Abstract
Disulfide-rich peptide venoms from animals such as snakes, spiders, scorpions, and certain marine snails represent one of nature's great diversity libraries of bioactive molecules. The various species of marine cone shells have alone been estimated to produce >50,000 distinct peptide venoms. These peptides have stimulated considerable interest because of their ability to potently alter the function of specific ion channels. To date, only a small fraction of this immense resource has been characterized because of the difficulty in elucidating their primary structures, which range in size between 10 and 80 aa, include up to 5 disulfide bonds, and can contain extensive posttranslational modifications. The extraordinary complexity of crude venoms and the lack of DNA databases for many of the organisms of interest present major analytical challenges. Here, we describe a strategy that uses mass spectrometry for the elucidation of the mature peptide toxin components of crude venom samples. Key to this strategy is our use of electron transfer dissociation (ETD), a mass spectrometric fragmentation technique that can produce sequence information across the entire peptide backbone. However, because ETD only yields comprehensive sequence coverage when the charge state of the precursor peptide ion is sufficiently high and the m/z ratio is low, we combined ETD with a targeted chemical derivatization strategy to increase the charge state of cysteine-containing peptide toxins. Using this strategy, we obtained full sequences for 31 peptide toxins, using just 7% of the crude venom from the venom gland of a single cone snail (Conus textile).
Keywords: conotoxins, cysteine derivatization, de novo sequencing, electron transfer dissociation, mass spectrometry
Venomous animals such as spiders, snakes, scorpions, and certain sea snails produce a vast array of bioactive peptides, many of which are rich in disulfide bonds that stabilize their 3-dimensional structures. The enormous size of this natural combinatorial library can be appreciated by considering just the ≈500 known species of cone snail, which are estimated to produce >50,000 distinct peptide toxins (conotoxins) (1, 2); and this number is dwarfed by the toxins present in the ≈38,000 known species of spider (3). Interest in these peptide toxins derives in part from their potent and specific interactions with ion channels (3, 4) and in part from their structural integrity as disulfide-rich miniproteins (5, 6) Consequently they have become important tools for studying ion channels (7) and have considerable potential as pharmaceuticals (8, 9).
One limit on the utilization of this rich resource of bioactive peptide toxins has been the difficulty in elucidating their primary structures, which range in size between 10 and 80 aa, include up to 5 disulfide bonds, and in certain cases (e.g., cone snail venom) can contain extensive posttranslational modifications. The extraordinary complexity of crude venom samples (often containing >100 components) (4) and the lack of DNA databases for many of the organisms of interest presents a major analytical challenge. Most of the currently known primary structures of peptide toxins have been obtained using Edman sequencing (10), comparison of tandem MS (MS/MS) data with cDNA sequences (11–17), or a combination of these techniques (18–21). Although MS reduces the requirement for exhaustive purification compared with Edman sequencing (22, 23), it is often difficult to obtain full length de novo sequence exclusively by MS/MS. To date there are relatively few such examples, where the unambiguous sequence of a toxin has been determined exclusively by MS/MS in the absence of cDNA data (24–29). A major problem encountered in such analyses is incomplete sequence information, because full length de novo sequencing requires almost always backbone cleavage between each adjacent amino acid. Unfortunately, the most widely used fragmentation technique—collision activated dissociation (CAD)—rarely yields complete sequence coverage across the entire peptide; it usually yields selective fragmentation, precluding complete sequence characterization from N to C termini—especially for longer peptides (30, 31). The presence of posttranslational modifications further complicates de novo sequencing by CAD. The recent development of electron capture dissociation (ECD) and electron transfer dissociation (ETD) has begun to alleviate these problems (32, 33) because they yield considerably less selective fragmentation along the peptide backbone. However, these newer dissociation techniques have their own set of limitations, perhaps the most critical being the requirement for relatively high charge state precursor ions with consequently low m/z ratios to produce the most informative fragmentation spectra (34).
We present a strategy aimed at rapidly and sensitively elucidating the peptide toxin components of crude venom samples. Part of the impetus for this work was the historically low rate for determining the primary sequences of mature conotoxins (≈1 sequence/year/species) (35) and the large amounts of sample required for these analyses. Key to our strategy is the use of ETD, with its potential to produce complete sequence coverage. However, because ETD only yields comprehensive sequence coverage when the m/z ratio is low and at the same time the charge state (z) sufficiently high (34), we combined ETD with a targeted chemical derivatization strategy to increase the charge state of cysteine-containing peptide toxins. Increasing the charge state lowers the m/z ratio, ensuring extensive fragmentation of the peptide toxins with ETD. Here, the Cys residues were converted to their dimethyl Lys analogs, using commercially available N,N-dimethyl-2-chloro-ethylamine (36). The resulting charge increase led to a striking enhancement of the ETD fragmentation. We term this procedure “ETD of charge enhanced precursors,” or “ETD with CEP.” Using this strategy, we obtained sequences for 31 peptide toxins from just 7% of the crude venom from the venom gland of a single cone snail (Conus textile).
Results
Overall Strategy.
