Abstract
Background
Previously, we not only reported that dopamine D1 receptor gene (DRD1) is associated with nicotine dependence (ND), but demonstrated that two alleles (A and G) of polymorphism rs686 in the 3′-untranslated region (3′UTR) of DRD1 are expressed differentially. However, the mechanism underlying the differential expression remains to be determined. We hypothesize that it is caused by miRNA targeting.
Methods
We first used the MicroInspector algorithm to identify microRNAs (miRNAs) potentially targeting the rs686 polymorphism in the DRD1 3′UTR, and then employed a luciferase reporter assay combined with site-directed mutagenesis to test the predicted miRNA targeting. We also examined the miRNA targeting of DRD1 with a gene expression assay.
Results
Of two miRNAs predicted by computational analyses, we found that miR-504, not miR-296, up-regulated reporter luciferase activity and increased DRD1 expression by targeting the DRD1 3′UTR, whereas inhibition of miR-504, not miR-296, had the opposite effect. Furthermore, we revealed that the direct binding of miR-504 to the DRD1 3′UTR, verified by site-directed mutagenesis, caused a significant expression difference between the two alleles.
Conclusion
miR-504 up-regulates DRD1 expression by direct binding to the 3′UTR, which leads to differential allele-specific expression of DRD1.
Keywords: dopamine D1 receptor (DRD1), 3′ untranslated region (3′ UTR), single nucleic polymorphism (SNP), microRNA (miRNA), gene expression, nicotine dependence (ND)
INTRODUCTION
Recently, a significant number of non-coding, but possibly functional, polymorphisms have been found to be associated with many complex diseases [e.g. (1–3)]. However, characterizing these non-coding polymorphisms remains challenging. The recognition that small non-coding RNAs regulate gene expression provides a novel means to explore the effects of these variants. MicroRNAs (miRNAs) are such a class of regulatory RNAs reported to modulate various biological processes (4) and predicted to regulate as many as 30% of human mRNAs (5). The miRNA targeting is determined by the nature and extent of the complementarity between an miRNA and its target sequence in the 3′ untranslated region (3′UTR) of mRNA. Thus, a non-coding polymorphism residing in the miRNA or the miRNA target sequence may play a role in regulating gene expression, with a concomitant alteration in phenotype.
Nicotine dependence (ND) is an inheritable complex disorder determined by both genetic and environmental factors as well as their interactions. Dopamine receptors, playing key roles in mediating dopamine actions in the central nervous system, represent plausible candidate genes for genetic study on ND. Recently, we reported a significant association of dopamine D1 receptor gene, DRD1, with ND, and also demonstrated that polymorphism rs686 in the 3′UTR of DRD1 affects DRD1 expression (6). Interestingly, it also reported that the variant is associated with alcohol dependence (7) and autism spectrum disorder (8), providing additional genetic evidence of DRD1’s importance in neuropsychiatric disorders. We proposed previously that differential allelic expression of DRD1 is caused by an miRNA(s) (6). To test this hypothesis experimentally, we examined miRNA targeting in the vicinity of the rs686 polymorphism using both luciferase reporter and gene expression assays.
METHODS AND MATERIALS
Cell culture and transfection
Human embryonic kidney cell line HEK293 was purchased from the American Type Culture Collection and cultured in Dulbecco’s modified Eagle’s medium with 10% fetal bovine serum (Invitrogen, Carlsbad, CA). Choosing HEK293 cells, instead of other neurotypic cells, is justified by the moderate expression of both DRD1 and miR-504 in HEK293 cells, which we think is critical and appropriate for measurements of expression modulation. Cell transfections were carried out with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. For each experiment, quadruplicate transfections were performed for both the dual-luciferase reporter assay and the gene expression assay. Three independent experiments were conducted for replication.
