Abstract
The extraordinary diversity of herbivorous beetles is usually attributed to coevolution with angiosperms. However, the degree and nature of contemporaneity in beetle and angiosperm diversification remain unclear. Here we present a large-scale molecular phylogeny for weevils (herbivorous beetles in the superfamily Curculionoidea), one of the most diverse lineages of insects, based on ≈8 kilobases of DNA sequence data from a worldwide sample including all families and subfamilies. Estimated divergence times derived from the combined molecular and fossil data indicate diversification into most families occurred on gymnosperms in the Jurassic, beginning ≈166 Ma. Subsequent colonization of early crown-group angiosperms occurred during the Early Cretaceous, but this alone evidently did not lead to an immediate and major diversification event in weevils. Comparative trends in weevil diversification and angiosperm dominance reveal that massive diversification began in the mid-Cretaceous (ca. 112.0 to 93.5 Ma), when angiosperms first rose to widespread floristic dominance. These and other evidence suggest a deep and complex history of coevolution between weevils and angiosperms, including codiversification, resource tracking, and sequential evolution.
Keywords: coevolution, Coleoptera, Curculionoidea, herbivory, phylogeny
Weevils [superfamily Curculionoidea; (Fig. 1)] are an extraordinarily successful radiation of herbivorous beetles. They reach their greatest diversity in the humid tropics, but also occur in subaquatic, subterranean, desert, tundra, and other environments at nearly all latitudes and altitudes with vegetation. Weevils collectively feed on nearly all plant taxa and all kinds of living, dead, dying, and decaying plant parts. The ≈62,000 described species are classified into 7 families and ≈5,800 genera. [Recent authors recognize between 6 and 22 weevil families, and between 10 and 100 subfamilies (1). Here we follow the classification of ref. 2 (see supporting information (SI) Table S1).] The likely total number of species, including those awaiting discovery or description, is conservatively estimated at more than 220,000 (2). More than 80% of living weevil species belong to the family Curculionidae, the diversity of which exceeds that of any other known family of animals (3). Today, as more than a half century ago, “the classification of Curculionidae into natural subfamilies and tribes probably remains the largest outstanding problem in the higher classification of Coleoptera” (2, 4). Consequently, relationships within Curculionidae are both of greatest interest for reconstructing the evolutionary history of weevil associations with plants, and most critical for achieving stability in weevil classification.
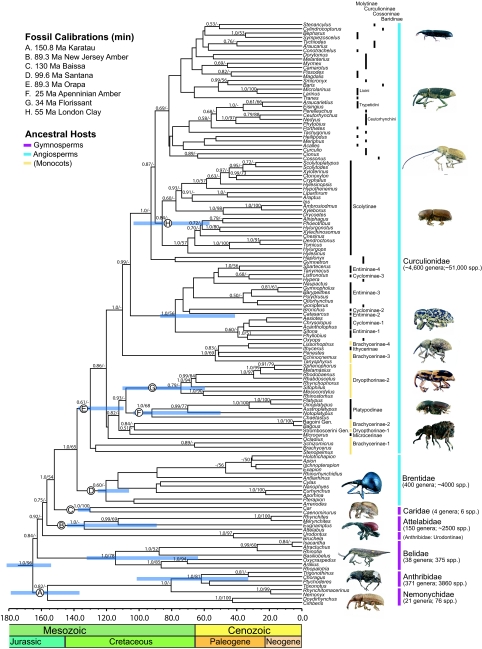
Fig. 1.
Curculio proboscideus (Curculionidae: Curculioninae) perched atop a flower of Helianthus sp. (Asteraceae). Note the elongation of the head to form the characteristic weevil rostrum or “snout.” In some groups, the rostrum is not only used for feeding, but also for preparing oviposition sites and placing eggs deep inside plant tissues (Photo credit: D. McKenna).