Our strategy for elucidating the primary structures of toxin components of crude venom samples involves 3 steps (Fig. 1): (I) off-line separation of the crude venom into fractions; (II) MS survey of each fraction to determine the accurate masses of the toxins and their disulfide content; and (III) MS/MS sequencing of toxins, using CAD, ETD, and ETD with CEP. This strategy was evaluated on the crude venom from a single specimen of C. textile, in which the toxins range in size between 1,000 and 4,000 Da, have up to 5 disulfide bonds, and are extensively modified (4, 37). To date, in Conus some 9 disulfide bonding patterns have been identified (35) across the 9 superfamilies that comprise at least 17 different pharmacologies; here we have made no attempt to map disulfide connectivities, but focus on gaining full N- to C-terminal sequence information. The present analysis used a total of 7% of the venom from a single cone snail specimen.
Fig. 1.
Overview of the de novo sequencing strategy. (I) UV trace of HPLC separation of crude venom extract from C. textile. (II) MALDI TOF MS of fraction i after no treatment, reduction, and alkylation. (III) On-line LC ESI-MS/MS using CAD and ETD on reduced and alkylated aliquots of fraction i. The final step shows the conversion of Cys residues to dimethylated Lys analogs followed by ETD fragmentation; MS/MS is shown for the (M + 5H)+5 ion of the 1,889.715 Da species in II. c ions are indicated by ⌉ and z· ions by L. Shell image Copyright 2005, Richard Ling.
Off-line separation of the crude venom into fractions.
To improve the dynamic range of our procedure and increase our chance of identifying the less abundant toxins, we separated the crude venom mixture into 16 fractions using off-line HPLC. These fractions each yielded between 1 and 15 different cysteine-containing toxins, resulting in the detection of a total of 92 distinct conotoxins (Table S1).
MS survey to determine the accurate masses of the toxins present in each fraction and their disulfide content.
MALDI-TOF MS of each fraction provided us with accurate molecular masses (<5 ppm) of the major toxin components. To determine the number of disulfide bonds in each toxin, we reduced, alkylated and analyzed the fractions by MALDI-TOF MS. The number of disulfide bonds present in each toxin was determined from the mass differences between the native and reduced toxin (i.e., a difference of 2 Da/disulfide bond). In addition, the number of cysteines was determined by examining the mass difference between the reduced and alkylated toxins (i.e., a difference of 57 Da/cysteine). In the example shown in Fig. 1, two dominant masses are observed in the untreated fraction at m/z 1,889.715 and 1,947.714, respectively. After reduction, these m/z's shift to respectively 1,895.769 and 1,953.769, indicating the presence of 3 disulfide bonds in each of these toxins. Subsequent to alkylation, the increase in the m/z value to 2,237.912 and 2,295.915, respectively, confirm the presence of 6 cysteines in each toxin. Table S1 provides the molecular masses and disulfide content of the 90 dominant species present in the venom sample (with signal-to-noise ratios >5). Of these components, 3 contain a single disulfide bond, 33 contain 2 disulfides, 48 contain 3 disulfides, 5 contain 4 disulfides, and 1 contains 5 disulfides. Their molecular masses range between 962 and 4,188 Da.
MS/MS sequencing of the toxins present in each fraction, using a combination of CAD, ETD, and ETD with CEP.
In the present work, we sought to develop an efficient de novo sequencing methodology, allowing complete N- to C-terminal sequence elucidation, which is applicable to a large variety of toxins. Thus, we first acquired LC-ESI-MS/MS data, using alternating CAD and ETD on each selected precursor ion. These dissociation methods are complementary in that CAD generally provides high sequence coverage for small (<1,500-Da) doubly-charged peptides, whereas ETD provides extensive coverage for larger peptides—especially when the charge state is high and the m/z ratio low (30, 34). To help locate the numerous Cys residues, we recorded MS/MS spectra on both the reduced (free thiol-containing cysteines) and alkylated (carboxyamidomethyl cysteines) forms of the toxins. In certain cases, the aforementioned steps did not yield complete sequence information. This was especially true for peptides with higher molecular masses (>1,500-Da) and lower charger states (≤ 3). To overcome this problem, we increased the charge state of the toxins by converting the Cys residues to their dimethyl lysine analogs (an addition of 71 Da/cysteine). The resulting ETD with CEP gave significantly improved fragmentation, further facilitating de novo sequencing of these higher molecular mass toxins.
De Novo Sequencing.
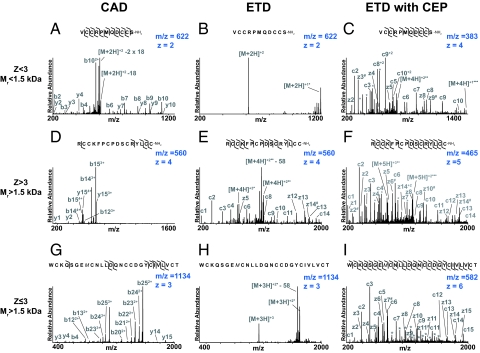
The characteristics of the fragmentation induced by CAD, and ETD are strongly dependent on the mass of the toxin and the charge state observed using ESI (Fig. 2). For lower masses and charge states (Mr < 1,500 Da and Z < 3), CAD frequently provides virtually complete sequence coverage (Fig. 2A), whereas ETD fragmentation is often sparse (Fig. 2B). Increasing the charge state by converting the Cys residues to their charged dimethyl lysine analogs (i.e., ETD with CEP) greatly enhances the ETD fragmentation (Fig. 2C), yielding virtually complete sequence coverage of the toxin. In the case shown, the ETD with CEP spectrum did not unambiguously resolve the order of the first 2 residues, whereas the CAD spectrum does—a complementarity that is very useful for comprehensive de novo sequencing. The complete sequence was determined to be VCCRPMQDCCS with an amidated C terminus (measured Mr of the native toxin = 1,238.416 Da, calculated Mr = 1,238.411 Da). A subsequent search of the National Center for Biotechnology Information (NCBI) database with our de novo sequence revealed the cDNA of a precursor containing this sequence, further confirming the present assignment.
