Abstract
MicroRNA-155 (miR-155) has emerged as a critical regulator of immune cell development, function, and disease. However, the mechanistic basis for its impact on the hematopoietic system remains largely unresolved. Because miRNAs function by repressing specific mRNAs through direct 3′UTR interactions, we have searched for targets of miR-155 implicated in the regulation of hematopoiesis. In the present study, we identify Src homology-2 domain-containing inositol 5-phosphatase 1 (SHIP1) as a direct target of miR-155, and, using gain and loss of function approaches, show that miR-155 represses SHIP1 through direct 3′UTR interactions that have been highly conserved throughout evolution. Repression of endogenous SHIP1 by miR-155 occurred following sustained over-expression of miR-155 in hematopoietic cells both in vitro and in vivo, and resulted in increased activation of the kinase Akt during the cellular response to LPS. Furthermore, SHIP1 was also repressed by physiologically regulated miR-155, which was observed in LPS-treated WT versus miR-155−/− primary macrophages. In mice, specific knockdown of SHIP1 in the hematopoietic system following retroviral delivery of a miR-155-formatted siRNA against SHIP1 resulted in a myeloproliferative disorder, with striking similarities to that observed in miR-155-expressing mice. Our study unveils a molecular link between miR-155 and SHIP1 and provides evidence that repression of SHIP1 is an important component of miR-155 biology.
Keywords: cancer, hematopoiesis, inflammation, miRNA, myeloid
In recent years, microRNAs have emerged as critical regulators of gene expression in a variety of mammalian cell types, including cells of the immune system (1–3). Through their ability to repress expression of specific target genes via direct 3′UTR interactions, several miRNAs have been shown to impact both physiological and pathological immune processes (4–12). Among the most prominent and well-studied immune system miRNAs to date, microRNA-155 (miR-155) clearly is involved in protective immunity when properly regulated, yet contributes to malignant conditions upon its dysregulated expression.
MiRNA-155 is expressed in a variety of immune cell types, including B cells (8, 9, 13–17), T cells (18), macrophages (19, 20), dendritic cells (8), and progenitor/stem cell populations (7, 21). Interestingly, miR-155 is found at low levels in most of these cells types until their activation by immune stimuli, such as antigen, Toll-like Receptor ligands, and inflammatory cytokines, which rapidly increase miR-155 expression (8, 9, 18–20). Consistent with its expression pattern, miR-155 appears to function in hematopoiesis and the immune response (7–9). For example, defective germinal center formation and antibody isotype class-switching have been observed in miR-155−/− mice following infection or vaccination (8, 9, 22). In T cells, miR-155 promotes skewing toward the Th1 subset (8, 9). In dendritic cells, miR-155 is necessary for proper activation of responder T cells in the context of antigen presentation (8).
Enhanced expression of miR-155 occurs constitutively in a subset of cancer cells of lymphoid (13–17) and myeloid origin (7, 23). We and others have recently demonstrated that sustained expression of miR-155 in the hematopoietic system leads to pathological outcomes. Our group expressed miR-155 ubiquitously in the hematopoietic compartment via bone marrow transfer of hematopoietic stem cells (HSCs) infected with a retroviral vector. This caused a myeloproliferative disorder (MPD) characterized by increased granulocyte/monocyte (GM) populations in the bone marrow, peripheral blood, and spleen, impaired erythropoiesis, and severe splenomegaly because of extramedullary hematopoiesis (7). Costinean et al. found that transgenic expression of miR-155 from a B-cell specific promoter can trigger a B-cell malignancy (4).
While much has been learned about miR-155-expression patterns and functions, the molecular basis underlying its biology is relatively uncharacterized. As a result, we have been searching for candidate targets of miR-155 implicated in overlapping biological processes. Using microarray technology, bioinformatics, and an extensive review of the literature, we have identified the inositol phosphatase SHIP1 as a target of miR-155. Through both gain and loss of function approaches, we demonstrate that miR-155 represses SHIP1 through direct 3′UTR interactions during both sustained and physiological expression of miR-155. Furthermore, specific knockdown of SHIP1 in the hematopoietic system using a miR-155-formatted siRNA against SHIP1 largely recapitulated the MPD phenotype we previously described in miR-155-expressing mice. Together, our data demonstrate a molecular link between miR-155 and SHIP1 in the immune system, and suggest that repression of SHIP1 is a critical aspect of miR-155 function.
Results
MicroRNA-155 Represses SHIP1 Through 3′UTR Interactions.
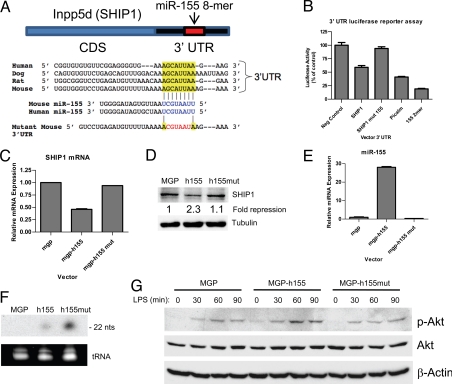
We previously performed a mRNA microarray analysis using RAW 264.7 macrophages stably expressing miR-155 to identify possible targets of miR-155 (7). Among the targets was SHIP1 (INPP5D), a gene that is repressed by miR-155 and which has a conserved 8-mer target “seed” in its 3′UTR (Fig. 1A). SHIP1 was of particular interest because miR-155 is the only miRNA with a highly conserved binding site located in the SHIP1 3′UTR according to the TargetScan algorithm (24), and because mice deficient in SHIP1 suffer from a myeloproliferative condition resembling that which we previously described for mice expressing miR-155 (7, 25–28).
