Abstract
The Retinoblastoma (Rb) protein is a conserved repressor of cell proliferation. In animals and plants, deregulation of Rb protein causes hyperproliferation and perturbs cell differentiation to various degrees. However, the primary developmental impact of the loss of Rb protein has remained unclear. In this study we investigated the direct consequences of Rb protein knockout in the Arabidopsis male germline using cytological and molecular markers. The Arabidopsis germ line derives from the unequal division of the microspore, producing a small germ cell and a large terminally differentiated vegetative cell. A single division of the germ cell produces the 2 sperm cells. We observed that the loss of Rb protein does not have a major impact on microspore division but causes limited hyperproliferation of the vegetative cell and, to a lesser degree, of the sperm cells. In addition, cell fate is perturbed in a fraction of Rb-defective vegetative cells. These defects are rescued by preventing cell proliferation arising from down-regulation of cyclin-dependent kinase A1. Our results indicate that hyperproliferation caused by the loss of Rb protein prevents or delays cell determination during plant male gametogenesis, providing further evidence for a direct link between fate determination and cell proliferation.
Keywords: male germline, pollen, cell cycle
In multicellular organisms, cell proliferation and cell differentiation are tightly coordinated both spatially and temporally. One key coordinator is the Rb-E2F pathway (1, 2). As the first identified tumor suppressor gene (3), Rb encodes the retinoblastoma (Rb) protein, which controls cell cycle progression from G1 into S phase (4). Upon phosphorylation by cyclin-dependent kinases (Cdks) at late G1 stage, the Rb protein loses its binding affinity for E2F family transcription factors. The released E2F transcription factors activate downstream cell cycle genes and commit cells to S phase. The Rb protein not only binds to E2F to repress transcription, but also recruits chromatin remodeling factors (5–10). Thus, the Rb protein exerts a broad range of cellular functions beyond cell cycle control, including differentiation (11), senescence (12), and apoptosis (13). Rb−/− knockout mice die from abnormal placenta development (14, 15). Mammalian genomes encode 2 other proteins related to the Rb protein: p107 and p130 (16–18), which further complicates the dissection of Rb function in mammals. It still remains unclear how the Rb protein coordinates cell proliferation and differentiation in animals (16).
In plants, the Rb-E2F pathway is conserved (19). The maize (Zea mays) genome contains 3 Rb genes, as in mammals (20–22). Arabidopsis thaliana contains a single Rb gene (RBR) (23). Loss of function of RBR completely impairs female gametogenesis, which precludes direct assessment of the role of the Rb protein in post-embryonic development (24, 25). Loss of RBR during female gametogenesis causes over-proliferation but does not appear to have a major effect on cell fate (25–28). To understand the role of RBR in development, different inducible systems disrupting RBR expression or over-expressing RBR were developed. Virus-induced gene silencing of NbRBR in Tobacco (Nicotiana benthamiana) caused deregulation of cell proliferation, differentiation, and endo-reduplication (29). RNA interference and inducible over-expression of Arabidopsis RBR impaired stem cell maintenance in roots (30). Inducible expression of a geminivirus RBR-binding protein in Arabidopsis leaves suggested that RBR prevents cell division and endoreduplication in a cell type-dependent manner (31).
As RBR represses MET1 expression (27) and likely recruits members of chromatin modifying complexes, the loss of RBR is expected to causes epigenetic modifications inherited through cell divisions (32, 33). Such modifications could impact on cell fate with secondary effects on proliferation. Alternatively deregulation of cell proliferation could impact directly on differentiation and cell fate as shown recently in Drosophila neuroblasts (34). It is thus difficult to analyze the direct effect of RBR on differentiation in experimental strategies perturbing RBR function during a large number of cell divisions before differentiation takes place.
In contrast to organogenesis of vegetative tissues, male gametogenesis comprises only 2 cell divisions. The first asymmetrical division of the meiotic microspore produces the larger vegetative cell and the smaller generative cell, which functions as a germ cell. The germ cell divides equally only once, producing 2 identical sperm cells. The differentiated vegetative cell produces the pollen tube, which delivers the 2 sperm cells to the 2 female gametes (35). In half of the haploid rbr microspores from heterozygous rbr/+ plants, the sudden deprivation of a functional RBR allele allows monitoring of the direct effect of the loss of RBR on cell proliferation and cell fate in the developing pollen.
We report that loss of RBR causes limited over-proliferation of the 2 pollen cell types. We further study the effect on cell fate using several markers and observe only a limited impact of rbr. The rbr phenotype is completely reversed in the absence of the cyclin dependent kinase A, leading to the hypothesis that rbr primarily targets cell cycle regulation with a secondary impact on cell fate.
Results and Discussion
We observed expression of RBR throughout pollen development in all cell types [supporting information (SI) Fig. S1]. Two mutant alleles rbr-1 (24, 26) and rbr-2, show reduced paternal transmission (Table S1) linked with reduced pollen viability (Fig. S2). We further characterized at the cellular and molecular levels the defects caused by rbr mutations during pollen development.
Limited Cell Over-Proliferation in rbr Pollen.