Fig. 2.
MS/MS spectra of 3 toxins studied with 3 different dissociation techniques [CAD (A, D, and G), ETD (B, E, and H), and ETD with CEP (C, F, and I)]. N-terminal fragment ions (b and c) are indicated by ⌉ and C-terminal fragment ions (y and z·) are indicated by L. Doubly charged ions are indicated with an asterisk. z· ions resulting from cleavage at cysteine and loss of the cysteine side chain are indicated with # (54). Charge reduced species are labeled in the spectrum with ∙, indicating the number of electrons transferred to the precursor ion.
As the masses of the toxins increase, the CAD spectra yield increasingly lower sequence coverage (Fig. 2 D and G), as do the ETD spectra of low charge state species (Z < 3) (Fig. 2H). By contrast, when Z > 3 the ETD fragmentation often yield close to complete sequence coverage (Fig. 2E). Although the last 2 C-terminal fragment ions were not observed in Fig. 2E, the sequence could nevertheless be unambiguously determined using our prior determination of the number of Cys residues in step II of our procedure (Fig. 1). In this way, the sequence was determined to be RCCKFPCPDSCRYLCC with an amidated C terminus (measured Mr of the native toxin = 1,888.707 Da, calculated Mr = 1,888.710 Da). Again, a subsequent search of the NCBI database with our de novo sequence revealed the cDNA of a precursor containing this sequence, confirming the present assignment.
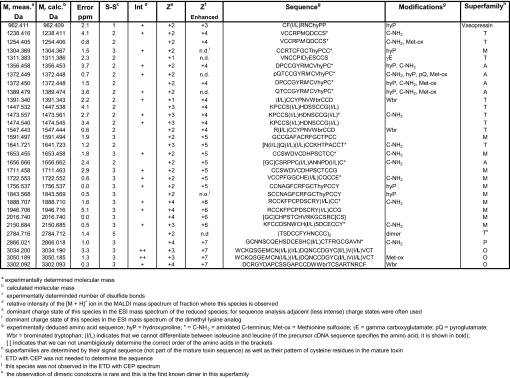
There is a large number of cases where neither CAD nor ETD yields complete sequence coverage. In such cases, ETD with CEP yields striking improvements in fragmentation efficiency and sequence coverage, especially when the charge state of the toxin before conversion of the cysteines to their dimethyl lysine analogs is ≤3. As an example, Fig. 2H shows the ETD spectrum of the highest observed charge state (3+) of a reduced and iodoacetamide alkylated toxin with Mr = 3,398.321 Da. The resulting spectrum is dominated by the unreacted precursor and charge-reduced species, with virtually no informative fragment ions. Increasing the charge state of the toxin from 3+ to 6+ via the aminoalkylation reaction leads to a striking improvement in the fragmentation efficiency (Fig. 2I)—enabling de novo sequence determination of this 27 residue toxin, except for the order of the 2 C-terminal residues. Because the toxin contains 6 cysteines (step II, Fig. 1), 1 of these 2 terminal residues is of necessity a cysteine; the second residue must consequently be a threonine to satisfy the mass requirement. We can infer the order of these 2 residues by the absence of a z1 ion, which implies that the terminal residue does not contain a charged sidechain and is therefore likely to be threonine rather than the dimethyl Lys analog (i.e., the modified Cys). Thus, we determined the sequence to be WCKQSGEMCNLLDQNCCDGYCIVLVCT with an oxidized methionine (measured Mr of the native toxin = 3,050.189 Da, calculated Mr = 3,050.185 Da). Our de novo sequence agrees with that determined by Edman sequencing for this so-called “King Kong” toxin (38), further validating the present approach. The cysteine derivatization described above successfully increased the charge state of all 31 toxins that we analyzed in detail, greatly facilitating their de novo sequence determination (Table 1).
Table 1.