Fig. 1.
MicroRNA-155 represses SHIP1 expression through 3′UTR interactions. (A) Schematic layout of the SHIP1 mRNA coding sequence (CDS) and 3′UTR, with the relative location of the miR-155 binding site. Depiction is not to scale. Sequence of mouse and human miR-155 and predicted interaction with conserved 8-mer miR-155 seeds found within the SHIP1 3′UTRs from different species (highlighted) are shown. The sequence of the SHIP1 3′UTR seed mutant used for reporter assays and predicted disruption of the miR-155 interaction is also shown. (B) Luciferase reporter assays were performed by transiently transfecting 293T cells with an empty plasmid (FUW) or miR-155-expressing plasmid (FUW-155), the indicated 3′UTR luciferase reporter plasmids, and a plasmid producing β-galactosidase. Luciferase values have been normalized to β-galactosidase, and the percent of luciferase activity in cells transfected with miR-155 is presented. Raw 264.7 cells stably infected with a retroviral vector expressing WT human miR-155 (MGP-h155), mutant seed human miR-155 (MGP-h155mut), or control (MGP) were assayed for SHIP1 levels by qPCR (C) and Western blotting (D). As a loading control for the Western blot, α-Tubulin was also assayed. The fold-repression of SHIP1 by the different constructs is shown. (E) Levels of mature human miR-155 in the different cell types were assayed by qPCR with primers that detect the WT mature miR-155 sequence. (F) RNA from the different cell types and a probe specific for the human miR-155 seed mutant sequence was used for Northern blotting. (G) The different cell types were stimulated with LPS (200 ng/ml) over the indicated time course and Ser-473-phosphorylated-Akt (p-Akt), total Akt, and β-Actin were assayed by Western blotting. Data represent at least 2 independent experiments.
To directly test whether miR-155 can repress SHIP1 through direct 3′UTR interactions, we cloned the 3′UTR of SHIP1 into a reporter plasmid downstream from luciferase and performed reporter assays using 293 T cells. While miR-155 produced from a cotransfected plasmid repressed expression of luciferase fused to the WT SHIP1 3′UTR, it failed to repress the SHIP1 3′UTR containing a mutated miR-155 seed sequence (Fig. 1B). As controls, miR-155 repressed the Picalm 3′UTR and 2-mer control constructs, but not the control UTR without miR-155 sites (see Fig. 1B). These data reveal that miR-155 directly targets the SHIP1 3′UTR leading to repressed expression.
To determine whether miR-155 can repress endogenous SHIP1, we assayed SHIP1 expression in RAW 264.7 cells expressing WT human miR155, human miR-155 containing a mutated seed region, or vector control. SHIP1 was measured at the mRNA and protein levels by quantitative PCR (qPCR) and Western blotting, respectively. WT miR-155 repressed SHIP1 mRNA and protein below control levels, while the miR-155 seed mutant had little impact on SHIP1 expression compared with the vector control (Fig. 1 C and D). The WT miR-155 was over-expressed in cells receiving the WT miR-155 vector (Fig. 1E) and the mature miR-155 seed mutant was produced in cells receiving the miR-155 seed-mutant vector (Fig. 1F), indicating that the specificity of the repression was determined by the seed region of miR-155.
SHIP1 is well known to be a negative regulator of the kinase Akt, a downstream target of the phosphatidylinositol-3-kinase (PI3K) pathway. Therefore, we assayed Akt activation following LPS treatment of the different RAW 264.7 derivatives. Consistent with reduced SHIP1 levels, cells expressing WT human miR-155 exhibited increased activation of Akt following LPS treatment, while Akt activation was similar in the control and human miR-155 seed mutant-expressing cells (Fig. 1G).
Enhanced Expression of SHIP1 in miR-155−/− Macrophages Following LPS Treatment.
