations in these samples are at least 2 orders of magnitude higher than the concentration of adventitious Zn(II) in the buffer. This approach also allowed us to monitor the role of each metal ion in catalysis.
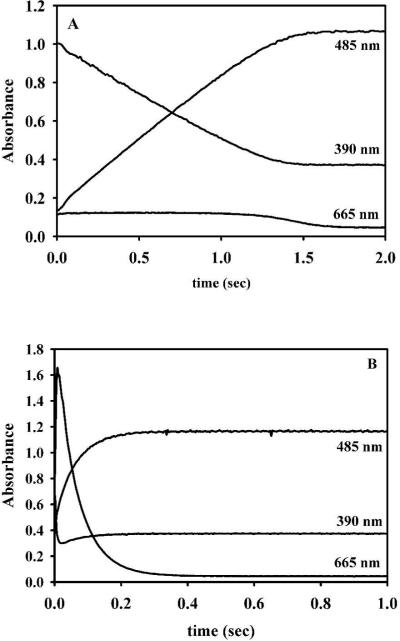
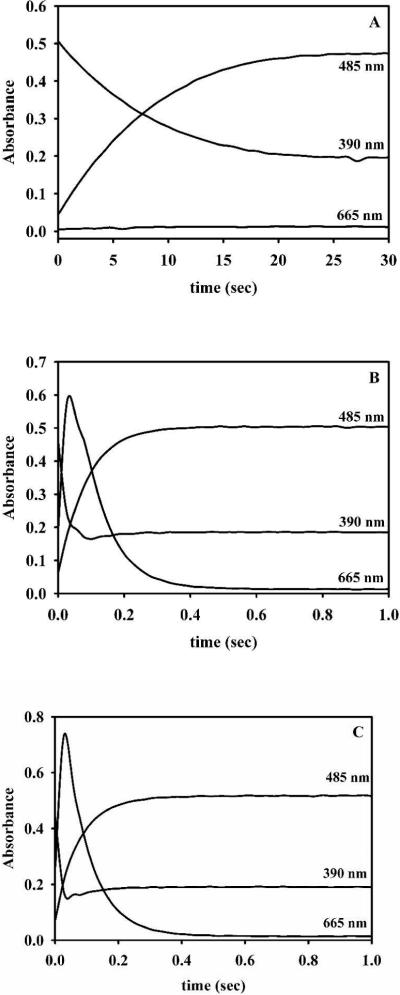
1Zn-L1 was prepared as described above. The stopped-flow traces for 1Zn-L1 showed that substrate (absorbs at 390 nm) was depleted within 1.3 seconds (Figure 3A) and that very little intermediate (absorbs at 665 nm) was observed. The stopped-flow traces were fitted to an exponential equation, and the rate of product formation was 0.92 ± 0.03 s−1 (Table 3). In comparison, the stopped-flow trace of ZnZn-L1 showed that substrate was depleted in 0.06 seconds and that significant amount of intermediate form (Figure 3B). The rate of product formation was 17 ± 1 s−1 (Table 3), which reflects an 18-fold increase in activity as compared to 1Zn-L1. Previous EXAFS studies on L1 demonstrated that there is sequential binding of Zn(II) to apo-L1 and that the first equivalent of Zn(II) binds to the Zn1 site28. This result coupled with the stopped-flow traces described above indicates that metal ions in both of the metal binding sites is required for the stabilization and observation of the reaction intermediate when nitrocefin is used as a substrate.
Figure 3. Stopped-flow traces of reaction of Zn(II)-containing L1 analogs and nitrocefin.
Stopped-flow traces of 50 μM 1Zn- (A) and ZnZn-L1 (B) analogs when reacted with 50 μM nitrocefin at 4 °C. The absorbance at 485 nm is due to the product, the absorbance at 390 nm is due to the substrate, and the absorbance at 665 nm is due to the intermediate 54.
Table 3.
Exponential fits to the stopped-flow kinetic data.
| Species | Rate of product formation (s−1)a |
|---|---|
| 1Zn-L1 | 0.92 ± 0.03 |
| ZnZn-L1 | 17 ± 1 |
| 1Co-L1 | 0.05 ± 0.01 |
| ZnCo-L1 | 12 ± 1 |
| ZnFe-L1 | 12 ± 1 |
| 1Fe-L1 | 0.12 ± 0.02 |
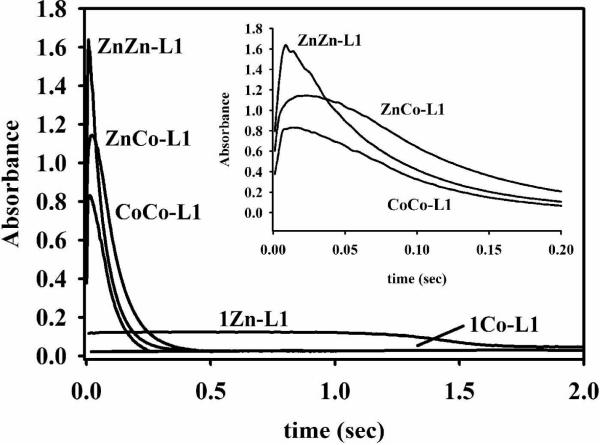
Stopped-flow studies were also conducted on the Co(II)-containing samples. The stopped-flow trace for 1Co-L1 showed that substrate decay took over 10 seconds and that no intermediate formed (Figure 4A). The rate of product formation was 0.05 ± 0.01 s−1 (Table 3). This result is not consistent with the steady-state kinetic results that showed that 1Co-L1 is very active (Table 2) and suggests that most of the activity observed in the steady-state kinetic studies was due to the ZnCo analog of L1. The stopped-flow trace for ZnCo-L1 (Figure 4B) showed that substrate depleted as fast as it did for ZnZn-L1 (Figure 3B), and the rate of product formation was 12 ± 1 s−1 (Table 3), which reflects a 240-fold increase in activity over that of 1Co-L1. There is a 1.4-fold decrease in the amount of intermediate formed for ZnCo-L1, as compared to ZnZn-L1; however, there is a 1.4-fold increase in the amount of intermediate formed for ZnCo-L1 as compared to CoCo-L1 (Figure 5). The intermediate decays faster in the reaction with ZnZn-L1, as compared with CoCo- and ZnCo-L1, and the rates of intermediate decay for CoCo- and ZnCo-L1 are very similar. This result, along with the results described above, strongly indicates that cobalt binds to the Zn2 site and that the Zn2 site is involved in stabilizing the intermediate.