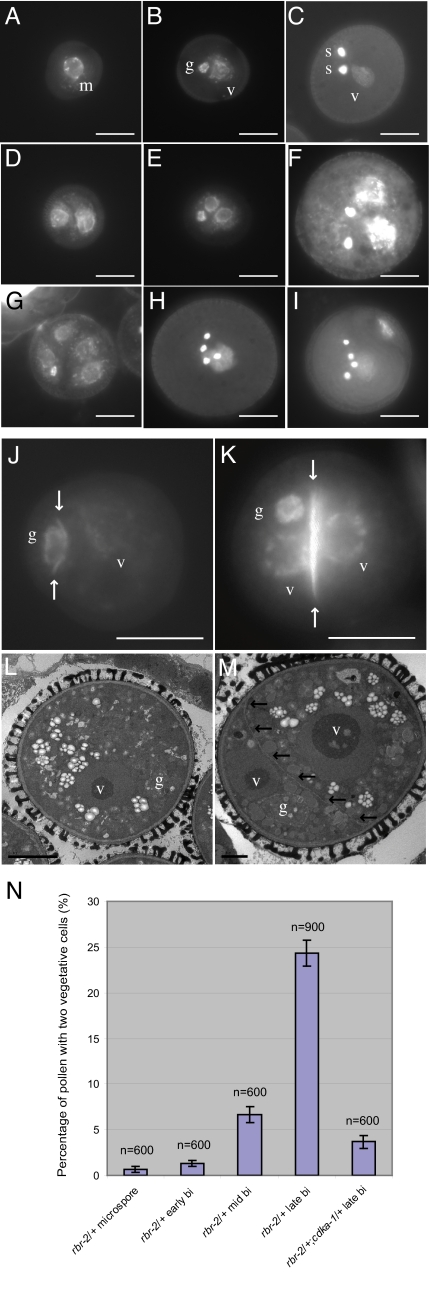
A recent study reported hyperproliferation of the vegetative nucleus of rbr pollen (26), but the origin of the supernumerary cells was not analyzed. We studied development of rbr pollen with nuclei stained by DAPI. WT microspores never divide equally (n = 1,000; Fig. 1A). In contrast, we observed in rbr/+ plants a very small fraction of microspores 0.67% (n = 600) that had divided equally into 2 cells (Fig. 1D). The very limited impact of rbr on microspore division might be explained by inheritance of residual RBR from the rbr/+ meiotic precursor.
Fig. 1.
Cell over-proliferation during pollen development in rbr/+ mutants. (A–C) WT pollen development. (A) The microspore with the undetermined cell fate undergoes an asymmetrical mitosis, leading to bicellular pollen (B). At that stage the pollen grain composes a large vegetative cell containing a small germ cell with a nucleus showing relatively higher chromatin compaction. The germ cell divides into 2 sperm cells with highly condensed chromatin, leading to the tricellular pollen grain (C). (D–I) rbr pollen development. Cell fates are determined on the basis of nuclear morphology. (D) At the microspore stage, rbr pollen grain with 2 undetermined cell nuclei. (E) Bi-cellular-stage rbr pollen grain with 2 vegetative cell nuclei and 1 germ cell nucleus. (F) Tri-cellular-stage rbr pollen with 2 vegetative cell nuclei and 2 sperm cell nuclei. (G) Tri-cellular-stage rbr pollen with 4 vegetative cell nuclei and 1 germ cell nucleus. (H) Tri cellular-stage rbr pollen with 1 vegetative cell nucleus and 4 germ cell nuclei. (I) Tri-cellular-stage rbr pollen with 2 vegetative cell nuclei and 4 germ cell nuclei. Nuclei are stained with DAPI. (Scale bars, 10 μm.) (J) Bi-cellular-stage WT pollen. The cell wall (arrows) is asymmetrically placed between the vegetative nucleus and the generative nucleus. (K) Bi-cellular-stage rbr pollen. The cell wall (arrows) is symmetrically placed between the 2 vegetative nuclei. Nuclei are stained with DAPI, and the cell walls are stained with aniline blue. (L and M) Transmission electron micrographs of bi-cellular-stage WT pollen (L) and rbr pollen (M). Note the internal wall indicated by arrows in rbr pollen. (Scale bars, 10 μm in J and K; 5 μm in L; 2 μm in M.) (N) Bar chart showing percentage of the pollen contains 2 vegetative cells in rbr-2/+ mutants at microspore, early bi-cellular, mid-bi-cellular, and late bi-cellular stages. At late bi-cellular stage, the over-proliferation in pollen from rbr-2/+;cdka-1/+ plants was reduced to one seventh of the over-proliferation in pollen from rbr-2/+ plants. Error bars correspond to SEs calculated on the basis of several samples of 100 pollen grains, and the size of total population analyzed (n) is indicated above each column. m, microspore nucleus; g, germ cell nucleus; v, vegetative cell nucleus; s, sperm cell nucleus.