Summary of characterized conotoxins
We now discuss how we overcome certain de novo sequencing ambiguities that commonly arise. Upon manual inspection of the ETD with CEP spectrum shown in Fig. 3, we were able to deduce the toxin sequence N [I/L] [Q/K] [I/L] [I/L] C C [Q/K] H T P A C C T with an amidated C terminus. However, this initial result contained several ambiguities, including an uncertainty concerning the 6 C-terminal residues—where the fragmentation pattern, mass and Cys content allowed for the alternative possible sequence A Q A C C V. Concerning the ambiguity of Val versus amidated Thr at the C terminus, it can be difficult to confidently distinguish between these residues because the mass difference is only 1 Da; this may be especially problematic in ETD/ECD spectra where H· rearrangement to/from z-type fragment ions is a common occurrence (39). Although the absence of a fragment ion between the 10th and 11th residue favors the interpretation TP (198 Da) over AQ (199 Da) (because Pro residues do not yield fragments at their N-termini in ETD), again the 1-Da difference prohibits an unequivocal call. Further confounding our ability to make an unambiguous call is our observation of a z-type ion consistent with cleavage between a putative AQ (albeit with low intensity). To resolve this ambiguity, we acquired high-resolution, high mass accuracy CAD spectra using a linear ion trap-Orbitrap mass spectrometer (see Fig. S1). The high mass accuracy (<3 ppm) allowed resolution of ambiguities arising from both the 1-Da differences (see above) and residues whose nominal masses are the same [Lys (128.095 Da) and Gln (128.059 Da)]. Here, the use of CAD allowed us to check for characteristic Pro cleavages (which produces a dominant ion corresponding to fragmentation at the N-terminal side of proline—i.e., just the opposite to that observed with ETD). This allows us to determine the sequence as N [I/L] Q [I/L] [I/L] C C K H T P A C C T (theoretical Mr = 1,641.723 Da), in agreement with our experimental determination (Mr = 1,641.721 Da). To the best of our knowledge, this toxin has not been described as a precursor or mature toxin.
Fig. 3.
ETD with CEP MS/MS spectrum of the (M + 5H)+5 ion of the indicated species after conversion of the Cys residues to dimethyl Lys analogs. Annotated as in Fig. 2.
Differentiation of the isomeric Ile and Leu residues (113.084 Da) requires fragmentation of their side chains by high energy collisions (25, 40, 41) or hot ECD (42), technologies not currently available to us. We note that hydroxyproline also has a nominal mass of 113 Da but can be easily differentiated from Ile and Leu by accurate mass measurements and its characteristic fragmentation pattern, which is similar to that of Pro (see Fig. S2).
Combined Use of de Novo Sequencing and Precursor cDNA Databases.
Although highly effective, the de novo sequencing strategy shown in Fig. 1 requires considerable skill and time in correctly interpreting the various fragmentation spectra. In an effort to further decrease the time required for spectral interpretation, we explored the possibility of using sequence tags to interrogate existing toxin cDNA sequence libraries. Using ETD with CEP, we can obtain sequence tags of ≈5 aa in just a couple of minutes from virtually any toxin. It is important to note that the majority of cDNA sequences code for toxin precursors—i.e., prepropeptides, comprising a signal-, pro- and mature-peptide (4). Although the active toxin sequence is located at the C terminus of the prepropeptides, it is generally not possible to predict the precise start and stop sites of the mature peptide. Additional complications arise from the large number of isoforms with closely related sequences and the plethora of posttranslational modifications, known to exist in Conus (37). Thus, even if we are able to find the experimental sequence tag in a cDNA library, it is still necessary to obtain the start and stop sites and any posttranslational modifications and amino acid variations that are present. This requires extensive high-quality MS/MS data.
To explore the utility of cDNAs in assisting toxin sequence determination, we devised a computer program called Toxfinder that searches existing cDNA databases, using as input parameters the experimentally determined molecular mass of the toxin, the number of cysteines present, and our de novo sequence tag. Because we do not always know the direction of the sequence tag, we search in both directions (from the N to C termini and from the C to N termini). The database is searched redundantly with isoleucine and leucine (nominal mass = 113); glutamine and lysine (nominal mass = 128) because we do not differentiate between these alternatives in sequence tags obtained from our low resolution experiments. Three types of Toxfinder outputs are possible: (i) Peptides that contain the specified sequence tag and satisfy the mass and number of cysteines specified in the input. These peptides are generated by in silico truncation of the precursor sequence about the de novo sequence tag to include the specified number of cysteines, while simultaneously fulfilling the mass requirement. Note that this procedure may yield more than one candidate that fulfills all of the input restraints. (ii) The sequence tag is found in a precursor, but the precursor cannot be processed in silico to produce a peptide that fulfills both the mass and cysteine-number input parameters. Here, the entire precursor sequence is provided as the output. (iii) The sequence tag cannot be found in the cDNA precursor database and no output is provided.