Vector construct and dual-luciferase assay
The 3′ UTR of DRD1 with the rs686 A or G allele was fused with the luciferase reporter gene in the pGL3-Promoter vector (Promega, Madison, WI) (6). Mutant (GGG→CCC) of the DRD1 3′UTR was obtained by site-directed mutagenesis using the QuikChange II XL mutagenesis kit (Stratagene, La Jolla, CA) and a pair of primers: 5′-caaaagctagaggagattgctctgccctttgctattaagaaactaaggtac-3′ (forward) and 5′-gtaccttagtttcttaatagcaaagggcagagcaatctcctctagcttttg-3′ (reverse). The mutation was confirmed by DNA sequencing. Vector pRL-SV40 (Promega), containing the same SV40 promoter and later polyadenylation signal as the pGL3-Promoter, was used as an internal control and co-transfected with the DRD1 3′ UTR-bearing constructs in a ratio of 1:100. miRNA mimics and inhibitors of miR-504 and miR-296 and their negative controls (Dharmacon, Lafayette, CO) were co-transfected with the reporter vectors at a final concentration of 20 nM (mimics) or 30 nM (inhibitors). After 24 hours of transient transfection, the activities of firefly and Renilla luciferases were analyzed using the Dual-Luciferase Reporter Assay kit with the 20/20n luminometer method (Promega). The data of relative luciferase units were normalized to the negative control and analyzed with Student’s two tailed t-test.
Gene expression assay
A concentration of 20 nM miRNA mimic or 40 nM miRNA inhibitor was used for transient transfection. After 24 hours, the total RNA of each sample was isolated with the TRIzol reagent (Invitrogen). Quantitative reverse transcriptase-polymerase chain reaction (RT-PCR) analysis was then used to assess DRD1 expression in the transfected HEK293 cells. A TaqMan probe for DRD1 (Hs00377719_g1) was used with the default settings in the ABI Prism 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). The final gene expression level was normalized to that of an internal control, glyceraldehyde 3-phosphate dehydrogenase (GAPDH). The expression data were analyzed with the comparative Ct method, also known as the 2−ΔΔCt method, normalized to the negative control and analyzed with Student’s two tailed t-test.
RESULTS
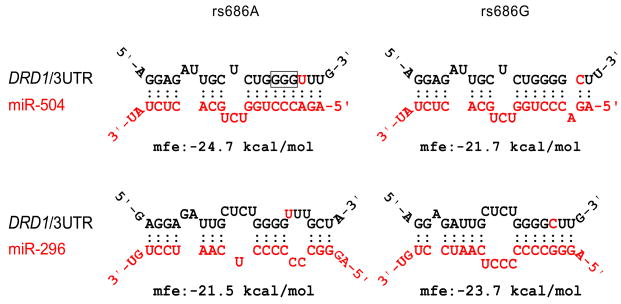
To identify miRNAs that likely target the vicinity of the rs686 polymorphism in the DRD1 3′UTR, we first utilized a computational algorithm, MicoInspector (9) (http://mirna.imbb.forth.gr/microinspector/), that yielded two candidate miRNAs, miR-504 and miR-296, whose seed sequences are complementary to the DRD1 mRNA sequence around the rs686 polymorphism (Figure 1). miR-504 has better base-pairing with the DRD1 3′UTR bearing rs686A, whereas miR-296 shows a more perfect match with that bearing rs686G. In the human genome, miR-504 is an intronic miRNA, hosted by the fibroblast growth factor 13 gene (FGF13) on chromosome Xq26.3, which is highly expressed in the brain and was previously associated with Börjeson-Forssman-Lehmann syndrome, an X-linked mental retardation (10). In contrast, miR-296 was identified in embryonic stem cells (11), and is an intergenic miRNA on human chromosome 20q13.32, close to the gene encoding Gαs, a G-protein coupled to the D1 dopamine receptor for cAMP signaling activation (12).
Figure 1.
Polymorphism rs686 in 3′UTR of DRD1 is predicted to be targeted by miR-504 and miR-296. miRNAs with a seed sequence base pairing on rs686 were identified with microInspector algorithm (http://mirna.imbb.forth.gr/microinspector/). Base pairing with rs686A/G and minimum free energy (mfe) calculation were performed with RNAhybrid algorithm (http://bibiserv.techfak.uni-bielefeld.de/rnahybrid/). The framed nucleotides, complementary to the seed sequence of miR-504, are the site of designed mutation (GGG→CCC) for the luciferase reporter assay.