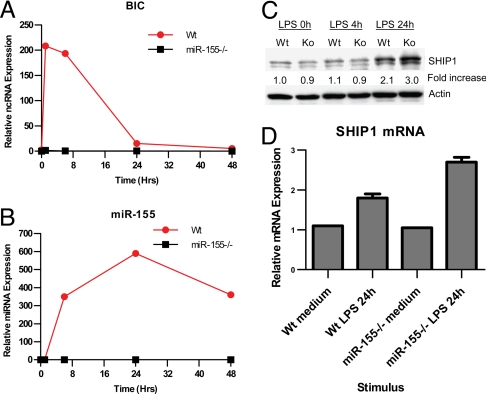
We next evaluated whether SHIP1 is regulated by miR-155 under physiological conditions. To achieve this, we generated bone marrow-derived macrophages (BMMs) from either WT or miR-155−/− mice. As LPS has been shown to be a potent inducer of miR-155 in macrophages (19), we stimulated these cells with LPS over a time course. In WT cells, rapid induction of the miR-155 precursor, BIC, was followed shortly after by elevated expression of mature miR-155 (Fig. 2 A and B), as previously seen with poly (I:C)-treated BMMs (19). We also noticed that while BIC levels fall considerably by 24 h, miR-155 expression peaks at this time point. Protein levels of SHIP1 were assayed at 0 h, 4 h, and 24 h after LPS treatment of both WT and miR-155−/− BMMs. We observed no change in SHIP1 protein levels up to 4 h following LPS treatment in cells of both genotypes (Fig. 2C). After 24 h, both WT and miR-155−/− BMMs demonstrated an increase in SHIP1 expression as compared with earlier time points. However, miR-155−/− BMMs had an enhanced level of SHIP1 protein compared to WT control cells at this time point (see Fig. 2C). SHIP1 mRNA levels reflected similar differences between WT and miR-155−/− BMMs following 24 h of LPS treatment (Fig. 2D). These results are consistent with miR-155 repressing SHIP1 expression after their induced coexpression by LPS, and demonstrate that this can occur under physiologically relevant conditions in primary cells.
Fig. 2.
Enhanced expression of SHIP1 in miR-155−/− macrophages following LPS treatment. Bone marrow-derived macrophages from WT or miR-155−/− mice were stimulated with 10-ng/ml LPS from Escherichia coli for the indicated periods of time. Expression of BIC (A) or mature miR-155 (B) was assayed by qPCR. Expression of SHIP1 was assayed in BMMs by Western blotting (C) and qPCR (D). β-Actin was assayed as a loading control for the Western blot, while qPCR data were normalized to L32. The fold-increase in SHIP1 expression versus the WT 0-h sample is shown. Data represent at least 2 independent experiments.
Knockdown of SHIP1 in Vivo Using siRNA in the Context of miR-155 Processing.
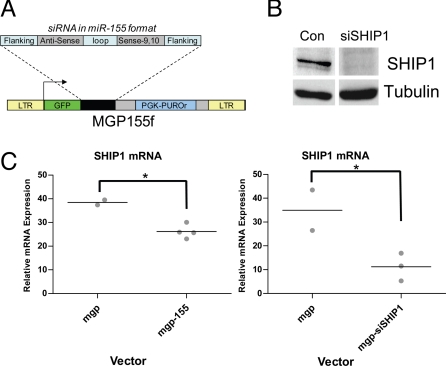
Having identified SHIP1 as a direct target of miR-155, we next determined if specific knockdown of SHIP1 levels could recapitulate the miR-155 MPD phenotype in mice that we recently described (7). Although this phenotype is predicted by the MPD observed in SHIP1−/− mice (26–28), we wanted to perform a specific reduction in SHIP1 using the same retroviral vector and bone marrow-reconstitution context as we used to promote sustained miR-155 expression in the hematopoietic system. To accomplish this, we built a retroviral vector that expresses a miR-155 formatted SHIP1 siRNA cassette (Fig. 3A). The cassette is driven by a RNA Polymerase II promoter and the hairpin arms and loop are comprised of mouse miR-155 sequences, while the stem structure contains an antisense sequence designed to target the SHIP1 coding sequence (see Fig. 3A). Knockdown of other genes in vitro has been shown using this approach (29). Following construction of this vector, we stably infected RAW 264.7 cells and assayed SHIP1 expression. SHIP1 protein levels were markedly reduced in cells expressing the siRNA cassette, compared to the control vector (Fig. 3B).
Fig. 3.
Knockdown of SHIP1 in vivo using siRNA in the context of miR-155 processing. (A) Schematic of the retroviral vector (MGP-155f) used to deliver siRNA against SHIP1 in miR-155 format. (B) Knockdown of SHIP1 was assayed in Raw 264.7 cells infected with MGP-siSHIP1 or control vector by Western blotting. α-Tubulin was assayed as a loading control. (C) Knockdown of SHIP1 in vivo by retroviral expression of miR-155 (MGP-155, n = 4 mice) or siSHIP1 (MGP-siSHIP1, n = 3 mice) in the hematopoietic compartment was assayed by qPCR using RNA isolated from total bone marrow following 2 months of hematopoietic reconstitution. Relative expression values have been normalized to L32 mRNA. A P-value of 0.05 or less using a Student's t test was considered statistically significant and indicated with an asterisk.
We next tested whether we could achieve knockdown of SHIP1 expression in vivo by expressing miR-155 or the siRNA against SHIP1. To this end, HSC-enriched bone marrow cells were infected with retroviral vectors encoding miR-155, siSHIP1 or controls and used to reconstitute lethally irradiated mice, as we previously described for delivery of miR-155. Following 2 months of reconstitution, we analyzed SHIP1 expression in the total bone marrow by qPCR. We observed a reduction in SHIP1 mRNA levels in mice expressing miR155 or siSHIP1 compared to control vectors (Fig. 3C).
Knockdown of SHIP1 in the Hematopoietic Compartment Causes a MPD Similar to that Observed in Mice Expressing miR-155.