Fig. 4 shows the ETD with CEP spectrum of a toxin with reduced Mr = 3,302.092 Da and 6 cysteines. Manual inspection of this spectrum quickly yielded the indicated sequence tag RGYDAP (indicated in blue). Although Toxfinder quickly identified a 70-residue toxin precursor sequence containing this tag (1MEKLTILLLV AAVLTSTQAL IQGGGDERQK AKINFLSRSD RDCRGYDAPC SSGAPCCDWW TCSARTNRCF70), it was unable to return a peptide candidate that also fulfills both the mass and cysteine-content restraints. This may be because the sequence is not in the database and the tag is simply a random match. More likely, the reason we were not able to find a match is because the mature toxin contains posttranslational modifications or is an isoform that is not present in the database. We thus tested these latter possibilities. Because we determined experimentally that the toxin contains 6 Cys residues, the mature toxin should at least include residues 43–69 of the precursor. Concentrating on this region, we looked for N- and C-terminal fragment ions that may allow us to determine the start and end residues of this toxin. The dominant singly-charged ions highlighted in blue (Fig. 4) are consistent with the N terminus starting at Asp-42. The remaining singly-charged ions in the low mass region are consistent with the C terminus ending at Phe-70. Then, assuming that our mature toxin corresponds to residues 42–70, its molecular mass of 3,224.186 Da is 77.906 Da less than our experimentally determined mass of 3,302.092 Da. This mass addition could be explained by bromination of a Trp residue (theoretical mass shift 77.910 Da) (43). Indeed, the sequence provided by Toxfinder contains 2 adjacent tryptophan residues. Inspection of the spectrum allowed us to assign this putative bromination specifically to Trp-19. In addition to the observed mass increase that is consistent with bromine addition, bromine has a diagnostic isotope pattern arising from its almost equally abundant isotopes, 79Br and 81Br. This pattern is seen for the fragment ion z11 (see Fig. 4 Inset) and subsequent z ions, further confirming bromination of Trp-19. Comparison of the fragmentation data with the cDNA sequence allowed experimental confirmation of 24 of the 29 residues (Fig. 4 and Fig. S3). However, although the accurate mass data defines the amino acid composition of the N-terminal 2 residues and C-terminal 3 residues, it does not specify their order. At present this order can only be inferred from the cDNA sequence. Nevertheless, this example demonstrates the facility of this combined de novo sequencing, database cDNA correlation approach for rapidly defining intact peptides (here a modified 29-residue toxin) from crude venom samples. This 29-residue toxin was the largest that we encountered from C. textile, although we have obtained similar information from even larger toxins (up to 37 residues) from other species (e.g., Leiurus quinquestriatus hebraeus; Fig. S4).
Fig. 4.
ETD with CEP MS/MS spectrum of the (M + 6H)+6 ion of the indicated species after conversion of the Cys residues to dimethyl Lys analogs. (Inset) The observed (Lower) and theoretical (Upper) isotope distribution for the z11 fragment ion, indicating the presence of a brominated Trp (WBr) residue. Annotated as in Fig. 2.
Discussion
We have described a versatile approach for de novo sequencing of disulfide-rich miniprotein toxins. Using reversed phase chromatography to partially separate the components of a crude venom sample from C. textile in combination with mass spectrometric analysis of the intact toxins, we sequenced 31 intact individual toxins, using just 7% of the contents of a single venom gland. These toxins ranged in size from 962 to 3,302 Da (9–29 amino acid residues) and were all analyzed without subdigestion. Critical to this analysis was our use of ETD with CEP to maximize the mass spectrometric sequence coverage of the toxins; we note that continuous coverage of all adjacent amino acids is imperative for an unambiguous sequence assignment. The complementary nature of CAD and ETD greatly facilitates de novo sequence analysis. The ubiquitous modifications observed in conotoxins are readily discerned with our procedure, although these may cause complications—e.g., hydroxyproline has the same nominal mass as Leu and Ile. We can resolve this potential ambiguity, using the distinctive fragmentation behavior of Pro in ETD and CAD, and the accurate mass difference.
As cDNA sequencing becomes faster and less costly, we expect the number of toxin precursor cDNA sequences to rapidly increase. In this regard, we have shown that the combined use of such cDNA sequences and ETD with CEP greatly facilitates the sequence analysis of mature toxins. However, the procedure still requires good mass spectrometric sequence coverage to (i) recognize the cDNA in a database via a sequence tag, (ii) define the N- and C-termini, (iii) elucidate posttranslational modifications and (iv) discern closely related isoforms.
Although the current approach has greatly improved our ability to rapidly obtain accurate toxin sequences, further improvements are desirable. Perhaps the most challenging portions of the toxins to sequence are the termini, where fragmentation is frequently not observed between the terminal and penultimate residues. Here, it may prove necessary to apply additional derivatization strategies or C-terminal digestion strategies that enhance detection of the terminal residues (44, 45). Another longstanding mass spectrometric sequencing problem is the ambiguity between the isobaric pair Leu and Ile. This sequencing ambiguity can be resolved in practice by sequencing the corresponding DNA (15–17, 22), Edman degradation of the purified toxin component, or through the use of peptide synthesis and functional assays (46). A different set of analytical problems are encountered as the mass of the toxins increase. Because large multiply-charged precursors produce fragmentation products with a range of different charge states, the fragmentation spectra become increasingly complex and difficult to analyze. This is especially relevant in low resolution ion trap mass analyzers, where it is challenging to assign charge states. In addition, the large number of fragmentation pathways leads to a concomitant decrease in signal-to-noise ratio of the fragment ion peaks. Such difficulties can be alleviated in 2 ways: (i) The spectrum can be simplified and the signal-to-noise increased by converting the multiply-charged fragment ions into their singly charge state, using proton transfer charge reduction (PTR) (47, 48). Because PTR was not available on our instrumentation, we reduced the charge of our fragment ions by extending the ion/ion reaction time. An undesired side effect of this approach is the production of internal fragment ions that =complicate spectral interpretation. (ii) The different charge states in the fragmentation spectra can be identified through the use of a high-resolution mass analyzer (e.g., Orbitrap or ICR-FTMS) (49, 50). High-resolution mass analysis also yields improved signal-to-noise, allowing the detection of low abundance fragment ions that may be indistinguishable from noise in low resolution devices. The use of PTR and high-resolution readout of ETD spectra should further facilitate spectral interpretation (47, 51) and increase the size of toxins that can be readily studied.