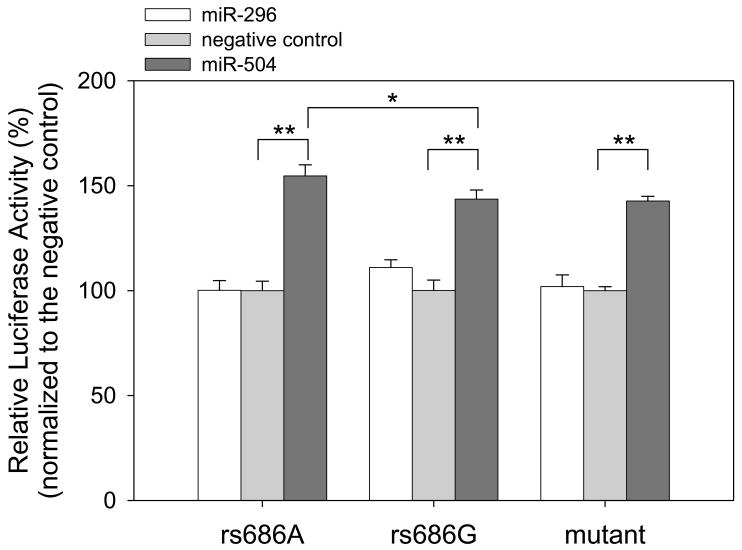
To determine predicted targeting of miR504 and miR-296 to the 3′UTR of DRD1, we co-transfected miRNA mimics of miR504 and miR-296 with the chimeric luciferase reporter bearing the rs686 A or G allele (6) into HEK293 cells in a comparison with a miRNA mimic negative control. Introduction of the miR-504 mimic significantly increased luciferase activity in the chimeric reporter-co-transfected cells (Figure 2), indicating that miR-504 up-regulates reporter expression. Furthermore, we observed a significantly greater increase of luciferase activity with the reporter bearing rs686A than that bearing rs686G (P < 0.05). In contrast, we found little significant change of luciferase activity in the co-transfection of the miR-296 mimic (Figure 2).
Figure 2.
Targeting of miR-504 to the 3′UTR of DRD1 up-regulates expression. miRNA mimics (20 nM) of miR-504, miR-296, and a negative control were co-transfected with the chimeric reporters (0.6 μg) and an internal transfection control vector (6 ng) into HEK293 cells in each well of 24-well plate. Chimeric reporters bear rs686A, rs686G, and a ‘seed’ mutant (GGG→CCC) of the DRD1 3′UTR (see Figure 1), respectively. After 24 hrs of transfection, luciferase activities in miR-504- or miR-296-transfected cells were measured and normalized to those in negative control-transfected cells. Representative data from triple independent experiments are shown with mean ± standard deviation (SD) (N = 4). * P < 0.05; ** P < 0.005; Student’s two-tailed t-test.
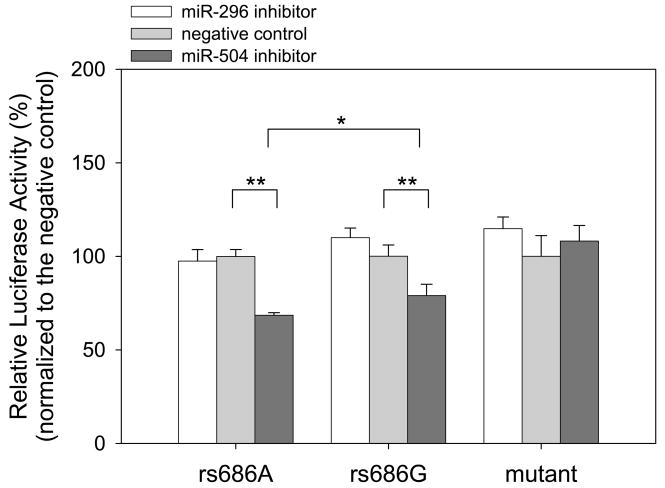
To block endogenous miRNA function, we next co-transfected miRNA inhibitors of miR-504 and miR296 with the chimeric reporters and compared the results with those of an miRNA inhibitor negative control. We found that introduction of miR-504 inhibitor significantly decreased luciferase activity (Figure 3), much more with the reporter bearing rs686A than that bearing rs686G (P < 0.05). However, we found no significant effect of miR-296 inhibitor on luciferase activity (Figure 3), just as with the miR-296 mimic (Figure 2).
Figure 3.