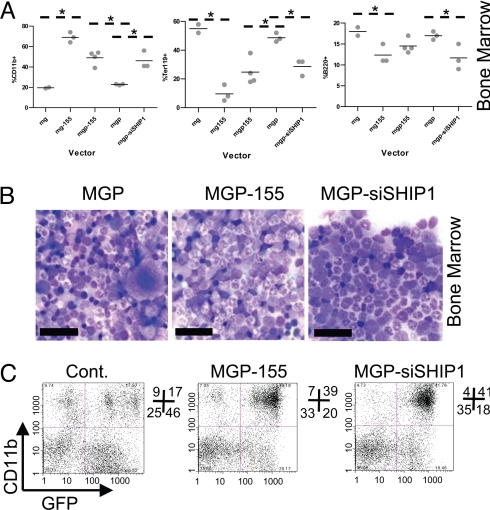
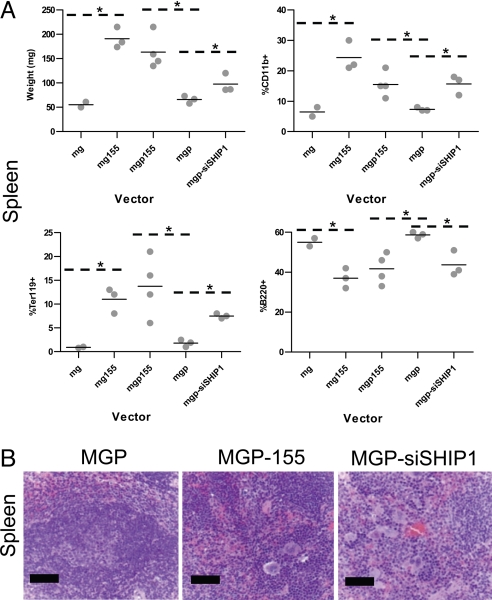
Mice expressing miR-155, siSHIP1, or control vectors were next studied to determine their impact on hematopoietic populations after 2 months of reconstitution. Both miR-155 (human and mouse sequences) and siSHIP1 caused similar MPD phenotypes in the bone marrow and spleen compared to control vectors (Figs. 4 and 5). Gross analysis revealed a miR-155 or siSHIP1-dependent splenomegaly and pale coloring of the bone marrow (see Fig. 5A and unpublished observations). Flow cytometry detected an increase in CD11b+ (Mac1+) myeloid populations in the bone marrow and spleen (see Figs. 4A and 5A). The percentage of Ter119+ erythroid precursor cells was increased in the spleen and decreased in the bone marrow, while the percentage of B220+ B cells was decreased in both the spleen and the bone marrow (see Figs. 4A and 5A).
Fig. 4.
Knockdown of SHIP1 or expression of miR-155 in the hematopoietic compartment cause similar MPDs in the bone marrow. (A) Bone marrow was extracted from mice expressing human miR-155 (MG-155, n = 3 mice), mouse miR-155 (MGP-155, n = 4 mice), siSHIP1 (MGP-siSHIP1, n = 3 mice), or control vectors (MG, n = 2 mice or MGP, n = 3 mice) 2 months following bone marrow reconstitution. Total bone marrow cells were assayed for expression of CD11b (Mac1), Ter119, or B220 using flow cytometry. Each dot represents an individual mouse. A P-value of 0.05 or less using a Student's t test was considered statistically significant and indicated with an asterisk. (B) Bone marrow from MGP, MGP-155, or MGP-siSHIP1 mice was smeared and Wright-stained. Photomicrographs are shown (1,000× magnification). (Scale bar, 20 μm.) (C) Representative flow cytometry plots from control, MGP-155 and siSHIP1 vector-containing mouse bone marrow analyzing GFP and CD11b expression.
Fig. 5.
Knockdown of SHIP1 or expression of miR-155 in the hematopoietic compartment causes splenomegaly and extramedullary hematopoiesis in the spleen. (A) Spleens were extracted from mice expressing human miR-155 (MG-155, n = 3 mice), mouse miR-155 (MGP-155, n = 4 mice), siSHIP1 (MGP-siSHIP1, n = 3 mice), or control vectors (MG, n = 2 or MGP, n = 3 mice) 2 months following bone marrow reconstitution. Spleens were weighed and RBC-depleted splenocytes subsequently assayed for expression of CD11b, Ter119, or B220 by FACS. Each dot represents an individual mouse. A P-value of 0.05 or less using a Student's t test was considered statistically significant and indicated with an asterisk. (B) Spleens from MGP, MGP-155, or MGP-siSHIP1 mice were fixed, sectioned, and H&E stained. Photomicrographs are shown (400× magnification). (Scale bar, 50 μm.)
Histological analyses of Wright-stained bone marrow smears confirmed the presence of pathological myeloproliferative conditions in miR-155- and siSHIP1-expressing mice, characterized by elevated numbers of GM progenitors at various stages of development compared to controls (see Fig. 4B). There was also a reduction in developing erythroid precursors and megakaryocytes in both miR-155 and siSHIP1 mice. Of note, miR-155 mice did exhibit a subtle increase in the number of dysplastic granulocytic cells compared with siSHIP1, possibly because of an additional miR-155 target. Flow cytometry also identified that both miR-155- and siSHIP1-expressing cells, which are GFP positive, are responsible for the increased myeloid populations (CD11b+) in the bone marrow (see Fig. 4C).