Rapid sensitive sequence analysis of peptide toxins, as described here, promises to massively expand the database of these fascinating bioactive molecules and facilitate many potentially informative avenues of study. These include studies of the variation in toxin repertoire in individual cone snails from a given species and rapid screening of crude venom from different animal venoms against different ion channels (52).
Materials and Methods
Detailed methods and materials are provided in SI Materials and Methods.
Crude Venom Preparation.
Crude venom duct contents were extracted with light sonication, using 30% acetonitrile/water with 0.1% TFA and centrifuged.
Off-Line HPLC Separation.
The crude venom extract from a single C. textile snail was dissolved in 0.5% acetic acid. Aliquots (2.3% of the total sample) were separated by HPLC and fifty 1-min fractions collected.
Sample preparation for MS.
Each fraction was reconstituted in 15 μL of 20% acetonitrile in 0.5% TFA, reduced and alkylated. For conversion of cysteines to dimethyl lysine analogs (36), untreated reconstituted fractions were mixed with twice the volume of 0.5 M Hepes buffer (pH = 7.8) and 1 μL of TCEP. After 30 min, 5 μL of freshly prepared 1 M (2-chloroethyl)-dimethylammonium chloride (Sigma–Aldrich) in 1 M Hepes buffer (pH = 7.8) was added and the reaction allowed to proceed for 3 h.
MALDI MS.
Aliquots of untreated, reduced and iodoacetamide alkylated fractions were mixed with saturated α-cyano-4-hydroxycinnamic acid in 50% water/50% acetonitrile and spotted onto a MALDI target (53). The fractions were analyzed by MALDI-MS using a prOTOF2000 mass spectrometer (PerkinElmer).
ESI MS.
Toxin-containing fractions were further analyzed by on-line HPLC-MS using either a LTQ XL or a LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific), operated in a data dependent mode cycling through a full scan (300–2,000 m/z, single μscan) followed by 8 MS/MS scans on each of the 4 most abundant ions.
Supplementary Material
Acknowledgments.
We thank A. Ruthenburg for helpfull discussions. This work was supported by the National Institutes of Health (NIH) Grant RR00862 (BTC).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900745106/DCSupplemental.
References
- 1.Olivera BM. Conus venom peptides: Reflections from the biology of clades and species. Annu Rev Ecol Syst. 2002;33:25–47. [Google Scholar]
- 2.Olivera BM. Conus peptides: Biodiversity-based discovery and exogenomics. J Biol Chem. 2006;281:31173–31177. doi: 10.1074/jbc.R600020200. [DOI] [PubMed] [Google Scholar]
- 3.Escoubas P. Molecular diversification in spider venoms: A web of combinatorial peptide libraries. Mol Divers. 2006;10:545–554. doi: 10.1007/s11030-006-9050-4. [DOI] [PubMed] [Google Scholar]
- 4.Terlau H, Olivera BM. Conus venoms: A rich source of novel ion channel-targeted peptides. Physiol Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 5.Kolmar H. Alternative binding proteins: Biological activity and therapeutic potential of cystine-knot miniproteins. FEBS J. 2008;275:2684–2690. doi: 10.1111/j.1742-4658.2008.06440.x. [DOI] [PubMed] [Google Scholar]
- 6.Hu SH, et al. The 1.1 angstrom crystal structure of the neuronal acetylcholine receptor antagonist, alpha-conotoxin PnIA from Conus pennaceus. Structure. 1996;4:417–423. doi: 10.1016/s0969-2126(96)00047-0. [DOI] [PubMed] [Google Scholar]
- 7.Armishaw CJ, Alewood PF. Conotoxins as research tools and drug leads. Curr Protein Pept Sci. 2005;6:221–240. doi: 10.2174/1389203054065437. [DOI] [PubMed] [Google Scholar]
- 8.Lewis RJ, Garcia ML. Therapeutic potential of venom peptides. Nat Rev Drug Discov. 2003;2:790–802. doi: 10.1038/nrd1197. [DOI] [PubMed] [Google Scholar]
- 9.Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: A model for concerted pharmacological discovery. Mol Interv. 2007;7:251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- 10.Craig AG. The characterization of conotoxins. J Toxicol Toxin Rev. 2000;19:53–93. [Google Scholar]
- 11.Wolfender JL, et al. Identification of tyrosine sulfation in Conus pennaceus conotoxins alpha-PnIA and alpha-PnIB: Further investigation of labile sulfo- and phosphopeptides by ESI, MALDI and AP MALDI MS. J Mass Spectrom. 1999;34:447–454. doi: 10.1002/(SICI)1096-9888(199904)34:4<447::AID-JMS801>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura T, Yu ZG, Fainzilber M, Burlingame AL. MS -based revision of the structure of a cysteine-rich peptide toxin with gamma-carboxyglutamic acid, TxVIIA, from the sea snail, Conus textile. Protein Sci. 1996;5:524–530. doi: 10.1002/pro.5560050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Quinton L, et al. New method for characterizing highly disulfide-bridged peptides in complex mixtures: Application to toxin identification from crude venoms. J Proteome Res. 2007;6:3216–3223. doi: 10.1021/pr070142t. [DOI] [PubMed] [Google Scholar]