The WT bicellular pollen comprises a vegetative cell with a large nucleus with de-condensed chromatin, and a smaller generative cell with a smaller nucleus (Fig. 1B). In contrast, rbr/+ plants produced 24.3% (n = 900) pollen containing 3 nuclei (Fig. 1E). One nucleus displayed the condensed chromatin typical of generative cells. The other 2 nuclei were larger with less condensed chromatin typical of vegetative cells (Fig. 1 B and E). Wild-type bicellular pollen is marked by a transient eccentric cell wall (Fig. 1 J and K). In contrast, the abnormal 3-celled rbr pollen observed at the bicellular WT stage showed an aberrant cell wall between the 2 vegetative cells (Fig. 1 K and M). The proportion of pollen containing 2 vegetative cells rose sharply during late bicellular stage, affecting half of the rbr pollen (Fig. 1N). We never observed any 3-celled pollen at that stage, suggesting that rbr causes an ectopic division of the vegetative cell.
Half of the pollen produced by rbr/+ plants inherits the rbr mutation. We estimated that 30% of the rbr pollen was dead at the bicellular stage (Fig. S2B; percentages are expressed relative to the estimated rbr pollen population and are thus twice as shown on Fig. S2B). Fifty percent of rbr pollen showed abnormal development and 20% showed WT morphology. At the tricellular stage, at least 60% of the rbr pollen was dead (Fig. S2B). As rbr-2 male transmission rate is of the order of 10% (Table S1), we could assume that 20% rbr pollen with normal morphology at bicellular stage underwent further development as WT. We thus estimated that, at the tricellular stage, less than 20% of the rbr pollen would derive from abnormal 3-celled pollen observed at bicellular stage. Corresponding to our estimate, we observed a total of 8% of abnormal pollen grains showing a complex array of phenotypes (n = 1000 pollen from rbr-2/+ plants). A predominant class of abnormal pollen contained 2 vegetative nuclei and 2 small sperm-like cells (4.6%; Fig. 1F). This class of abnormal pollen likely originated from the class shown in Fig. 1E in which either the generative cell divided into 2 sperm-like cells or the additional vegetative cell divided again, producing a generative cell. Several other types of pollen were observed (Fig. 1 G–I). Some pollen contained 4 vegetative nuclei and 1 sperm-like nucleus (1.6%; Fig. 1G). This pollen class likely results from an additional division of the 2 vegetative cells followed by 1 unequal division of 1 of the 4 vegetative cells producing a generative-like cell. We also observed pollen containing 4 sperm nuclei, either associated with 2 vegetative-like nuclei (1.2%; Fig. 1I) or inside 1 vegetative cell (3.4%; Fig. 1H). The latter class probably originates from a supernumerary division in the germ lineage. We did not observe any of the aforementioned phenotypes among WT pollen (n > 300 for each stage).
We targeted partial down-regulation of RBR in each pollen cell type by the expression of RBR hairpin RNAi constructs. Transgenic lines expressing the RBR RNAi construct under the control of the germ line-specific promoters of HTR10 (33,36,37) (47 lines observed) and GEX2 (38) (34 lines observed) did not show any defect in pollen viability or phenotype. In contrast, RBR RNAi expression restricted to the vegetative cell using the LAT52 promoter (39) caused a distinct increase in vegetative nuclear DNA fluorescence (Fig. S3) in 10%–25% of pollen, reflecting increased DNA synthesis. However we did not observe ectopic division of the vegetative cell. Hence, RBR RNAi expression under the LAT52 promoter caused a limited reduction of RBR activity leading to defects milder than the complete loss of RBR in rbr mutant alleles.
We conclude that rbr loss of function mostly affects the vegetative lineage and prevents arrest of cell division typical of vegetative cell fate. The loss of rbr function does not cause more than 2 additional rounds of cell division in comparison to WT. Further hyperproliferation in rbr pollen may be prevented by the limited supply of nutrients during pollen development leading to developmental arrest or death.
Cell Fate in rbr Pollen.
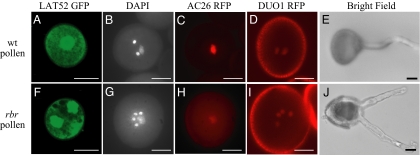
The nuclear morphology in rbr pollen suggested that cell over-proliferation in rbr pollen grains was associated with correct vegetative and germ cell fates. To address this question, we analyzed the expression of 6 cell fate markers in rbr pollen (Figs. 2 and 3). In the pollen displaying the rare phenotypic classes with duplication of the vegetative or germ cell lineages (Fig. 1I), the rbr pollen expressed the vegetative cell markers pLAT52-GFP (40) (Fig. 2F; n = 42) and pAC26-H2B-mRFP1 (41) (Fig. 2H; n = 14) in the large vegetative-like cells and the germ line marker pDUO1-DUO1-mRFP1 (42) in the small germ-like cells (Fig. 2I; n = 9). These observations suggested that rbr does not affect cell fate in this class of pollen. Accordingly we observed that 0.3% (n = 1,327) of pollen grains from rbr mutant germinated 2 pollen tubes likely originating from 2 vegetative cells (Fig. 2J). We concluded that, despite cell over-proliferation in rbr pollen, the vegetative cell fate and sperm cell fate are not affected when pollen experiences a complete duplication.