Figure 4. Stopped-flow traces of the reaction of Co(II)-containing L1 analogs with nitrocefin.
50 μM 1Co- (A) and ZnCo-L1 (B) analogs were reacted with 50 μM nitrocefin at 4 °C. The absorbance at 485 nm is due to the product, the absorbance at 390 nm is due to the substrate, and the absorbance at 665 nm is due to the intermediate.
Figure 5. Intermediate formation by L1 analogs.
The absorbance at 665 nm arises from the presence of intermediate. Each reaction contained 50 μM L1 analog and 50 μM nitrocefin at 4 °C in 50 mM cacodylate, pH 7.0. Inset: Intermediate formation for ZnZn-, ZnCo-, and CoCo-L1 analogs over 200 ms.
RFQ-EPR studies
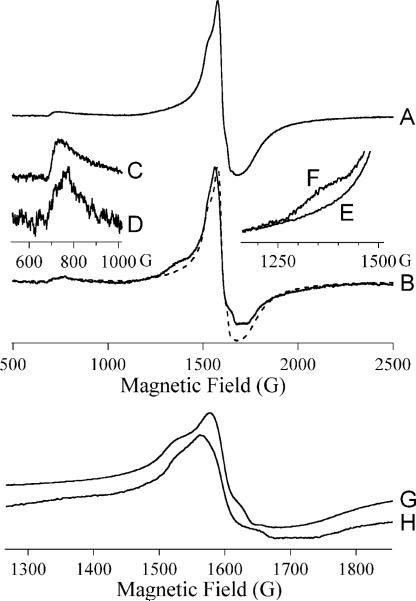
EPR spectra recorded on ZnCo-L1 during an RFQ-EPR experiment are shown in Figure 6. The resting signals from ZnCo-L1 recorded at 10 K, 2 mW (Figure 6A) and at 7 K, 80 mW (Figure 6B) were very similar and are due to two isolated S = 3/2, MS = ± ½ systems. These systems are in turn due to Co(II) in either of the binding sites in singly-occupied L1. Upon reaction with nitrocefin for 10 ms, the color of the sample became bright blue, and the EPR spectra shown in Figure 6C – E were observed. At 10 K, 2 mW (Figure 6C), the inflections in the spectrum (1600 − 2000 G), due to the presence of the isolated rhombic species of Figure 2A, were no longer observable, and instead, a small but distinct sharp peak at 1025 G (geff. = 6.65) was observed. At successively higher microwave power and lower temperature, this signal became more prominent as other features were lost to saturation and rapid-passage effects (Figure 6D, E), characteristic of an MS = ± 3/2 system and of tetrahedral Co(II). Upon further reaction, the sample turned red, indicating the hydrolysis of nitrocefin, and new EPR signals were observed (Figure 6F, G) that are presumably due to a product complex. These signals showed no evidence of an MS = ± 3/2 component but were unusual in that the gz feature at 2650 G (geff. = 2.6) was very well-resolved, indicative of constrained geometry and consistent with binding of Co(II) to a more rigid ligand than water 32.
Figure 6. RFQ-EPR of ZnCo-L1 with nitrocefin.
Spectra (A) and (B) are from resting ZnCo-L1. Spectra (C – E) are from ZnCo-L1 after reaction with nitrocefin for 10 ms at 3 °C. Spectra (F) and (G) are from ZnCo-L1 after incubation with nitrocefin for 2 min (at which time all of the added nitrocefin has been hydrolyzed). Spectra (A), (C) and (F) were recorded at 10 K, 2 mW, spectra (B), (D) and (G) at 7 K, 80 mW, and spectrum (E) at 5 K, 126 mW. Spectra are shown with arbitrary intensities.
Preparation and characterization of the ZnFe-analog of L1
Our ability to prepare a mixed-metal analog of L1 by using a biological incorporation method led us to speculate whether a ZnFe analog of L1 could also be prepared. Recently, our studies using the over-expression plasmid that results in L1 being folded in the cytoplasm allowed us to prepare an iron-containing analog of L118. However, the FeFe-L1 analog was catalytically-inactive. In an effort to prepare a ZnFe analog of L1, we over-expressed L1 in minimal medium containing 100 μM Fe(II). The purified enzyme was green in color, contained 0.9 equivalents of Fe and 0.2 equivalents of Zn(II), and exhibited a kcat of 2.6 s−1 and a Km of 53 μM, when using nitrocefin as the substrate. The addition of 0.8 equivalents of Zn(II) resulted in a sample that exhibited a kcat of 24 s−1 and a Km of 4 μM (Table 2).