Weevils first appear unequivocally in the Late Jurassic fossil record (Karatau, Oxfordian-Kimmeridgian, 161.2−150.8 Ma) (5). These early weevils belong to the family Nemonychidae (2) and most likely developed in the reproductive structures of conifers in a manner similar to living nemonychids (6, 7). While early weevils most likely fed on conifers, most living weevil species are specialist herbivores on flowering plants (angiosperms; >250,000 living species). Shifts to feeding on angiosperms are associated with enhanced taxonomic diversification in weevils (6), and weevils underwent considerable diversification during the Cretaceous (145.5–65.5 Ma) (2, 5, 6), a period when angiosperms also flourished (8–10). Consequently, the extraordinary taxonomic diversity of weevils is often attributed to coevolution with angiosperms (2, 6, 11). However, because of uncertainties about higher-level relationships and divergence times in weevils, the evolutionary history of weevil-angiosperm interactions remains unclear.
To gain insight into the degree and nature of contemporaneity in weevil and angiosperm diversification, we used a large-scale temporally calibrated phylogeny for weevils estimated from combined molecular and fossil data. Our molecular data set included up to 8 kilobases (kb) of DNA sequence data (4 nuclear and 2 mitochondrial genes) from a worldwide sample of 135 weevil genera representing all families and subfamilies and 8 outgroups.
Results
Weevil Relationships.
Overall, we recovered moderate to strong bootstrap (BS) or posterior probability (PP) support for ≈53% (71 out of 134) of ingroup internodes (Fig. 2).* Family-level relationships were mostly compatible with recent concepts (2, 11, 14–16); however, several new and intriguing relationships were recovered at the subfamily-level. A clade including Nemonychidae and Anthribidae [<50% ML BS; 0.82 Bayesian PP] was sister to all other weevils. Nemonychidae was rendered paraphyletic by the family Anthribidae (minus Urodontinae) (<50% BS; 1.0 PP), consistent with other authors who have noted this possibility based on similarities in hind wings and other adult and larval features. Most Anthribidae use angiosperm-dependent fungi as hosts, consistent with their first appearance in the fossil record coeval with a rapidly diversifying angiosperm flora in the late Early Cretaceous (2), and much later than the largely conifer-associated Nemonychidae. Placement of the subfamily Urodontinae separate from other Anthribidae (and Nemonychidae) was unexpected, and conflicts with morphology. The hypothesis of monophyly for Anthribidae plus Urodontinae was rejected under BI (PP = 0), but not under ML inference (Kishino-Hasegawa test or KH, P = 0.47). The belid subfamilies Oxycoryninae (including Aglycyderini) and Belinae were recovered as sister groups within a monophyletic family Belidae (78% BS; 1.0 PP), consistent with recent analyses of morphological characters (11, 14, 16–18). We recovered moderate-to-strong support for monophyly of the families Attelabidae (<50% BS; 1.0 PP) and Caridae (100% BS; 1.0 PP) under ML and/or BI. The placement of Caridae as sister group of the megaclade Brentidae plus Curculionidae received strong support under BI (54% BS; 1.0 PP), in agreement with morphology (11). The subfamilies Apioninae, Brentinae, Eurhynchinae, and Nanophyinae (but not the enigmatic Ithycerinae and Microcerinae) together comprised a monophyletic but poorly supported family Brentidae (<50% BS; 0.60 PP). The hypothesis of monophyly for Brentidae, including Microcerinae and Ithycerinae, was rejected under BI (PP = 0), but not under ML (KH, P = 0.22).
Fig. 2.
Maximum clade credibility tree for weevils based on the minimum age Bayesian analysis. Bayesian PP ≥ 0.50 and maximum likelihood BS values ≥ 50% are shown on the tree (PP/BS). Ninty-five percent confidence intervals for the ages of family- and subfamily-level clades, and for the ingroup, are indicated with blue bars. Letters correspond to fossil calibration points used in the molecular dating analysis. Numbers of described species are from ref. 2. Images of weevil exemplars are not to scale. Outgroups have been removed.