Fig. 2.
Cell fate specification in rbr pollen. (A–E) WT pollen grains. (F–J) rbr pollen grains. (A and F) Bi-cellular pollen grains expressing the vegetative cell marker pLAT52-GFP. (B and G) Fluorescence images of tri-cellular pollen grains stained with DAPI (C and H) The same pollen grains as B and G, respectively, expressing the vegetative cell marker pAC26-H2B-mRFP. WT pollen grain with 1 vegetative cell and 2 sperm cells (B) has only the vegetative cell nucleus expressing pAC26-H2B-mRFP (C). rbr pollen with 1 vegetative cells and 4 germ cells (G). Only the vegetative cell nucleus expresses pAC26-H2B-mRFP (H). (D and I) Tri-cellular pollen grains expressing the germ cell marker pDUO1-DUO1-mRFP. (E and J) In vitro pollen germination. WT pollen produces only 1 pollen tube germinated (E), whereas 2 pollen tubes germinated from the same rbr pollen grain (J). (Scale bars, 10 μm.)
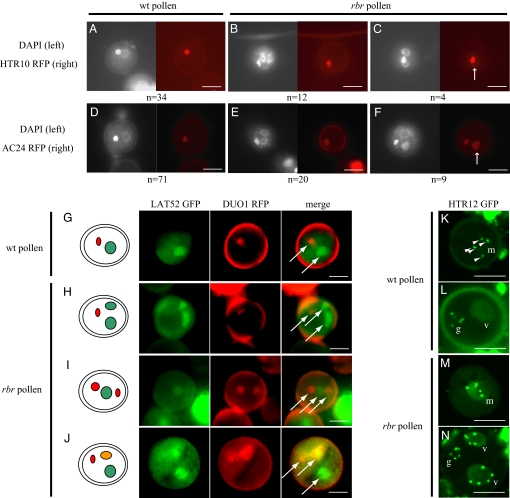
Fig. 3.
Mis-specification of cell fate in rbr pollen. (A–C) Fluorescence images of bi-cellular-stage pollen grains stained with DAPI (Left) and expressing pHTR10-HTR10-mRFP (Right). (D–F) Fluorescence images of bi-cellular-stage pollen grains stained with DAPI (Left) and expressing pAC24-H2B-mRFP (Right). Below each figure, n indicates the number of each case observed. (G–I) Fluorescence images of bi-cellular-stage pollen grains co-expressing the vegetative marker pLAT52-GFP and the germline marker pHTR10-HTR10-mRFP. Panels (Left to Right) are schematic representation of pollen co-expressing the 2 markers, GFP channel, RFP channel, and merged image. Arrows indicate the positions of cell nuclei. (K–N) Fluorescence images of microspores (K andM) and bi-cellular-stage pollen grains (L and N) expressing the centromeric Histone 3 variant fused to GFP (HTR12-GFP). In WT (K and L), HTR12-GFP accumulates at the 5 chromocenters (arrowheads) of the microspore nucleus (m) (K) and the germ cell nucleus (g) (L), but it is not possible to distinguish chromocenters in the vegetative cell nucleus (v). In contrast, in rbr pollen, HTR12-GFP is detected at chromocenters in microspores (M) and in both cell types at bi-cellular stage (N). (Scale bars, 10 μm.)
We further studied the cell fates in the 3-celled rbr pollen, most representative of the rbr phenotype at WT bicellular stage (Fig. 3). In WT bi-cellular pollen, the germ cell expresses the markers pHTR10-HTR10-mRFP (Fig. 3A) and pAC24-mRFP (41) (Fig. 3D). In two thirds of rbr pollen with 1 germ cell nucleus and 2 vegetative cell nuclei, the markers were correctly expressed (Fig. 3 B and E). However, in a third of 3-celled rbr pollen, both the germ cell nucleus and one of the vegetative-like nuclei expressed the germline markers (Fig. 3 C and F). Such ectopic expression was never observed in WT pollen (n > 300 for each marker). In addition, when we observed the co-expression of the vegetative marker pLAT52-GFP and the germline marker pDUO1-DUO1-mRFP (Fig. 3 G–J), a quarter of rbr pollen expressed the germline fate marker incorrectly. The additional vegetative-like cell expressed either the germline marker (n = 13 of 76; Fig. 3I) or both markers simultaneously (n = 7 of 76; Fig. 3J). We never observed mis-expression of the vegetative marker in the rbr germline (n = 76).