The EPR signal of 1Fe-L1 (Figure 2F) consisted of two types of signals. A rhombic S = 5/2 signal was observed with resonances at geff. ∼ 9 and ∼ 4.3, and with some structure in the g ∼ 4.3 region indicative of protein-bound Fe(III). The other contribution was from two very similar and largely overlapping signals with geff. < 2 (3400 − 4000 G) and indicative of an antiferromagnetically coupled Fe(II)-Fe(III) dinuclear site33, 34. Spin quantitation35 of these signals suggests that 50% of iron is from a mononuclear Fe(III) center, while 50% is from Fe(III)Fe(II) or Fe(II)Fe(III) centers. The amount of signal corresponding to Fe(III)Fe(II) (or Fe(II)Fe(III)) varied among samples, and the amount of the antiferromagnetically-coupled centers was greatest in the sample corresponding to Figure 2F. More typically, we obtained spectra that contained only ca. 10% of the signal due to the spin-coupled center, perhaps due to oxidation of the center to the EPR-inactive Fe(III)Fe(III) center. Addition of Zn(II) generated ZnFe-L1, though the EPR signal varied from sample to sample. In all cases, there were small but reproducible changes in the Fe(III) signal, perhaps indicative of formation of an Zn(II)Fe(III) center in some molecules, and the intensity of the Fe(II)Fe(III) signal diminished, sometimes by a rather modest amount, as in Figure 2G, and sometimes almost completely. Spin quantitation35 of the signals in Figure 2G were consistent with a 25−30% contribution from a spin-coupled center and a 70−75% contribution from Fe(III) in a single site. Further addition of iron, to form FeFe-L1, consistently abolished the Fe(II)Fe(III) signal (Figure 2H), as did additions of Ni(II) (Figure 2I) and Co(II) (Figure 2J). Additionally, marked changes in the Fe(III) signals in these bimetallic forms of L1 were observed. The g ∼ 9 and 4.3 regions of the spectrum of FeFe-L1 differ from those of 1Fe-L1 and ZnFe-L1. Additional transitions were observed flanking the g ∼ 4.3 region of the spectrum of FeNi-L1, indicating of a narrowing of the distribution of E/D due to lowering of strain terms and a more constrained Fe(III) environment. The shape and intensity change of the g ∼ 9 feature suggests changes in both strains and in D. In FeCo-L1, transitions due to Fe(III) and Co(II) in the region 800 − 3000 G could not be deconvoluted with confidence, but the very sharp nature of the g ∼ 4.3 resonance from Fe(III) again indicates changes in the zero-field splitting parameters of Fe(III).
In an effort to further probe which site (Zn1 or Zn2) that the Fe binds, we attempted to obtain a 1H NMR spectrum of this sample; however, no peaks were observed between −200 to +200 ppm. We believe that the inability to observe any peaks in this sample is due, in part, to the relatively slow electron spin relaxation rate (T1e) of high-spin Fe(III) and the large size (118 kDa) of L1, both of which result in significant broadening of 1H NMR peaks36. Our inability to observe paramagnetically-shifted resonances due to the spin-coupled Fe(III)Fe(II) centers in the L1 samples is most likely due to the presence of a single bridging group (hydroxide) in this analog and different relaxation properties as compared to similar centers in GLX2−533.
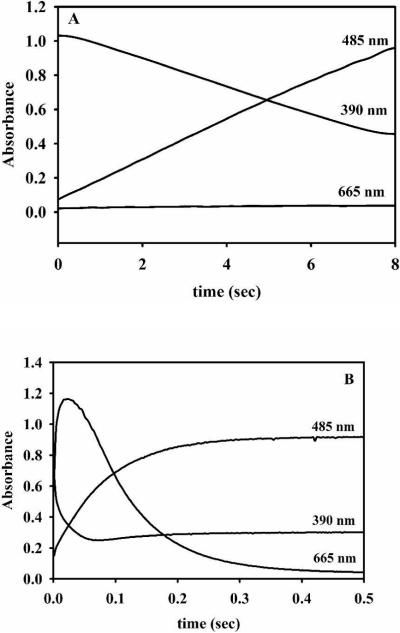
The ZnFe-analog of L1 was also prepared by adding 1 equivalent of Fe(II) directly to 1Zn-L1, which was made by adding 1 equivalent of Zn(II) directly to apo-L1. This sample exhibited almost identical steady-state kinetic constants as the sample described above (Table 2). Similar to the results on cobalt-containing samples of L1, the 1Fe-L1 analog exhibited little or no activity and produced no intermediate (Figure 7), suggesting the steady-state kinetic data for this enzyme was due to small amounts of ZnFe-L1. The rate of product formation for 1Fe-L1 was 0.12 ± 0.02 (Table 3). The stopped-flow traces for ZnFe-L1 showed substrate depletion occurred during the first 0.08 seconds and 2.6-fold less intermediate formed for this enzyme as compared to ZnZn-L1 (Figure 7). The rate of product formation for ZnFe-L1 (made by adding Zn(II) to 1Fe-L1 or by adding Fe(II) to 1Zn-L1) was 12 ± 1, which reflects a 100-fold increase in activity as compared to 1Fe-L1 (Table 3).