We recovered a monophyletic family Curculionidae (<50% BS; 0.61 PP; including Ithycerinae and Microcerinae). Basal positions in Curculionidae were occupied by weevils with the ancestral “pedotectal” (19) type of male genitalia: Brachycerinae, Microcerinae, Platypodinae, Dryophthorinae, and Ithycerinae, followed by “higher” Curculionidae (<50% BS; 0.99 PP), comprised of groups with the derived “pedal” type of male genitalia. Brachycerinae (which in the concept adopted here also includes the Erirhininae of authors) occupied basal positions in Curculionidae, forming a paraphyletic grade that also included the subfamilies Dryophthorinae and Platypodinae. Ithycerinae and Microcerinae were intermingled among these “basal” curculionids in our analyses, in contrast with morphological studies, which increasingly favor their placement in the family Brentidae (2). The enigmatic subfamily Platypodinae (ambrosia beetles) was recovered in a position sister to Dryophthorinae (<50% BS; 0.93 PP), a relationship first proposed and supported by larval morphology (15), but not previously recovered in molecular phylogenetic studies, which mostly recover Platypodinae as apomorphic derivatives of Scolytinae. While the hypothesis of monophyly for Platypodinae plus Scolytinae was rejected under BI (PP = 0), it could not be rejected under ML (KH, P = 0.49). Dryophthorinae was monophyletic (<50% BS; 0.79 PP) minus Stromboscerini.
We recovered several groups among higher Curculionidae with moderate-to-strong internodal support (under ML or BI), comprising what are usually treated as subfamilies or tribes, [e.g., Baridinae: Ceutorhynchini (97% BS, 1.0 PP), Molytinae: Lixini (100% BS, 1.0 PP), and Scolytinae (<50% BS, 0.84 PP)]. However, a more thorough understanding of relationships and timing and patterns of diversification in Curculionidae will apparently require additional sampling. The limited resolution obtained herein reinforces the idea that clarifying curculionid relationships is a difficult task (2, 4, 11). Nevertheless, several groups emerge from our analyses that were suspected on the basis of similar morphological features, life histories, or habits, but whose relationships had otherwise been obscured by homoplasy.
Cyclominae, Entiminae, Gonipterini, and Hyperini together comprised a single clade (56% BS, 1.0 PP) in a position between Brachycerinae: Erirhinini (minus Stenopelmus) and the remaining higher weevils. The phylogenetic position of Scolytinae was particularly notable. Scolytinae are usually considered close relatives of Platypodinae and Cossoninae (14, 20). However, there is little support for a close relationship between Scolytinae and Platypodinae under BI (see above), and a sister-group relationship between Scolytinae and all or a subset of cossonine genera, including Araucariini, the ostensible link between Cossoninae and Scolytinae (21), also appears unlikely, implying multiple origins of the gallery-forming habit, but the alternative scenario of a single origin cannot be ruled out under ML (Scolytinae + Cossoninae PP = 0, KH P = 0.49; Scolytinae + Araucariini PP = 0, KH P = 0.34). Sister to Scolytinae we recovered a poorly supported clade comprised of most other higher Curculionidae, including taxa classified in the subfamilies Baridinae, Cossoninae, Curculioninae, and Molytinae. Many workers have noted the difficulty of separating Molytinae from the traditional “Cryptorhynchinae,” Cossoninae, and “Lixinae,” using morphological characters (2, 22), an observation consistent with our results. All subfamilies of higher Curculionidae, except Scolytinae, were poly- or paraphyletic in our analyses, but Scolytinae were relatively well sampled, and we expect that including more taxa or characters from other curculionid subfamilies will contribute additional well-supported resolution.
Weevils on Plants in Evolutionary Time.