The rbr vegetative cell appears to behave like a microspore attempting imperfectly to reiterate an unequal division, producing an additional cell with vegetative fate, germ cell fate, or mixed fate identity. According to this hypothesis, genes expressed in the microspore but not later in the vegetative cell should be expressed in the vegetative cells of rbr pollen. Immunolocalization of the centromeric histone 3 variant HTR12 in WT tri-cellular pollen had shown that this protein marks only sperm cell nuclei (43). Accordingly, the centromeric histone HTR12 fused to GFP (HTR12-GFP) placed under the control of its own promoter (44) was expressed in the WT microspore (Fig. 3K) but was no longer detected in the vegetative cell nucleus after bi-cellular stage (Fig. 3L). HTR12-GFP expression was observed in all microspores from rbr-2/+ plants (n = 100; Fig. 3M), suggesting that RBR did not have a major impact on HTR12-GFP expression at that stage. In contrast to WT bicellular pollen, rbr 3-celled pollen showed ectopic expression of HTR12-GFP in vegetative cells (n = 24; Fig. 3N). This observation supported our hypothesis that the rbr vegetative cell retains the undetermined identity of the microspore. We thus concluded that rbr prevents cell fate establishment in the vegetative cell. A non-exclusive alternative explanation is that increased DNA methylation activity caused by increased MET1 expression in rbr background (27) impacts on heterochromatin organization, causing HTR12 recruitment. Our results thus led us to propose that loss of retinoblastoma function prevents cell fate establishment during male gametogenesis with an impact that depends on the cell type.
rbr Pollen Defects Are Rescued by Deregulation of the Cell Cycle.
Perturbation of the RBR pathway by over-expression of cyclin D3 impacts on cell proliferation and the timing of endo-reduplication in leaves and other vegetative tissues (45, 46). As endoreduplication usually marks differentiation in vegetative tissues, it was proposed that the cyclin D pathway controls cell differentiation (45). Although the impact on cell fate was not directly established in these studies, it is possible that the cyclin D pathway associated with cyclin-dependent kinase A (CDKA) regulates RBR function (47) and mediates the transition toward differentiation via the promotion of endo-reduplication in plants.
We further hypothesized that, if rbr directly prevents cell commitment to differentiate, preventing hyperproliferation in an rbr background should not rescue the defective cell fate in rbr pollen. To prevent cell proliferation without affecting cell fate, we choose to manipulate the Cyclin Dependent Kinase A (CDKA), which controls RBR licensing of the entry to S phase but presumably not the involvement of RBR in chromatin remodeling complexes. In animals, a few reports have shown involvement of CDKA homologues in cell fate in Drosophila (48) and in C. elegans (49). However, the mechanisms involving Cdks in cell polarity remain unclear. In Arabidopsis the function of the major Cdk CDKA has been solely linked to the control of the cell cycle in vegetative tissues (50) and during male gametogenesis (51–53). We thus rationalized that antagonizing RBR regulation of the cell cycle by CDKA manipulation would allow us to uncouple RBR functions in cell cycle regulation from other functions related to chromatin regulation. We tested in rbr-2 pollen the effect of hypo-proliferation caused by the loss-of-function cdka mutant allele. In the rbr-2/+;cdka-1/+ double mutant, we studied the transmission of rbr-2 and the phenotype of the pollen. The presence of cdka-1 almost completely rescued the paternal transmission efficiency of rbr-2 (Table S1), in agreement with the prediction of a complete viability of the rbr-2; cdka-1 (z = 32.52, P < 0.000001, 2-tailed test if no complementation; z = −1.24, P = 0.1075, 2-tailed test if full complementation). Accordingly, pollen lethality (Fig. S4) and over-proliferation (Fig. 1N) were greatly decreased in rbr-2/+;cdka-1/+ plants. The percentage of defective pollen was decreased by more than half in rbr-2/+; cdka-1/+ plants in comparison to that from rbr-2/+ plants, both at bicellular and tricellular stages (Fig. S2B), leading to full rescue of pollen death in rbr-2/+; cdka-1/+ plants.
It thus appears that restoring proliferation to WT levels in an rbr background rescues the defects in cell fate establishment observed in the rbr mutant. We propose that the primary effect of the loss of function of RBR in male gametogenesis is mediated by its role in cell proliferation.
Conclusions
Our study suggests that the control of the degree of proliferation by RBR is essential for proper cell fate establishment during male gametogenesis. One scenario is that the loss of retinoblastoma function primarily promotes hyperproliferation with secondary effects on commitment to cell fate during early development. It is not clear how cell fate is established in the bicellular pollen, but gradients of fate determinants have been hypothesized (35, 55). The additional cells produced by the rbr pollen might be positioned improperly relative to developmental cues, causing anomalous or mixed-cell fate. An alternative scenario proposes that RBR directly coordinates cell division and cell fate commitment. This could be mediated directly by the cell cycle machinery as suggested by a role of CDKA homologues in cell fate reported in a few cell types (48,49,56). A third non-exclusive hypothesis relates to the role of the Rb protein in chromatin modifications. In mammals it was shown that the Rb protein interacts with several chromatin remodeling complexes (6–9,11). These complexes might be conserved in plants. Cell fate establishment would then require chromatin modifications dependent on DNA duplication. We propose that, in the absence of RBR function, hyperproliferation coupled to the absence of recruitment of chromatin modifying complexes prevents this cell fate establishment.
Experimental Procedures
Plant Strains and Growth Conditions.