Figure 7. Stopped-flow traces of Fe-containing L1 analogs reacted with nitrocefin.
50 μM 1Fe- (A), ZnFe-L1 (made by adding Fe(II) to 1Zn-L1) (B), and ZnFe-L1 (made by adding Zn(II) to 1Fe-L1) (C) were reacted with 50 μM nitrocefin at 4 °C. The absorbance at 485 nm is due to the product, the absorbance at 390 nm is due to the substrate, and the absorbance at 665 nm is due to the intermediate.
RFQ-EPR studies with FeZn-L1 and nitrocefin
As with ZnCo-L1, the EPR spectrum of ZnFe-L1 was observed to change upon incubation with nitrocefin for 10 ms at 3 °C (Figure 8). The various transitions that make up the g ∼ 4.3 line in ZnFe-L1 (Figure 8A) are due to the mean E/D being slightly less than 1/3 and the strain-dependent distribution in E/D not being large enough to broaden out all of the transitions. Upon reaction with nitrocefin, the resonance positions of these partially-resolved transitions change, indicative of a change in E/D and, hence, in the ligand field at Fe(III) (Figure 8B, G, H). Further change in the g ∼ 9 resonance was observed (Figure 8C, D), and a shoulder was observed at g ∼ 5 (1350 G Figure 8E, F) upon reaction of ZnFe-L1 with nitrocefin.
Figure 8. RFQ-EPR of ZnFe-L1 with nitrocefin.
Trace A shows the 500 − 2500 G region of the spectrum of resting ZnFe-L1. Trace B shows the 500 − 2500 G region of the spectrum of ZnFe-L1 upon reaction with nitrocefin for 10 ms at 3 °C (solid) overlaid with that of resting ZnFe-L1 (dashed). The inserts show more detailed comparisons between the spectra over particular field ranges; traces (C), (F) and (G) are from resting ZnFe-L1 and traces (D), (E) and (H) are from ZnFe-L1 after reaction with nitrocefin for 10 ms at 4 °C.
DISCUSSION
Zn(II) plays an essential catalytic role in enzymes from all of the major classes of enzymes and a structural role in a large number of other proteins37-39. Due to its valence electronic configuration of [Ar]3d10, Zn(II) is silent to most spectroscopic techniques. Fortunately, Zn(II) can be substituted with Co(II), and the resulting enzymes are catalytically-active and contain metal binding sites nearly identical to those of the Zn(II)-containing analogs32,40. For mononuclear Zn(II)-containing enzymes such as carbonic anhydrase, the Co(II)-substituted analog yields unambiguous results regarding the function of the metal site in catalysis41. However in the case of dinuclear Zn(II)-containing enzymes, the interpretation of kinetic/spectroscopic results are more complicated due to the presence of up to three distinct species, [M1_], [_M2] and [M1M2], that can interact with substrates in distinct ways and that can display overlapping spectroscopic signatures. Nonetheless, previous studies on dinuclear metal ion-containing aminopeptidase from Aeromonas proteolytica demonstrated that mixed-metal ion containing analogs of the enzyme could be used to probe the role of each metal in catalysis/binding31,32,42. The metal binding mode of this enzyme is sequential, which allowed for the preparation and characterization of the ZnZn, ZnCo (or CoZn), 1Zn, and 1Co analogs. For other enzymes such as BcII however, the binding constants of the two metal binding sites are similar, leading to mixtures of enzyme containing mononuclear, dinuclear, and even trinuclear metal ion containing analogs13,14. The interpretation of kinetic and spectroscopic results on such mixtures is difficult if not impossible to accomplish.
Since the metal binding Kd1 and Kd2 for Zn(II) binding to L1 was reported to be 2.6 and 6.0 nM 9, respectively, we did not initially believe that we could prepare enzyme samples containing 1Zn-, 1Co-, or ZnCo-centers by simply adding the metal ion to metal-free enzyme. Therefore, our first attempt to prepare these analogs involved the use of site-directed mutagenesis. The rationale for these studies was to introduce a mutation in one of the metal binding sites and to weaken metal binding to this site. Since Asp120 is essential for catalysis in L1 23, we decided to substitute the metal binding histidines in the enzyme. Five site-directed mutants, with single point mutations, were generated; however surprisingly, only two of the resulting mutants were soluble. Fortunately, there was one HXXC mutation in each of the metal binding sites (H116C for Zn1 site, H121C for Zn2 site). Steady-state kinetic studies showed that the catalytic activities of both mutants were low (Table 1), which is consistent with the low observed Zn(II) incorporation. After incubation with excess Zn(II) and dialysis to remove loosely-bound or nonbound Zn(II), both mutants were shown to bind nearly 1 equivalent of Zn(II), which suggests that the one amino acid substitution did impair metal binding as expected. Steady-state kinetic studies conducted in the presence of 100 μM Zn(II) demonstrated that H121C exhibits similar activity (kcat = 33 ± 3 s−1) as wild-type L1, although the mutant exhibited a much higher value for Km. In contrast, the H116C mutant exhibited almost no activity even in the presence of added Zn(II). Since His161 is in the Zn1 site, this result suggests that the Zn1 site is important for catalysis; however, we cannot rule out the possibility that the point mutation did not alter the substrate binding site. These results also demonstrate that a mutation to one of the metal binding histidines results in an enzyme that requires excess metal ion to saturate the mutated site. Since the excess metal ions would undoubtedly complicate subsequent spectroscopic analyses, we concluded that this strategy cannot be used to prepare the mixed-metal analogs of L1.