- 14.Escoubas PQL, Nicholson GM. Venomics: Unravelling the complexity of animal venoms with MS. J Mass Spectrom. 2008;43:279–295. doi: 10.1002/jms.1389. [DOI] [PubMed] [Google Scholar]
- 15.Jakubowski JA, et al. Determining sequences and post-translational modifications of novel conotoxins in Conus victoriae using cDNA sequencing and MS. J Mass Spectrom. 2004;39:548–557. doi: 10.1002/jms.624. [DOI] [PubMed] [Google Scholar]
- 16.Gowd KH, Dewan KK, Iengar P, Krishnan KS, Balaram P. Probing peptide libraries from Conus achatinus using MS and cDNA sequencing: Identification of delta and omega-conotoxins. J Mass Spectrom. 2008;43:791–805. doi: 10.1002/jms.1377. [DOI] [PubMed] [Google Scholar]
- 17.Escoubas P, Sollod B, King GF. Venom landscapes: Mining the complexity of spider venoms via a combined cDNA and MS approach. Toxicon. 2006;47:650–663. doi: 10.1016/j.toxicon.2006.01.018. [DOI] [PubMed] [Google Scholar]
- 18.Diego-Garcia E, et al. The Brazilian scorpion Tityus costatus Karsch: Genes, peptides and function. Toxicon. 2005;45:273–283. doi: 10.1016/j.toxicon.2004.10.014. [DOI] [PubMed] [Google Scholar]
- 19.Corpuz GP, et al. Definition of the M-conotoxin superfamily: Characterization of novel peptides from molluscivorous Conus venoms. Biochemistry. 2005;44:8176–8186. doi: 10.1021/bi047541b. [DOI] [PubMed] [Google Scholar]
- 20.Favreau P, et al. The venom of the snake genus Atheris contains a new class of peptides with clusters of histidine and glycine residues. Rapid Commun Mass Spectrom. 2007;21:406–412. doi: 10.1002/rcm.2853. [DOI] [PubMed] [Google Scholar]
- 21.Czerwiec E, et al. Novel gamma-carboxyglutamic acid-containing peptides from the venom of Conus textile (vol 273, pg 2779, 2006) FEBS J. 2006;273:3118. doi: 10.1111/j.1742-4658.2006.05294.x. [DOI] [PubMed] [Google Scholar]
- 22.Quinton L, et al. Characterization of toxins within crude venoms by combined use of FTMS and cloning. Anal Chem. 2005;77:6630–6639. doi: 10.1021/ac050575k. [DOI] [PubMed] [Google Scholar]
- 23.Jakubowski JA, Sweedler JV. Sequencing and mass profiling highly modified conotoxins using global reduction/alkylation followed by MS. Anal Chem. 2004;76:6541–6547. doi: 10.1021/ac0494376. [DOI] [PubMed] [Google Scholar]
- 24.Braga MCV, et al. MS and HPLC profiling of the venom of the Brazilian vermivorous mollusk Conus regius: Feeding behavior and identification of one novel conotoxin. Toxicon. 2005;45:113–122. doi: 10.1016/j.toxicon.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 25.Mandal AK, et al. Sequencing of T-superfamily conotoxins from Conus virgo: Pyroglutamic acid identification and disulfide arrangement by MALDI MS. J Am Soc Mass Spectrom. 2007;18:1396–1404. doi: 10.1016/j.jasms.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 26.Steen H, Mann M. Analysis of bromotryptophan and hydroxyproline modifications by high-resolution, high-accuracy precursor ion scanning utilizing fragment ions with mass-deficient mass tags. Anal Chem. 2002;74:6230–6236. doi: 10.1021/ac025994l. [DOI] [PubMed] [Google Scholar]
- 27.Nair SS, et al. De novo sequencing and disulfide mapping of a bromotryptophan-containing conotoxin by FTICR MS. Anal Chem. 2006;78:8082–8088. doi: 10.1021/ac0607764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wermelinger LS, et al. Fast analysis of low molecular mass compounds present in snake venom: Identification of ten new pyroglutamate-containing peptides. Rapid Commun Mass Spectrom. 2005;19:1703–1708. doi: 10.1002/rcm.1973. [DOI] [PubMed] [Google Scholar]
- 29.Samgina TY, et al. De novo sequencing of peptides secreted by the skin glands of the Caucasian Green Frog Rana ridibunda. Rapid Commun Mass Spectrom. 2008;22:3517–3525. doi: 10.1002/rcm.3759. [DOI] [PubMed] [Google Scholar]
- 30.Zubarev RA, Zubarev AR, Savitski MM. Electron capture/transfer versus collisionally activated/induced dissociations: Solo or duet? J Am Soc Mass Spectrom. 2008;19:753–761. doi: 10.1016/j.jasms.2008.03.007. [DOI] [PubMed] [Google Scholar]