The WT ecotype Columbia (Col-0) was provided by the Nottingham Arabidopsis Stock Centre. The A. thaliana rbr mutant alleles (Columbia accession) used in this study were rbr-2 (SALK_002946; SALK collection), and rbr-3 (GABI_170G02; GABI-Kat collection) (23). Marker lines for cell identity were pDUO1-DUO1-mRFP (C24) (42), pAC24-H2B-mRFP, pAC26-H2B-mRFP (C24), and pHTR10-HTR10m-RFP (Col) (36). pLAT52-GFP (Col) was a gift from Alice Cheung (Amherst, MA).
RT-PCR.
Pollen at different stages of development were isolated and RNA extracted as described previously (41). Total RNA was prepared using the RNeasy mini kit (Qiagen) followed by DNase treatment (Ambion). Reverse transcription was performed by M-MuLV reverse transcriptase (New England Biolabs) with RNA ribonuclease inhibitor (Promega).
RBR Hairpin Interference Plasmid Construction and Transformation.
To express hairpin dsRNA targeted to RBR transcripts specifically in the vegetative cell, 500 bp of RBR coding sequence was cloned in sense and antisense orientations into a modified Gateway expression vector pK7LAT52RNAi harboring the vegetative cell-specific LAT52 promoter. A 495-bp LAT52 promoter fragment was amplified using KOD HiFi DNA Polymerase (Novagen) with primers containing restriction sites for HindIII and XhoI. The LAT52 promoter fragment was cloned into a Gateway RNAi destination vector pK7gwiwgL using the HindIII and XhoI sites to generate the pK7LAT52hpRNAi vector. A 500-bp RBR fragment was amplified by PCR and cloned by recombination using the Gateway cloning system according to manufacturer's instructions (Invitrogen) to generate the pLAT52hpRBR construct. Verified plasmid was transformed into Agrobacterium tumefaciens strain GV3101 and used to generate transgenic lines in A. thaliana ecotype Col-0 using the floral dip method. Transgenic progeny were selected for kanamycin resistance.
Microscopy and Image Processing.
Alexander staining and DAPI fluorescence in pollen grains were visualized as described previously (41). Light microscopy was performed on a stereomicroscope (DM6000; Leica). Images were recorded with a monochrome digital camera (Photometrics; Roper Scientific). Fluorescence was imaged using laser scanning confocal microscopy (LSM 510 META upright; Zeiss). Figures were composed with Adobe Photoshop 7.0.1 and Illustrator 10.0.3 (Adobe Systems). Transmission electron microscopy was performed with 85-nm thin sections were prepared on a Leica Ultracut UCT ultramicrotome. Samples were observed at 120 kV under a JEM-1230 transmission electron microscope (JEOL).
Supplementary Material
Acknowledgments.
This work was funded by Temasek LifeSciences Laboratory and the Singapore Millenium Foundation (F.B., Z.C., S.H.P.); and by the University of Leicester and the United Kingdom Biotechnology and Biological Sciences Research Council (D.T., S.H.). Additional support was received from the Department of Biological Sciences at National University of Singapore.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0810992106/DCSupplemental.
References
- 1.Harbour JW, Dean DC. The Rb/E2F pathway: expanding roles and emerging paradigms. Genes Dev. 2000;14:2393–2409. doi: 10.1101/gad.813200. [DOI] [PubMed] [Google Scholar]
- 2.Korenjak M, Brehm A. E2F-Rb complexes regulating transcription of genes important for differentiation and development. Curr Opin Genet Dev. 2005;15:520–527. doi: 10.1016/j.gde.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Friend SH, et al. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature. 1986;323:643–646. doi: 10.1038/323643a0. [DOI] [PubMed] [Google Scholar]
- 4.Weinberg RA. The retinoblastoma protein and cell cycle control. Cell. 1995;81:323–330. doi: 10.1016/0092-8674(95)90385-2. [DOI] [PubMed] [Google Scholar]
- 5.Brehm A, Kouzarides T. Retinoblastoma protein meets chromatin. Trends Biochem Sci. 1999;24:142–145. doi: 10.1016/s0968-0004(99)01368-7. [DOI] [PubMed] [Google Scholar]
- 6.Harbour JW, Dean DC. Chromatin remodeling and Rb activity. Curr Opin Cell Biol. 2000;12:685–689. doi: 10.1016/s0955-0674(00)00152-6. [DOI] [PubMed] [Google Scholar]
- 7.Dahiya A, Wong S, Gonzalo S, Gavin M, Dean DC. Linking the Rb and polycomb pathways. Mol Cell. 2001;8:557–569. doi: 10.1016/s1097-2765(01)00346-x. [DOI] [PubMed] [Google Scholar]
- 8.Nielsen SJ, et al. Rb targets histone H3 methylation and HP1 to promoters. Nature. 2001;412:561–565. doi: 10.1038/35087620. [DOI] [PubMed] [Google Scholar]
- 9.Vandel L, et al. Transcriptional repression by the retinoblastoma protein through the recruitment of a histone methyltransferase. Mol Cell Biol. 2001;21:6484–6494. doi: 10.1128/MCB.21.19.6484-6494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sharif J, et al. The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA. Nature. 2007;450:908–912. doi: 10.1038/nature06397. [DOI] [PubMed] [Google Scholar]