Consequently, we utilized a biological incorporation strategy in an attempt to prepare L1 analogs containing only one equivalent of Co(II) and the mixed metal ion containing analogs. Over-expression of L1 in minimal medium containing cobalt resulted in an enzyme that binds ca. 0.9 equivalents of Co(II). Spectroscopic studies strongly suggest that Co(II) is not delocalized between the two metal binding sites and that it binds to the consensus Zn2 site (Figures 1 and 2). A similar metal content is obtained when L1 is over-expressed in the presence of Zn(II) using this same technique. Our previous EXAFS studies28 and recent crystallographic studies by Dideberg43 demonstrate that Zn(II) preferentially binds to the Zn1 site. In agreement with the model of Co(II) binding to the Zn2 site and Zn(II) binding to the Zn1 site, the addition of 1 equivalent of Zn(II) to 1Co-L1 results in an enzyme with almost identical EPR properties as 1Co-L1 (Figure 2B, C); however, the two analogs exhibit significantly different pre-steady state kinetic behaviors (Figure 4). Surprisingly, a FeZn analog of L1 can also be prepared by using the same strategy. EPR studies show that the resulting 1Fe-L1 contains a predominant ZnFe center; however, samples also sometimes contained some antiferromagnetically-coupled Fe(III)Fe(II) (Figure 2G). The formation of an ZnFe center was reflected in differences between the Fe(III) EPR spectra of 1Fe-L1 and ZnFe-L1, and the narrowing of the spectrum upon incorporation of Zn(II) suggests increased conformational rigidity of the active site in the dimetallic form. While the effect of Zn(II) on the EPR signal was quite subtle, much more dramatic effects were observed with Ni(II) and Co(II). In both cases a significant reduction in the structural microheterogeneity of the Fe(III) environment was revealed by EPR, giving rise to resolved E/D < 1/3 transitions with Ni(II) and a very sharp E/D = 1/3 g = 4.3 line with Co(II). Interestingly, no spin-spin exchange coupling was detected in FeCo-L1. Both metal ions in L1 are required for maximum catalytic activity. Thus, the binding of the second metal ion fine tunes the electronic structure of the first ion via a structural, rather than electronic, mechanism. We were unable to obtain 1H NMR spectra of the Fe-containing analogs of L1 due to the relatively slow T1e of Fe(III)44 and presumably due to the low concentration of Fe(II)Fe(III) in the sample. Nonetheless, we hypothesize that Fe(III) is binding to the Zn2 site since the H-H-D motif is a common Fe(III) binding site in biology45. Fe(II) can bind at the Zn1 site, but the addition of Zn(II) to 1Fe-L1 results in a reduction of the signal corresponding to the mixed-valent, dinuclear iron center (Figure 2G). The ZnFe analog can be prepared either by adding Fe to 1Zn-L1 or Zn(II) to 1Fe-L1, since the resulting enzymes exhibit the same steady state and pre-steady state kinetic characteristics (Table 2 and Figure 7). Taken together, these results demonstrate that mixed-metal analogs of L1 can be generated and used in mechanistic studies to probe the role of each metal in catalysis.
Stopped-flow kinetic studies on 1Zn-, 1Co-, ZnZn-, and ZnCo-L1 were used to probe the role of the metal ions. 1Zn-L1, with Zn(II) in the Zn1 site, exhibited some activity (ZnZn-L1 is 18-fold more active than 1Zn-L1, Table 3); however, very little intermediate was detected in these studies (Figure 3). On the other hand, 1Co-L1, with Co(II) in the Zn2 site, is almost completely inactive (ZnZn-L1 is 340-fold more active than 1Co-L1, Table 3). It is likely that the small activity exhibited by 1Co-L1 in the stopped-flow studies (0.29% as compared to ZnZn-L1) is due to small amounts of Zn(II) in 1Co-L1 preparations (Table 1) and in the buffer (estimated to be 100 nM, which is 0.2% of the concentration of enzyme in the stopped-flow studies). The 1Fe-L1 analog was >140-fold less active than ZnZn-L1 (Figures 4 and 7; Table 3), and this higher activity, as compared to that of 1Co-L1, is mostly due to the higher amounts of Zn(II) in the 1Fe-L1 samples (Table 2). We cannot unambiguously rule out that one of the Fe-containing analogs of L1 is active; although, our studies indicate that FeFe-L1 is inactive (Table 2)18. These results indicate that both metal ions are required to detect intermediate in the reaction of nitrocefin with the ZnCo- and ZnFe-analogs of L1 (Figures 3, 4, 5, and 7). These results also indicate that an analog of L1 with metal (Co(II) or Fe) only in the Zn2 site is inactive. In contrast, an analog of L1 with Zn(II) in the Zn1 site does exhibit some activity, albeit very small (compare rates of product formation for 1Zn-L1 with those of 1Co-L1 and 1Fe-L1; Table 3), and this analog does allow for the formation of a small (4%) amount of intermediate (Figure 3A). Taken together, these results demonstrate that both metal ions in L1 are required for maximum catalytic activity. The Zn1 site “prefers” Zn(II) over any other metal ion, and the role of this metal ion is presumably to provide the reactive nucleophile during catalysis. L1 analogs with metal ion only in the Zn2 are not catalytically-active. The Zn2 site can bind a number of metal ions including Co(II) and Fe(III)/Fe(II). The role of this site is to stabilize the reaction intermediate during catalysis. This result is consistent with previous suggestions on CcrA46 and on model complex-catalyzed hydrolysis of nitrocefin47,48. It is not absolutely essential to have the Zn2 filled in order that L1 be active since 1Zn-L1 does exhibit some catalytic activity. Based on previous studies on CcrA46,49,50, the roles of the metal ions are most like the same in this subgroup 3A β-lactamase. The results presented above can not necessarily be applied to BcII, since there is considerable controversy presently regarding whether the mononuclear Zn(II)-containing enzyme is active10,12-14. In addition, Vila and coworkers have reported that no ring-opened, nitrogen anionic intermediate is observed when BcII is reacted with nitrocefin51.