H&E staining of fixed spleen sections from miR-155 or siSHIP1 mice revealed expanded interfollicular regions containing developing myeloid populations, erythroid precursors, and megakaryocytes compared to control mice (see Fig. 5B). The normal follicular architecture of the spleen was disrupted by these expanded myeloid populations in both cases. Thus, miR-155 expression and specific SHIP1 knockdown in the hematopoietic system triggers marked extramedullary hematopoiesis, a likely consequence of the dysregulated blood cell development in the bone marrow.
Discussion
Similar to miR-155, many proteins have evolved to regulate immune cell function and cause disease upon their dysregulated expression. Among such proteins, the inositol phosphatase SHIP1 is expressed in the hematopoietic system and has a broad impact on the biology of different hematopoietic cell types (30). SHIP1 functions at the molecular level by hydrolyzing the 5′ phosphate of Phosphatidylinositol (PI)-3, 4, 5-P3 to generate PI-3,4-P2, a process that blocks PI3K-mediated membrane localization of certain PH domain-containing signaling molecules, such as Akt and PLCγ (31–33). Consequently, mice with a global SHIP1 deficiency develop a MPD characterized by increased GM populations, and decreased B-lymphocyte numbers. This condition is thought to occur as a result of its role as a negative regulator of signaling by growth factors and other immune receptors (30, 34). Furthermore, knockout of SHIP1 in B lymphocytes causes spontaneous germinal center formation and antibody class-switching (30, 35, 36), while a SHIP deficiency in T cells skews peripheral T lymphocytes toward Th1 and away from Th2 in response to an immune challenge (37). Thus, SHIP1 impacts the same cell types that express miR-155, and plays an opposing role in many cases. In the present study, we identify and characterize a direct link between miR-155 and SHIP1, whereby miR-155 can directly repress expression of SHIP1 and thereby impede its function.
The connection between miR-155 and SHIP1 has implications for normal immune physiology, as described above, as well as pathological conditions, such as cancer. Our present findings demonstrate a strong correlation between the MPDs caused by miR-155 expression or specific knockdown of SHIP1. Both perturbed the hematopoietic process, resulting in increased GM cell populations, reduced lymphocyte numbers, impaired erythropoiesis, and extramedullary hematopoiesis in the spleen. Therefore, miR-155 repression of SHIP1 may prove to be a contributing factor to human MPDs and myeloid leukemias, where miR-155 has been shown to be over-expressed (7, 23). Of note, SHIP1 is mutated in some acute myeloid leukemia patients, where loss of function has been implicated in the oncogenic process (38, 39). As miR-155 levels are elevated in certain B-cell lymphomas (13–17), and because SHIP1 is a negative regulator of B-cell activation and survival (30, 35, 36), the miR-155-SHIP1 axis should also prove to be of relevance to B-cell malignancies. Two recent abstracts suggest that this is the case (40, 41). It is also plausible that virally encoded orthologs of miR-155 (42–44), or miR-155 induction by viruses such as Epstein-Barr virus (45), can decrease SHIP1 expression en route to B-cell activation and transformation. Interestingly, the seed region of these orthologs is identical to that of miR-155, while the flanking regions have diverged dramatically from the mature mammalian miR-155 sequence. This would indicate that certain viral miRNAs have specifically evolved to repress seed-dependent targets of miR-155, including SHIP1.
An important question in the field is whether miRNAs function through repression of a single or a few targets, or via the cumulative impact of repressing large sets of targets. To date, several putative targets of miR-155 have been predicted through bioinformatic and proteomic approaches (46), indicating that there may be great complexity underlying miR-155 function. It may be that the MPD caused by miR-155 also involves other targets than SHIP1, but our studies show that knockdown of SHIP1 alone can phenocopy many of the effects of miR-155. MiR-155 has various roles in different cell types and physiological situations and analysis of particular specific targets, such as PU.1 (22), AID (47, 48), SOCS1 (49), and now SHIP1, suggest that individual targets likely make significant contributions to miR-155 function in a context-dependent manner. For example, PU.1 is repressed by miR-155 and its over-expression has been shown to recapitulate the Ig class-switching defect observed in miR-155−/− B cells (22). AID is repressed by miR-155 in B lymphocytes, which has been elegantly demonstrated via germline mutation of the miR-155 seed in the 3′UTR of AID (47, 48). The AID studies also provide evidence that miR-155 targeting of AID impacts Ig class-switching and the rate of c-myc translocations in B lymphocytes. SOCS1 targeting by miR-155 has just recently been shown to impact T-regulatory cell homeostasis (49). Similar observations have been made for specific targets of miR-150 (11, 50), miR-223 (6), and miR-17–92 (10, 12), suggesting a common theme of a few significant targets making dominant contributions to miRNA function. Ultimately, germline mutation of miRNA target-seed sequences within the 3′UTR regions of specific targets, as has been done for AID (47, 48), will provide the strongest argument for relevance.
As both miR-155 and SHIP1 regulate critical and overlapping functions of a variety of cell types of the immune system, therapeutic manipulation of this molecular interaction may prove to be useful in the treatment of diverse pathological conditions, including infection, cancer, and autoimmunity.
Materials and Methods
Cell Culture.