Inhibition of endogenous miR-504 expression down-regulates expression. miRNA inhibitors (30 nM) of miR-504, miR-296, and a negative control were co-transfected with the chimeric reporters (0.6 μg) and an internal transfection control vector (6 ng) into HEK293 cells in each well of the 24-well plate. The chimeric reporters bear rs686A, rs686G, and a ‘seed’ mutant (GGG→CCC) of the DRD1 3′UTR (see Figure 1), respectively. After 24 hrs of transfection, luciferase activities in miR-504-or miR-296-transfected cells were measured and normalized to those in negative control-transfected cells. Representative data from triple independent experiments are shown with mean ± SD (N = 4). * P < 0.05; ** P < 0.005; Student’s two-tailed t-test.
To verify that miR-504 indeed targets the DRD1 3′UTR through direct base pairing, we generated a mutation in the DRD1 3′UTR to disrupt its complementarity to the miR-504 seed sequence (GGG→CCC mutation in Figure 1). With the same assays, we found that the mutant showed significantly less alteration of luciferase activity than the reporter bearing the rs686A allele in co-transfection of miR-504 mimic (P < 0.05), similar to the one bearing the rs686G allele (Figure 2, right). Additionally, introduction of miR-504 inhibitor caused little effect on luciferase activity of the mutant (Figure 3, right). This indicates that direct base-pairing of miR-504 with the DRD1 3′UTR is requisite for miR-504 regulatory function in expression up-regulation.
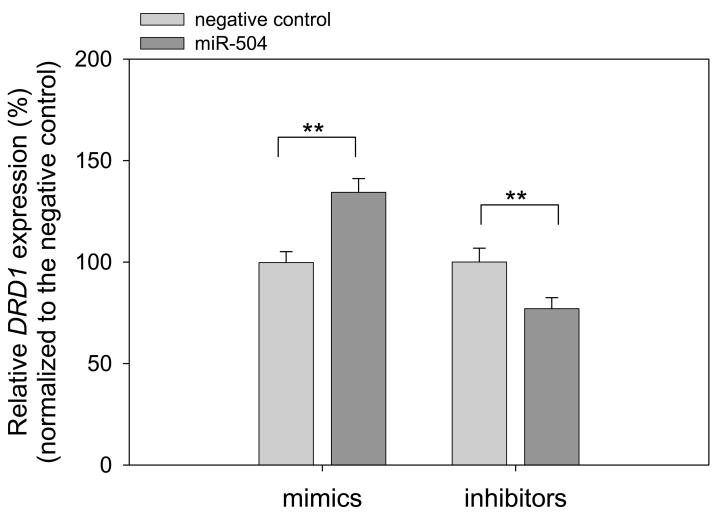
Finally, we assessed expression alteration of endogenous DRD1 in transfection of miR-504 mimic and inhibitor. Using real-time quantitative RT-PCR assay, we revealed that introduction of miR-504 mimic increased the amount of DRD1 mRNA, whereas transfection of miR-504 inhibitor decreased DRD1 expression (Figure 4). These findings are consistent with those from the reporter luciferase assays. Together, our results indicate that miR-504 up-regulates DRD1 expression by direct binding to the 3′UTR and that miR-504 targeting can cause allelic expression imbalance of DRD1.
Figure 4.
miR-504 targets the 3′UTR of DRD1. Expressions of DRD1 in HEK293 cells transfected with miR-504 mimic (20 nM) and inhibitor (40 nM) were assessed by quantitative real-time RT-PCR. Effects of miR-504 mimic and inhibitor in DRD1 expression were first normalized with internal control GAPDH, and then compared separately with those of their negative controls. Data are shown with mean ± SD (N = 4). **P < 0.005; Student’s two-tailed t-test.
DISCUSSION
Previously, we reported a polymorphism rs686 in the DRD1 3′UTR, functional in affecting DRD1 expression, that is significantly associated with ND (6). In this report, we reveal that the differential expressions of DRD1 bearing rs686 variants may be attributable to miR-504 targeting. Although our bioinformatics analyses predicted both miR-504 and miR-296 as potential candidates targeting the DRD1 3′UTR(Figure 1), only miR-504 was suggested to bind the DRD1 3′UTR directly (Figures 2–4). Little evidence was found for miR-296 (Figures 2 and 3). Further, we showed that miR-504 increased DRD1 mRNA (Figure 4) and up-regulated expression of the reporter bearing the DRD1 3′UTR with the rs686A allele to a greater extent than that with the rs686G allele (Figure 2), whereas inhibition of miR-504 attenuated expression of the reporters (Figure 3). These results are in accordance with the computational prediction that miR-504 has better base pairing with the DRD1 3′UTR bearing the rs686A allele. Moreover, our data provide new evidence of miRNA function in expression up-regulation, a novel gene modulation of miRNA identified recently (13), rather than wide-spread translation repression.