The successful preparation of a heterometallic analog of L1 that contained a paramagnetic metal ion in one metal binding site allowed us to directly probe the reaction mechanism of L1 with RFQ EPR studies. The EPR spectrum of ZnCo-L1 was consistent with Co(II) being 5-coordinate in the resting form of the enzyme (Figure 6A and B). Within 10 ms reaction time, a 4-coordinate tetrahedral intermediate, not seen at all in any of the resting spectra from Co(II)-containing L1, was formed. This species decayed as substrate was exhausted and a higher coordination product complex remained. This result confirms our previous work that showed that substrate, intermediate, and product coordinate the metal ion(s) in L120. RFQ-EPR of ZnFe-L1 also showed catalytically-competent changes in the EPR spectrum, here due to Fe(III).
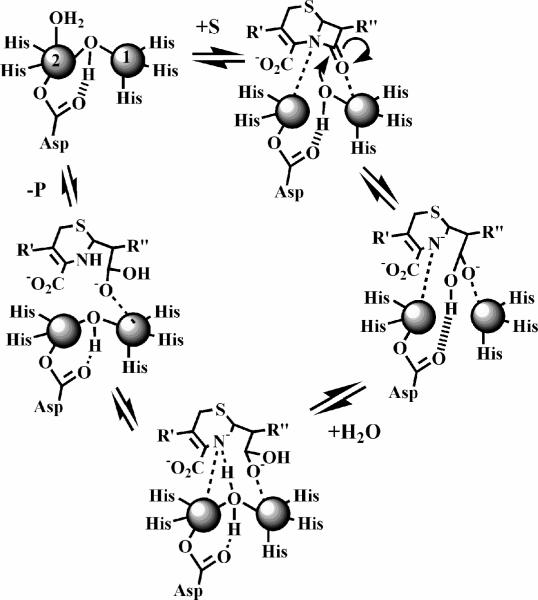
Based on all of the data on L1 presented to date, we are in position to propose a reaction mechanism of nitrocefin hydrolysis by L1 (Figure 9). When nitrocefin binds, the terminally-bound water molecule on Zn2 releases and the β-lactam carbonyl interacts with the metal ion in the Zn1 site while the nitrogen lone pair on the nitrogen of the β-lactam interacts with Zn227,52. The binding of substrate results in the loss of the Zn2-bridging hydroxide bond, thereby generating a four-coordinate metal ion in the Zn2 site and the reactive nucleophile that is directed for attack by Asp12023. The resulting, very short-lived tetrahedral species is converted to the ring-opened, nitrogen anionic intermediate after the loss of the β-lactam bond. At this time it is not clear if one metal ion or both are involved in the stabilization of the intermediate, but the data in this work clearly shows that the metal ion in the Zn2 site is essential for stabilization. The breakdown of the intermediate involves a protonation, which likely occurs during the concerted formation of a new bridging water/hydroxide. Our previous kinetic studies strongly suggested that Asp120 plays a role in orienting the acidic proton on the solvent molecule for protonation of intermediate23. When other substrates are used, there is evidence that the reaction intermediate does not accumulate53, suggesting that ring opening and protonation of the β-lactam nitrogen is concerted. Regardless of substrate, the EP complex is in equilibrium with the resting enzyme, and in both cases, the coordination number at the Zn2 site is 5.
Figure 9.
Proposed reaction mechanism of L1 for the hydrolysis of nitrocefin.
The successful preparation of mononuclear metal ion containing and heterometallic analogs of L1 has allowed us for the first time to probe the roles of the metal ions in this enzyme. It is clear that the metal ion in the Zn1 site is essential for activity and that the most active form of the enzyme requires both metal ions. The metal ion in the Zn2 site appears to be involved in the stabilization of a reaction intermediate and possibly in orienting the β-lactam nitrogen for protonation. These results demonstrate that potential inhibitors can be designed to target the Zn1 site only or both sites, although compounds that bind to the Zn2 site and that block the Zn1 site may also be effective inhibitors.
Acknowledgment
The authors thank the Volwiler Professorship (MWC) and the National Institutes of Health (EB001980 and AI056231 to BB) for funding this work.