Raw 264.7 macrophage and 293T cells were cultured in complete DMEM with 10% FBS, 100 units/ml penicillin, 100 units/ml streptomycin. For generation of BMMs, bone marrow cells were isolated from the tibias and femurs of mice as previously described (19). All cells were cultured in a humidified incubator with 5% CO2 at 37 °C. Primary macrophages were stimulated using fresh DMEM containing 10 ng/ml 055-B5 LPS (Sigma), while Raw 264.7 cells were treated with LPS at 200 ng/ml.
Sequence Alignments.
SHIP1 3′UTR sequences from human, mouse, rat and dog were obtained and aligned with each other and with the miR-155 seed region using TargetScan (24).
DNA Constructs.
Retroviral constructs MG and MG-155 (human sequence) were described previously (19). Oligonucleotide sequences used to generate new constructs are provided as supplemental data [supporting information (SI) Table S1]. The MGP-155 expression cassette containing the mouse miR-155 hairpin sequence and flanking regions was cloned from cDNA made from LPS-treated BMMs. The cassette was subcloned into MGP. MGP is a modified pMSCV vector (Clontech) where GFP was placed downstream of the 5′ LTR, and the miR-155 expression cassette was cloned downstream of the GFP stop codon (detailed cloning strategy available upon request). The h155, h155mut, and siSHIP1 oligonucleotides, which produce mature human miR-155, human miR-155 seed mutant, and siRNA against mouse SHIP1, respectively, were designed using the Invitrogen Block-iT pol II miR RNAi strategy and PCR amplified using Fw NotI- and Rev XhoI- containing primers. The Invitrogen Block-iT RNAi Designer was used to predict the siRNA sequence against mouse SHIP1. For reporter assays, the Picalm 3′UTR was cloned as described previously (7). The mouse SHIP1 3′UTR was amplified by PCR from cDNA derived from mouse RAW 264.7 cells. This PCR product was cloned into pmiReport (Ambion) using SpeI and HindIII. Assembly PCR was used to mutate the 6-nucleotide miR-155 seed region. A 2-mer control insert and the IRAK1 3′UTR (used as negative control) were described previously (7).
Luciferase-Beta Gal Reporter Assays.
Experiments were performed as previously described using FUW, FUW-155, β-gal expression vector, and pmiReport vectors transfected into 293T cells (7). Transfections were carried out with TransIT 293 (Mirus). Data were normalized for transfection efficiency using a β-gal reporter and is represented as the ratio of luciferase activity of the transfection containing FUW-155 to that of the transfection containing FUW.
RAW 264.7 Stable Cell Lines.
To generate VSV-G-pseudotyped MSCV retroviruses, 2 × 106 293T cells were transfected with pGag-Pol, pVSV-G, and either MGP, MGP-h155, MGP-h155mut, or MGP-siSHIP1. Transfection was performed with TransIT 293 as per manufacturer's instructions. After 48 h, viral supernatant was harvested and used to infect 5 × 105 RAW 264.7 cells for 8 h in the presence of polybrene at 10 μg/ml. After 48 h, stably transduced cells were selected using puromycin at 10 μg/ml for 7 to 10 days.
Mice.
Wt mice on a C57BL/6 genetic background were bred and housed in the Caltech Office of Laboratory Animal Resources facility. Mice deficient in miR-155 and on a C57BL/6 genetic background were obtained from Alan Bradley at the Wellcome Trust Sanger Institute, Cambridge, U.K. All experiments were approved by the Caltech Institutional Animal Care and Use Committee.
Bone Marrow Reconstitution.
Experiments were performed as previously described (7), with the following modifications: HSC-enriched bone marrow was cultured for 48 h before the first spin infection using the respective retroviral vector. Transfection of retroviral constructs was performed using TransIT 293. Cells were subjected to 2 spin infections, and transduced cells were delivered to recipient mice through retro-orbital injection.
RNA Quantification.
SYBR Green-based quantitative real-time PCR was conducted using the 7300 Real-time PCR system (Applied Biosystems) to assay BIC, miR-155, 5s, SHIP1 mRNA, and L32 mRNA levels as described previously (7). Mature miR-155 and 5s RNA were assayed using a mirVana miRNA detection kit as per manufacturer's instructions (Ambion). Mouse BIC, SHIP1, and L32 mRNA were detected using specific primers (see Table S1). Northern blotting was performed as described (19) using a probe reverse complementary to the human miR-155 seed mutant (see Table S1).
Western Blotting.
Cell extracts were size-fractionated by SDS/PAGE and transferred to a nitrocellulose membrane using a semidry transfer apparatus (Bio-Rad). Western blotting was performed using the following antibodies: SHIP1 V-19 (sc-1963), SHIP1 M-14 (sc-1964), α-Tubulin B-7 (sc-5286), donkey anti-goat HRP-conjugated (sc-2020), goat anti-rabbit HRP-conjugated (sc-2004), goat anti-mouse HRP-conjugated (sc-2005) (Santa Cruz Biotechnology); α-Actin (A2066), β-Actin (A1978) (Sigma); SHIP1 (D1163), Akt1 (C73H10), Phospho-AKT (Ser-473) (Cell Signaling). Protein-expression intensities were determined using Scion Image software.
Flow Cytometry.