Polymorphism rs686 of DRD1 resides in the 3′UTR complementary to the seed sequence of miR-504, the base pairing of which is critical for miRNA binding and function. Genetic variation of rs686 from ‘A’ to ‘G’ in the DRD1 3′UTRcould significantly decrease DRD1 expression by miR-504 targeting. This miRNA-mediated differential modulation of DRD1 expression may alter the density of the D1 receptor in the brain. Because of the significant role of the D1 receptor in mediating dopamine action, the predisposition of DRD1 expression may contribute to the molecular mechanisms underlying ND. Furthermore, because miR-504 is chromosome X associated, the modulation of DRD1 expression by miR-504 may play a role in the sex influences on ND.
Acknowledgments
This project was funded in part by National Institutes of Health grants DA-12844 and DA-13783 (to M.D.L.).
Footnotes
FINANCIAL DISCLOSURE
The authors report no biomedical financial interests or potential conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Amos CI, Wu X, Broderick P, Gorlov IP, Gu J, Eisen T, et al. Genome-wide association scan of tag SNPs identifies a susceptibility locus for lung cancer at 15q25.1. Nature genetics. 2008;40:616–622. doi: 10.1038/ng.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frayling TM, Timpson NJ, Weedon MN, Zeggini E, Freathy RM, Lindgren CM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haiman CA, Le Marchand L, Yamamato J, Stram DO, Sheng X, Kolonel LN, et al. A common genetic risk factor for colorectal and prostate cancer. Nature genetics. 2007;39:954–956. doi: 10.1038/ng2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 6.Huang W, Ma JZ, Payne TJ, Beuten J, Dupont RT, Li MD. Significant association of DRD1 with nicotine dependence. Human genetics. 2008;123:133–140. doi: 10.1007/s00439-007-0453-9. [DOI] [PubMed] [Google Scholar]
- 7.Batel P, Houchi H, Daoust M, Ramoz N, Naassila M, Gorwood P. A haplotype of the DRD1 gene is associated with alcohol dependence. Alcoholism, clinical and experimental research. 2008;32:567–572. doi: 10.1111/j.1530-0277.2008.00618.x. [DOI] [PubMed] [Google Scholar]
- 8.Hettinger JA, Liu X, Schwartz CE, Michaelis RC, Holden JJ. A DRD1 haplotype is associated with risk for autism spectrum disorders in male-only affected sib-pair families. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:628–636. doi: 10.1002/ajmg.b.30655. [DOI] [PubMed] [Google Scholar]
- 9.Rusinov V, Baev V, Minkov IN, Tabler M. MicroInspector: a web tool for detection of miRNA binding sites in an RNA sequence. Nucleic acids research. 2005;33:W696–700. doi: 10.1093/nar/gki364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gecz J, Baker E, Donnelly A, Ming JE, McDonald-McGinn DM, Spinner NB, et al. Fibroblast growth factor homologous factor 2 (FHF2): gene structure, expression and mapping to the Borjeson-Forssman-Lehmann syndrome region in Xq26 delineated by a duplication breakpoint in a BFLS-like patient. Human genetics. 1999;104:56–63. doi: 10.1007/s004390050910. [DOI] [PubMed] [Google Scholar]
- 11.Suh MR, Lee Y, Kim JY, Kim SK, Moon SH, Lee JY, et al. Human embryonic stem cells express a unique set of microRNAs. Developmental biology. 2004;270:488–498. doi: 10.1016/j.ydbio.2004.02.019. [DOI] [PubMed] [Google Scholar]
- 12.Missale C, Nash SR, Robinson SW, Jaber M, Caron MG. Dopamine receptors: from structure to function. Physiological reviews. 1998;78:189–225. doi: 10.1152/physrev.1998.78.1.189. [DOI] [PubMed] [Google Scholar]
- 13.Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]