References
- 1.Fisher JF, Meroueh SO, Mobashery S. Chem. Rev. 2005;105:395–424. doi: 10.1021/cr030102i. [DOI] [PubMed] [Google Scholar]
- 2.Perez F, Endimiani A, Hujer KM, Bonomo RA. Curr. Opin. Pharmacol. 2007;7(5):459–469. doi: 10.1016/j.coph.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh TR, Toleman MA, Poirel L, Nordmann P. Clin. Microbiol. Rev. 2005;18(2):306–325. doi: 10.1128/CMR.18.2.306-325.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Crowder MW, Spencer J, Vila AJ. Acc. Chem. Res. 2006;39(10):721–728. doi: 10.1021/ar0400241. [DOI] [PubMed] [Google Scholar]
- 5.Moran-Barrio J, Gonzalez JM, Lisa MN, Costello AL, Dal Peraro M, Carloni P, Bennett B, Tierney DL, Limansky AS, Viale AM, Vila AJ. J. Biol. Chem. 2007;282:18286–18293. doi: 10.1074/jbc.M700467200. [DOI] [PubMed] [Google Scholar]
- 6.Carfi A, Pares S, Duee E, Galleni M, Duez C, Frere JM, Dideberg O. EMBO J. 1995;14(20):4914–4921. doi: 10.1002/j.1460-2075.1995.tb00174.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabiane SM, Sohi MK, Wan T, Payne DJ, Bateson JH, Mitchell T, Sutton BJ. Biochemistry. 1998;37:12404–12411. doi: 10.1021/bi980506i. [DOI] [PubMed] [Google Scholar]
- 8.Davies AM, Rasia RM, Vila AJ, Sutton BJ, Fabiane SM. Biochemistry. 2005;44:4841–4849. doi: 10.1021/bi047709t. [DOI] [PubMed] [Google Scholar]
- 9.Wommer S, Rival S, Heinz U, Galleni M, Frere JM, Franceschini N, Amicosante G, Rasmussen B, Bauer R, Adolph HW. J. Biol. Chem. 2002;277(27):24142–24147. doi: 10.1074/jbc.M202467200. [DOI] [PubMed] [Google Scholar]
- 10.Badarau A, Page MI. Biochemistry. 2006;45:11012–11020. doi: 10.1021/bi0610146. [DOI] [PubMed] [Google Scholar]
- 11.Badarau A, Page MI. Biochemistry. 2006;45:10654–10666. doi: 10.1021/bi060934l. [DOI] [PubMed] [Google Scholar]
- 12.Badarau A, Damblon C, Page MI. Biochem. J. 2007;401:197–203. doi: 10.1042/BJ20061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Llarrull LI, Tioni MF, Kowalski J, Bennett B, Vila AJ. J. Biol. Chem. 2007;282(42):30586–30595. doi: 10.1074/jbc.M704613200. [DOI] [PubMed] [Google Scholar]
- 14.Gonzalez JM, Medrano Martin FJ, Costello AL, Tierney DL, Vila AJ. J. Mol. Biol. 2007;373:1141–1156. doi: 10.1016/j.jmb.2007.08.031. [DOI] [PubMed] [Google Scholar]
- 15.Heinz U, Adolph HW. CMLS, Cell. Mol. Life Sci. 2004;61:2827–2839. doi: 10.1007/s00018-004-4214-9. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning - A Laboratory Manual. Second ed. Vol. 1. Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Crowder MW, Walsh TR, Banovic L, Pettit M, Spencer J. Antimicro. Agents Chemo. 1998;42(4):921–926. doi: 10.1128/aac.42.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu Z, Gunasekera TS, Spadafora L, Bennett B, Crowder MW. Biochemistry. 2008;47(30):7947–7953. doi: 10.1021/bi8004768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma NP, Hajdin C, Chandrasekar S, Bennett B, Yang KW, Crowder MW. Biochemistry. 2006;45:10729–10738. doi: 10.1021/bi060893t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garrity JD, Bennett B, Crowder MW. Biochemistry. 2005;44:1078–1087. doi: 10.1021/bi048385b. [DOI] [PubMed] [Google Scholar]
- 21.Carenbauer AL, Garrity JA, Periyannan G, Yates RB, Crowder MW. BMC Biochemistry. 2002;3:4–10. doi: 10.1186/1471-2091-3-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Garrity JD, Pauff JM, Crowder MW. J. Biol. Chem. 2004;279(38):39663–39670. doi: 10.1074/jbc.M406826200. [DOI] [PubMed] [Google Scholar]
- 23.Garrity JD, Carenbauer AL, Herron LR, Crowder MW. J. Biol. Chem. 2004;279(2):920–927. doi: 10.1074/jbc.M309852200. [DOI] [PubMed] [Google Scholar]
- 24.Crowder MW, Yang KW, Carenbauer AL, Periyannan G, Seifert MA, Rude NE, Walsh TR. J. Biol. Inorg. Chem. 2001;6:91–99. doi: 10.1007/s007750000173. [DOI] [PubMed] [Google Scholar]
- 25.Garmer DR, Krauss M. J. Am. Chem. Soc. 1993;115:10247–10257. [Google Scholar]