Fluorophore-conjugated monoclonal antibodies specific to CD11b (Mac1), Ter-119, or B220 (eBioscience) were used to stain RBC-lysed splenocytes and RBC-containing bone marrow cells that were washed and fixed with paraformaldehyde (1% final). Stained cells were assayed using a BD FACSCalibur flow cytometer (BD) and further analyzed with FlowJo software.
Morphological Assessment of Hematolymphoid Tissues.
Histological and cytological samples were prepared and analyzed as described previously (7).
Statistical Tests.
All statistical tests were performed using Microsoft Excel.
Supplementary Material
Acknowledgments.
We thank Alan Bradley for providing us with miR155−/− animals. This study was funded in part by the Irvington Institute Fellowship Program of the Cancer Research Institute (R.M.O.), the Graduate Research Fellowship Program of the National Science Foundation (A.A.C.), a clinical fellowship training grant from the California Institute of Regenerative Medicine and the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at University of California Los Angeles (D.S.R.), and by National Institute of Health Grant 1R01AI079243–01.
Footnotes
Conflict of interest: D.B. is on the Board of Directors and Chairs the Scientific Advisory Board of Regulus Therapeutics, a microRNA company. R.M.O has consulted for Regulus Therapeutics.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902636106/DCSupplemental.
References
- 1.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 2.Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 3.Lodish HF, Zhou B, Liu G, Chen CZ. Micromanagement of the immune system by microRNAs. Nat Rev Immunol. 2008;8:120–130. doi: 10.1038/nri2252. [DOI] [PubMed] [Google Scholar]
- 4.Costinean S, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Johnnidis JB, et al. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- 7.O'Connell RM, et al. Sustained expression of microRNA-155 in hematopoietic stem cells causes a myeloproliferative disorder. J Exp Med. 2008;205:585–594. doi: 10.1084/jem.20072108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez A, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thai TH, et al. Regulation of the germinal center response by microRNA-155. Science. 2007;316:604–608. doi: 10.1126/science.1141229. [DOI] [PubMed] [Google Scholar]
- 10.Ventura A, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132:875–886. doi: 10.1016/j.cell.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xiao C, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 12.Xiao C, et al. Lymphoproliferative disease and autoimmunity in mice with increased miR-17–92 expression in lymphocytes. Nat Immunol. 2008;9:405–414. doi: 10.1038/ni1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eis PS, et al. Accumulation of miR-155 and BIC RNA in human B cell lymphomas. Proc Natl Acad Sci USA. 2005;102:3627–3632. doi: 10.1073/pnas.0500613102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fulci V, et al. Quantitative technologies establish a novel microRNA profile of chronic lymphocytic leukemia. Blood. 2007;109:4944–4951. doi: 10.1182/blood-2006-12-062398. [DOI] [PubMed] [Google Scholar]
- 15.Kluiver J, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207:243–249. doi: 10.1002/path.1825. [DOI] [PubMed] [Google Scholar]
- 16.Tam W, Ben-Yehuda D, Hayward WS. BIC, a novel gene activated by proviral insertions in avian leukosis virus-induced lymphomas, is likely to function through its noncoding RNA. Mol Cell Biol. 1997;17:1490–1502. doi: 10.1128/mcb.17.3.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Berg A, et al. High expression of B-cell receptor inducible gene BIC in all subtypes of Hodgkin lymphoma. Genes Chromosomes Cancer. 2003;37:20–28. doi: 10.1002/gcc.10186. [DOI] [PubMed] [Google Scholar]
- 18.Haasch D, et al. T cell activation induces a noncoding RNA transcript sensitive to inhibition by immunosuppressant drugs and encoded by the proto-oncogene, BIC. Cell Immunol. 2002;217:78–86. doi: 10.1016/s0008-8749(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 19.O'Connell RM, Taganov KD, Boldin MP, Cheng G, Baltimore D. MicroRNA-155 is induced during the macrophage inflammatory response. Proc Natl Acad Sci USA. 2007;104:1604–1609. doi: 10.1073/pnas.0610731104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci USA. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgantas RW, 3rd, et al. CD34+ hematopoietic stem-progenitor cell microRNA expression and function: a circuit diagram of differentiation control. Proc Natl Acad Sci USA. 2007;104:2750–2755. doi: 10.1073/pnas.0610983104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vigorito E, et al. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Garzon R, et al. Distinctive microRNA signature of acute myeloid leukemia bearing cytoplasmic mutated nucleophosmin. Proc Natl Acad Sci USA. 2008;105:3945–3950. doi: 10.1073/pnas.0800135105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 25.Harder KW, et al. Perturbed myelo/erythropoiesis in Lyn-deficient mice is similar to that in mice lacking the inhibitory phosphatases SHP-1 and SHIP-1. Blood. 2004;104:3901–3910. doi: 10.1182/blood-2003-12-4396. [DOI] [PubMed] [Google Scholar]