- 31.Mikesh LM, et al. The utility of ETD MS in proteomic analysis. Biochim Biophys Acta. 2006;1764:1811–1822. doi: 10.1016/j.bbapap.2006.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zubarev RA, Kelleher NL, McLafferty FW. ECD of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 33.Syka JEP, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by ETD MS. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Good DM, Wirtala M, McAlister GC, Coon JJ. Performance characteristics of ETD MS. Mol Cell Proteomics. 2007;6:1942–1951. doi: 10.1074/mcp.M700073-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Kaas Q, Westermann JC, Halai R, Wang CKL, Craik DJ. ConoServer, a database for conopeptide sequences and structures. Bioinformatics. 2008;24:445–446. doi: 10.1093/bioinformatics/btm596. [DOI] [PubMed] [Google Scholar]
- 36.Simon MD, et al. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buczek O, Bulaj G, Olivera BM. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol Life Sci. 2005;62:3067–3079. doi: 10.1007/s00018-005-5283-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hillyard DR, et al. A molluscivorous conus toxin—Conserved frameworks in conotoxins. Biochemistry. 1989;28:358–361. doi: 10.1021/bi00427a049. [DOI] [PubMed] [Google Scholar]
- 39.Savitski MM, Kjeldsen F, Nielsen ML, Zubarev RA. Hydrogen rearrangement to and from radical z fragments in ECD of peptides. J Am Soc Mass Spectrom. 2007;18:113–120. doi: 10.1016/j.jasms.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 40.Biemann K. Sequencing of peptides by tandem MS and high-energy collision-induced dissociation. Methods Enzymol. 1990;193:455–479. doi: 10.1016/0076-6879(90)93433-l. [DOI] [PubMed] [Google Scholar]
- 41.Medzihradszky KF, Burlingame AL. The advantages and versatility of a high-energy collision-induced dissociation-based strategy for the sequence and structural determination of proteins. Methods (Orlando) 1994;6:284–303. [Google Scholar]
- 42.Kjeldsen F, Haselmann KF, Sorensen ES, Zubarev RA. Distinguishing of Ile/Leu amino acid residues in the PP3 protein by (hot) ECD in FTICR MS. Anal Chem. 2003;75:1267–1274. doi: 10.1021/ac020422m. [DOI] [PubMed] [Google Scholar]
- 43.Jimenez EC, et al. Bromocontryphan: Post-translational bromination of tryptophan. Biochemistry. 1997;36:989–994. doi: 10.1021/bi962840p. [DOI] [PubMed] [Google Scholar]
- 44.Russo A, Chandramouli N, Zhang LQ, Deng HT. Reductive glutaraldehydation of amine groups for identification of protein N-termini. J Proteome Res. 2008;7:4178–4182. doi: 10.1021/pr800224v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakazawa T, et al. Terminal proteomics: N- and C-terminal analyses for high-fidelity identification of proteins using MS. Proteomics. 2008;8:673–685. doi: 10.1002/pmic.200700084. [DOI] [PubMed] [Google Scholar]
- 46.Loughnan ML, Alewood PF. Physico-chemical characterization and synthesis of neuronally active alpha-conotoxins. Eur J Biochem. 2004;271:2294–2304. doi: 10.1111/j.1432-1033.2004.04146.x. [DOI] [PubMed] [Google Scholar]
- 47.Coon JJ, et al. Protein identification using sequential ion/ion reactions and tandem MS. Proc Natl Acad Sci USA. 2005;102:9463–9468. doi: 10.1073/pnas.0503189102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stephenson JL, McLuckey SA. Ion/ion proton transfer reactions for protein mixture analysis. Anal Chem. 1996;68:4026–4032. doi: 10.1021/ac9605657. [DOI] [PubMed] [Google Scholar]
- 49.Frank AM, Savitski MM, Nielsen ML, Zubarev RA, Pevzner PA. De novo peptide sequencing and identification with precision MS. J Proteome Res. 2007;6:114–123. doi: 10.1021/pr060271u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.McAlister GC, et al. A proteomics grade ETD-enabled hybrid linear ion trap-orbitrap MS. J Proteome Res. 2008;7:3127–3136. doi: 10.1021/pr800264t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hubler SL, et al. Valence parity renders z·-type ions chemically distinct. J Am Chem Soc. 2008;130:6388–6394. doi: 10.1021/ja7099985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacKinnon R, Cohen SL, Kuo AL, Lee A, Chait BT. Structural conservation in prokaryotic and eukaryotic potassium channels. Science. 1998;280:106–109. doi: 10.1126/science.280.5360.106. [DOI] [PubMed] [Google Scholar]
- 53.Blethrow JD, Tang C, Deng C, Krutchinsky AN. Modular mass spectrometric tool for analysis of composition and phosphorylation of protein complexes. PLoS ONE. 2007;2:e358. doi: 10.1371/journal.pone.0000358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chalkley RJ, Brinkworth CS, Burlingame AL. Side-chain fragmentation of alkylated cysteine residues in ECD MS. J Am Soc Mass Spectrom. 2006;17:1271–1274. doi: 10.1016/j.jasms.2006.05.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.