- 11.Macaluso M, Montanari M, Giordano A. Rb family proteins as modulators of gene expression and new aspects regarding the interaction with chromatin remodeling enzymes. Oncogene. 2006;25:5263–5267. doi: 10.1038/sj.onc.1209680. [DOI] [PubMed] [Google Scholar]
- 12.Funayama R, Ishikawa F. Cellular senescence and chromatin structure. Chromosoma. 2007;116:431–440. doi: 10.1007/s00412-007-0115-7. [DOI] [PubMed] [Google Scholar]
- 13.Harbour JW, Dean DC. Rb function in cell-cycle regulation and apoptosis. Nat Cell Biol. 2000;2:E65–67. doi: 10.1038/35008695. [DOI] [PubMed] [Google Scholar]
- 14.de Bruin A, et al. Identification and characterization of E2F7, a novel mammalian E2F family member capable of blocking cellular proliferation. J Biol Chem. 2003;278:42041–42049. doi: 10.1074/jbc.M308105200. [DOI] [PubMed] [Google Scholar]
- 15.Wu L, et al. Extra-embryonic function of Rb is essential for embryonic development and viability. Nature. 2003;421:942–947. doi: 10.1038/nature01417. [DOI] [PubMed] [Google Scholar]
- 16.Wikenheiser-Brokamp KA. Retinoblastoma family proteins: insights gained through genetic manipulation of mice. Cell Mol Life Sci. 2006;63:767–780. doi: 10.1007/s00018-005-5487-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giordano A, Rossi A, Romano G, Bagella L. Tumor suppressor pRb2/p130 gene and its derived product Spa310 spacer domain as perspective candidates for cancer therapy. J Cell Physiol. 2007;213:403–406. doi: 10.1002/jcp.21225. [DOI] [PubMed] [Google Scholar]
- 18.Stevaux O, Dyson NJ. A revised picture of the E2F transcriptional network and RB function. Curr Opin Cell Biol. 2002;14:684–691. doi: 10.1016/s0955-0674(02)00388-5. [DOI] [PubMed] [Google Scholar]
- 19.Shen WH. The plant E2F-Rb pathway and epigenetic control. Trends Plants Sci. 2002;7:505–511. doi: 10.1016/s1360-1385(02)02351-8. [DOI] [PubMed] [Google Scholar]
- 20.Gordon-Kamm W, et al. Stimulation of the cell cycle and maize transformation by disruption of the plant retinoblastoma pathway. Proc Natl Acad Sci USA. 2002;99:11975–11980. doi: 10.1073/pnas.142409899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grafi G, et al. A maize cDNA encoding a member of the retinoblastoma protein family: involvement in endoreduplication. Proc Natl Acad Sci USA. 1996;93:8962–8967. doi: 10.1073/pnas.93.17.8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sabelli PA, et al. RBR3, a member of the retinoblastoma-related family from maize, is regulated by the RBR1/E2F pathway. Proc Natl Acad Sci USA. 2005;102:13005–13012. doi: 10.1073/pnas.0506160102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vandepoele K, et al. Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell. 2002;14:903–916. doi: 10.1105/tpc.010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ebel C, Mariconti L, Gruissem W. Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature. 2004;429:776–780. doi: 10.1038/nature02637. [DOI] [PubMed] [Google Scholar]
- 25.Ingouff M, Jullien PE, Berger F. The female gametophyte and the endosperm control cell proliferation and differentiation of the seed coat in Arabidopsis. Plant Cell. 2006;18:3491–3501. doi: 10.1105/tpc.106.047266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston AJ, Matveeva E, Kirioukhova O, Grossniklaus U, Gruissem W. A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr Biol. 2008;18:1680–1686. doi: 10.1016/j.cub.2008.09.026. [DOI] [PubMed] [Google Scholar]
- 27.Jullien PE, et al. Retinoblastoma and Its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 2008;6:e194. doi: 10.1371/journal.pbio.0060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ingouff M, et al. The two male gametes share equal ability to fertilize the egg cell in Arabidopsis thaliana. Curr Biol. 2009;19:R19–R20. doi: 10.1016/j.cub.2008.11.025. [DOI] [PubMed] [Google Scholar]
- 29.Park JA, et al. Retinoblastoma protein regulates cell proliferation, differentiation, and endoreduplication in plants. Plant J. 2005;42:153–163. doi: 10.1111/j.1365-313X.2005.02361.x. [DOI] [PubMed] [Google Scholar]
- 30.Wildwater M, et al. The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell. 2005;123:1337–1349. doi: 10.1016/j.cell.2005.09.042. [DOI] [PubMed] [Google Scholar]
- 31.Desvoyes B, Ramirez-Parra E, Xie Q, Chua NH, Gutierrez C. Cell type-specific role of the retinoblastoma/E2F pathway during Arabidopsis leaf development. Plant Physiol. 2006;140:67–80. doi: 10.1104/pp.105.071027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. doi: 10.1038/nrg1601. [DOI] [PubMed] [Google Scholar]