- 26.Helgason CD, Antonchuk J, Bodner C, Humphries RK. Homeostasis and regeneration of the hematopoietic stem cell pool are altered in SHIP-deficient mice. Blood. 2003;102:3541–3547. doi: 10.1182/blood-2002-12-3939. [DOI] [PubMed] [Google Scholar]
- 27.Helgason CD, et al. Targeted disruption of SHIP leads to hemopoietic perturbations, lung pathology, and a shortened life span. Genes Dev. 1998;12:1610–1620. doi: 10.1101/gad.12.11.1610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu Q, et al. SHIP is a negative regulator of growth factor receptor-mediated PKB/Akt activation and myeloid cell survival. Genes Dev. 1999;13:786–791. doi: 10.1101/gad.13.7.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chung KH, et al. Polycistronic RNA polymerase II expression vectors for RNA interference based on BIC/miR-155. Nucleic Acids Res. 2006;34:e53. doi: 10.1093/nar/gkl143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leung WH, Tarasenko T, Bolland S. Differential roles for the inositol phosphatase SHIP in the regulation of macrophages and lymphocytes. Immuol Res. 2009 doi: 10.1007/s12026-008-8078-1. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Backers K, Blero D, Paternotte N, Zhang J, Erneux C. The termination of PI3K signalling by SHIP1 and SHIP2 inositol 5-phosphatases. Adv Enzyme Regul. 2003;43:15–28. doi: 10.1016/s0065-2571(02)00043-2. [DOI] [PubMed] [Google Scholar]
- 32.Lioubin MN, et al. p150Ship, a signal transduction molecule with inositol polyphosphate-5-phosphatase activity. Genes Dev. 1996;10:1084–1095. doi: 10.1101/gad.10.9.1084. [DOI] [PubMed] [Google Scholar]
- 33.Sly LM, Rauh MJ, Kalesnikoff J, Buchse T, Krystal G. SHIP, SHIP2, and PTEN activities are regulated in vivo by modulation of their protein levels: SHIP is up-regulated in macrophages and mast cells by lipopolysaccharide. Exp Hematol. 2003;31:1170–1181. doi: 10.1016/j.exphem.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 34.Kalesnikoff J, et al. The role of SHIP in cytokine-induced signaling. Rev Physiol Biochem Pharmacol. 2003;149:87–103. doi: 10.1007/s10254-003-0016-y. [DOI] [PubMed] [Google Scholar]
- 35.Helgason CD, et al. A dual role for Src homology 2 domain-containing inositol-5-phosphatase (SHIP) in immunity: aberrant development and enhanced function of b lymphocytes in SHIP−/− mice. J Exp Med. 2000;191:781–794. doi: 10.1084/jem.191.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Q, et al. The inositol polyphosphate 5-phosphatase ship is a crucial negative regulator of B cell antigen receptor signaling. J Exp Med. 1998;188:1333–1342. doi: 10.1084/jem.188.7.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tarasenko T, et al. T cell-specific deletion of the inositol phosphatase SHIP reveals its role in regulating Th1/Th2 and cytotoxic responses. Proc Natl Acad Sci USA. 2007;104:11382–11387. doi: 10.1073/pnas.0704853104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo JM, et al. Possible dominant-negative mutation of the SHIP gene in acute myeloid leukemia. Leukemia. 2003;17:1–8. doi: 10.1038/sj.leu.2402725. [DOI] [PubMed] [Google Scholar]
- 39.Luo JM, et al. Mutation analysis of SHIP gene in acute leukemia. Zhongguo Shi Yan Xue Ye Xue Za Zhi. 2004;12:420–426. [PubMed] [Google Scholar]
- 40.Chen L, Cui B, Zhang S, Chen G, Croce CM, Kipps TJ. Association between the proficiency of B-cell receptor signaling and the relative expression levels of ZAP-70, SHIP-1, and Mir-155 in chronic lymphocytic leukemia. Blood. 2008;112:3155. [Google Scholar]
- 41.Pedersen IM, et al. Onco-Mir-155 targets SHIP to regulate TNF-dependent B-cell lymphoma growth. Blood. 2008;112:604. [Google Scholar]
- 42.Gottwein E, et al. A viral microRNA functions as an orthologue of cellular miR-155. Nature. 2007;450:1096–1099. doi: 10.1038/nature05992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skalsky RL, et al. Kaposi's sarcoma-associated herpesvirus encodes an ortholog of miR-155. J Virol. 2007;81:12836–12845. doi: 10.1128/JVI.01804-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao Y, et al. A functional MicroRNA-155 ortholog encoded by the oncogenic Marek's disease virus. J Virol. 2009;83:489–492. doi: 10.1128/JVI.01166-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yin Q, et al. MicroRNA-155 is an Epstein-Barr virus-induced gene that modulates Epstein-Barr virus-regulated gene expression pathways. J Virol. 2008;82:5295–5306. doi: 10.1128/JVI.02380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Selbach M, et al. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 47.Dorsett Y, et al. MicroRNA-155 suppresses activation-induced cytidine deaminase-mediated Myc-Igh translocation. Immunity. 2008;28:630–638. doi: 10.1016/j.immuni.2008.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Teng G, et al. MicroRNA-155 is a negative regulator of activation-induced cytidine deaminase. Immunity. 2008;28:621–629. doi: 10.1016/j.immuni.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lu LF, et al. Foxp3-dependent microRNA155 confers competitive fitness to regulatory T cells by targeting SOCS1 protein. Immunity. 2009;30:80–91. doi: 10.1016/j.immuni.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu J, et al. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.