- 26.Hu Z, Periyannan GR, Crowder MW. Anal. Biochem. 2008;378:177–183. doi: 10.1016/j.ab.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ullah JH, Walsh TR, Taylor IA, Emery DC, Verma CS, Gamblin SJ, Spencer J. J. Mol. Biol. 1998;284:125–136. doi: 10.1006/jmbi.1998.2148. [DOI] [PubMed] [Google Scholar]
- 28.Costello A, Periyannan G, Yang KW, Crowder MW, Tierney DL. J. Biol. Inorg. Chem. 2006;11:351–358. doi: 10.1007/s00775-006-0083-z. [DOI] [PubMed] [Google Scholar]
- 29.Outten CE, O'Halloran TV. Science. 2001;292(5526):2488–2492. doi: 10.1126/science.1060331. [DOI] [PubMed] [Google Scholar]
- 30.Bennett B, Antholine WE, D'Souza VM, Chen GJ, Ustinyuk L, Holz RC. J. Am. Chem. Soc. 2002;124(44):13025–13034. doi: 10.1021/ja026341p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bennett B, Holz RC. Biochemistry. 1997;36:9837–9846. doi: 10.1021/bi970735p. [DOI] [PubMed] [Google Scholar]
- 32.Bennett B. Curr. Topics Biophys. 2002;26(1):49–57. [Google Scholar]
- 33.Marasinghe GPK, Sander IM, Bennett B, Periyannan G, Yang KW, Makaroff CA, Crowder MW. J. Biol. Chem. 2005;280(49):40668–40675. doi: 10.1074/jbc.M509748200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schilling O, Wenzel N, Naylor M, Vogel A, Crowder M, Makaroff C, Meyer-Klaucke W. Biochemistry. 2003;42(40):11777–11786. doi: 10.1021/bi034672o. [DOI] [PubMed] [Google Scholar]
- 35.Bou-Abdallah F, Chasteen ND. J. Biol. Inorg. Chem. 2008;13(1):15–24. doi: 10.1007/s00775-007-0304-0. [DOI] [PubMed] [Google Scholar]
- 36.Bertini I, Turano P, Vila AJ. Chem. Rev. 1993;93:2833–2932. [Google Scholar]
- 37.Auld DS. Zinc Catalysis in Metalloproteases. In: Hill HAO, Sadler PJ, Thomson AJ, editors. Metal Sites in Proteins and Models: Phosphatases, Lewis Acids, and Vanadium. Vol. 89. Springer-Verlag; New York: 1997. pp. 29–50. [Google Scholar]
- 38.Auld DS. Biometals. 2001;14(3−4):271–313. doi: 10.1023/a:1012976615056. [DOI] [PubMed] [Google Scholar]
- 39.Auld DS. Cocatalytic zinc sites. In: Messerschmidt A, editor. Handbook of Metalloproteins. Vol. 3. John Wiley & Sons; New York: 2004. pp. 416–431. [Google Scholar]
- 40.Bertini I, Luchinat C. Adv. Inorg. Biochem. 1984;6:72–111. [PubMed] [Google Scholar]
- 41.Bertini I, Luchinat C, Scozzafava A. Struct. Bonding. 1982;48:45–92. [Google Scholar]
- 42.Bennett B, Holz RC. J. Am. Chem. Soc. 1997;119(8):1923–1933. [Google Scholar]
- 43.Nauton L, Kahn R, Garau G, Hernandez JF, Dideberg O. J. Mol. Biol. 2008;375:257–269. doi: 10.1016/j.jmb.2007.10.036. [DOI] [PubMed] [Google Scholar]
- 44.Bertini I, Luchinat C. NMR of Paramagnetic Molecules in Biological Systems. Benjamin Cummings Publishing Company; Menlo Park, CA: 1986. p. 317. [Google Scholar]
- 45.Hegg EL, Que L. Eur. J. Biochem. 1997;250:625–629. doi: 10.1111/j.1432-1033.1997.t01-1-00625.x. [DOI] [PubMed] [Google Scholar]
- 46.Wang Z, Fast W, Benkovic SJ. Biochemistry. 1999;38:10013–10023. doi: 10.1021/bi990356r. [DOI] [PubMed] [Google Scholar]
- 47.Kaminshaia NV, Spingler B, Lippard SJ. J. Am. Chem. Soc. 2000;122:6411–6422. [Google Scholar]
- 48.Kaminskaia NV, Spingler B, Lippard SJ. J. Am. Chem. Soc. 2001;123(27):6555–6563. doi: 10.1021/ja002699e. [DOI] [PubMed] [Google Scholar]
- 49.Wang Z, Benkovic SJ. J. Biol. Chem. 1998;273(35):22402–22408. doi: 10.1074/jbc.273.35.22402. [DOI] [PubMed] [Google Scholar]
- 50.Wang Z, Fast W, Benkovic SJ. J. Am. Chem. Soc. 1998;120(41):10788. [Google Scholar]
- 51.Rasia RM, Vila AJ. ARKIVOC. 2003;3:507–516. [Google Scholar]
- 52.Spencer J, Read J, Sessions RB, Howell S, Blackburn GM, Gamblin SJ. J. Am. Chem. Soc. 2005;127:14439–14444. doi: 10.1021/ja0536062. [DOI] [PubMed] [Google Scholar]
- 53.Spencer J, Clark AR, Walsh TR. J. Biol. Chem. 2001;276(36):33638–33644. doi: 10.1074/jbc.M105550200. [DOI] [PubMed] [Google Scholar]
- 54.McMannus-Munoz S, Crowder MW. Biochemistry. 1999;38:1547–1553. doi: 10.1021/bi9826512. [DOI] [PubMed] [Google Scholar]