- 33.Mathieu O, Reinders J, Caikovski M, Smathajitt C, Paszkowski J. Transgenerational stability of the Arabidopsis epigenome is coordinated by CG methylation. Cell. 2007;130:851–862. doi: 10.1016/j.cell.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 34.Chia W, Somers WG, Wang H. Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis. J Cell Biol. 2008;180:267–272. doi: 10.1083/jcb.200708159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McCormick S. Control of male gametophyte development. Plant Cell. 2004;16(suppl):S142–S153. doi: 10.1105/tpc.016659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ingouff M, Hamamura Y, Gourgues M, Higashiyama T, Berger F. Distinct dynamics of HISTONE3 variants between the two fertilization products in plants. Curr Biol. 2007;17:1032–1037. doi: 10.1016/j.cub.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 37.Okada T, Endo M, Singh MB, Bhalla PL. Analysis of the histone H3 gene family in Arabidopsis and identification of the male-gamete-specific variant AtMGH3. Plant J. 2005;44:557–568. doi: 10.1111/j.1365-313X.2005.02554.x. [DOI] [PubMed] [Google Scholar]
- 38.Engel ML, Holmes-Davis R, McCormick S. Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiol. 2005;138:2124–2133. doi: 10.1104/pp.104.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Twell D. Use of a nuclear-targeted ß-glucuronidase fusion protein to demonstrate vegetative cell-specific gene expression in developing pollen. Plant J. 1992;2:887–892. [Google Scholar]
- 40.Cheung AY, et al. Regulation of pollen tube growth by Rac-like GTPases. J Exp Bot. 2003;54:73–81. doi: 10.1093/jxb/erg044. [DOI] [PubMed] [Google Scholar]
- 41.Chen Z, Tan JL, Ingouff M, Sundaresan V, Berger F. Chromatin assembly factor 1 regulates the cell cycle but not cell fate during male gametogenesis in Arabidopsis thaliana. Development. 2008;135:65–73. doi: 10.1242/dev.010108. [DOI] [PubMed] [Google Scholar]
- 42.Rotman N, et al. A novel class of MYB factors controls sperm-cell formation in plants. Curr Biol. 2005;15:244–248. doi: 10.1016/j.cub.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fang Y, Spector DL. Centromere positioning and dynamics in living Arabidopsis plants. Mol Biol Cell. 2005;16:5710–5718. doi: 10.1091/mbc.E05-08-0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dewitte W, et al. Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell. 2003;15:79–92. doi: 10.1105/tpc.004838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dewitte W, et al. Arabidopsis CYCD3 D-type cyclins link cell proliferation and endocycles and are rate-limiting for cytokinin responses. Proc Natl Acad Sci USA. 2007;104:14537–14542. doi: 10.1073/pnas.0704166104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meijer M, Murray JAH. The role and regulation of D-type cyclins in the plant cell cycle. Plant Mol Biol. 2000;43:621–633. doi: 10.1023/a:1006482115915. [DOI] [PubMed] [Google Scholar]
- 48.Tio M, Udolph G, Yang X, Chia W. cdc2 links the Drosophila cell cycle and asymmetric division machineries. Nature. 2001;409:1063–1067. doi: 10.1038/35059124. [DOI] [PubMed] [Google Scholar]
- 49.Kostic I, Li S, Roy R. cki-1 links cell division and cell fate acquisition in the C. elegans somatic gonad. Dev Biol. 2003;263:242–252. doi: 10.1016/j.ydbio.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 50.Beemster GT, De Vusser K, De Tavernier E, De Bock K, Inze D. Variation in growth rate between Arabidopsis ecotypes is correlated with cell division and A-type cyclin-dependent kinase activity. Plant Physiol. 2002;129:854–864. doi: 10.1104/pp.002923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwakawa H, Shinmyo A, Sekine M. Arabidopsis CDKA;1, a cdc2 homologue, controls proliferation of generative cells in male gametogenesis. Plant J. 2006;45:819–831. doi: 10.1111/j.1365-313X.2005.02643.x. [DOI] [PubMed] [Google Scholar]
- 52.Nowack MK, et al. A positive signal from the fertilization of the egg cell sets off endosperm proliferation in angiosperm embryogenesis. Nat Genet. 2006;38:63–67. doi: 10.1038/ng1694. [DOI] [PubMed] [Google Scholar]
- 53.Kim HJ, et al. Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature. 2008;455:1134–1137. doi: 10.1038/nature07289. [DOI] [PubMed] [Google Scholar]
- 54.Boxem M, van den Heuvel S. lin-35 Rb and cki-1 Cip/Kip cooperate in developmental regulation of G1 progression in C. elegans. Development. 2001;128:4349–4359. doi: 10.1242/dev.128.21.4349. [DOI] [PubMed] [Google Scholar]
- 55.Eady C, Lindsey K, Twell D. The Significance of Microspore Division and Division Symmetry for Vegetative Cell-Specific Transcription and Generative Cell Differentiation. Plant Cell. 1995;7:65–74. doi: 10.1105/tpc.7.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cowan CR, Hyman AA. Cyclin E-Cdk2 temporally regulates centrosome assembly and establishment of polarity in Caenorhabditis elegans embryos. Nat Cell Biol. 2006;8:1441–1447. doi: 10.1038/ncb1511. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.