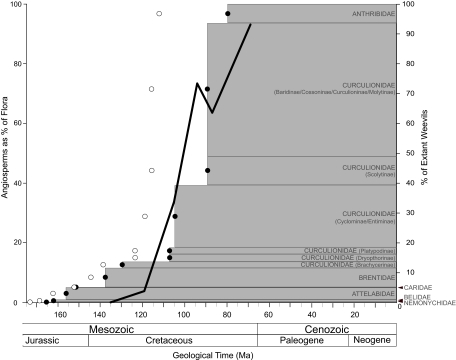
We recover an Early to early Middle Jurassic origin of modern weevils (ca. Aalenian-Bathonian, 175.6–164.7 Ma), with all families and most subfamilies arising by the end of the Cretaceous (Figs. 2 and 3; Table S2). The ancestrally angiosperm-associated sister groups Brentidae and Curculionidae diverged during the earlier Early Cretaceous (ca. Valanginian–Hauterivian, 140.2–130 Ma). Crown diversification of both families appears to have commenced by the late Early Cretaceous (Aptian, 125.0–112.0 Ma), and all subfamilies appeared by the early Late Cretaceous (Turonian, 93.5–89.3 Ma). The family Curculionidae shows sustained higher-level diversification throughout the Late Cretaceous (99.6 Ma to 65.5 Ma) and Paleogene (particularly Paleocene–Eocene, 65.5–33.9 Ma) (see Fig. 2). Superimposed plots of stem-group divergence times for major weevil clades, and for angiosperm dominance over the course of the Cretaceous (see Fig. 3), reveal evidence for a major increase in diversity of angiosperm-associated weevils approximately concurrent with the mid-Cretaceous rise of angiosperms to widespread floristic dominance (ca. Albian–Cenomanian, 112.0 to 93.5 Ma) (8, 9).
Fig. 3.
Superimposed plots of stem-group divergence times for the earliest representatives of major weevil clades (maximum fossil age analysis, white circles; minimum fossil age analysis, black circles) and angiosperm dominance over the course of the Cretaceous [black curve (adapted from ref. 9)], reveal evidence for an increase in weevil diversity beginning during the mid Cretaceous, concurrent with the rise of angiosperms to widespread floristic dominance, and well after the first appearance of crown-group angiosperms [fossils ≈132–141 Ma (24), molecules ≈140–180 Ma (25)]. Note that Anthribidae is shown here separate from Nemonychidae (from which it is derived in our analyses), in order to accurately illustrate the disparate timing of origin and magnitude of extant diversity in these 2 groups (the species-poor anthribid subfamily Urodontinae is not shown separately from other Anthribidae). We propose that this temporal lag in the diversification of angiosperm-associated weevils is evidence for the combined major role of ecological-evolutionary opportunity and intrinsic traits (fine-tuned, elaborated, and accumulated over the course of a long history of association with living, dead, dying, and decaying plants and plant organs and tissues) in the evolutionary radiation of weevils. Patterns of weevil diversification during the time interval between the origin of each major weevil clade and the present remain unclear.
Discussion
The larvae and adults of most extant Nemonychidae feed on conifers, and primarily on pollen. Most Anthribidae feed on ascomycete fungi growing on or in angiosperm wood, a habit proposed to have evolved from phytophagy in decaying cones of conifers or possibly cycads (23). This scenario is compatible with our results, which support a Late Cretaceous origin of Anthribidae (excluding Urodontinae) from within the largely conifer-associated Nemonychidae. Conifers are abundantly represented in the Mesozoic fossil record (8, 9), and are also thought to be the ancestral hosts of weevils in the families Attelabidae (2), Belidae (18), and Caridae (2), consistent with estimated divergence times for stem-group representatives of these families (ca. 150–170 Ma) (see Fig. 3), which predate most estimates for the timing of first appearance of crown-group angiosperms [fossils ≈132–141 Ma (24), molecules ≈140–180 Ma (25)]. Conifers are also thought to be the ancestral hosts of the weevil sister group, superfamily Chrysomeloidea (long-horned beetles, leaf-beetles, and allies) (6, 26, 27).
The ancestrally angiosperm-associated sister groups Brentidae and Curculionidae (2) diverged during the earlier Early Cretaceous (ca. Berriasian–Valanginian, 145.5–130 Ma). Brentidae noticeably lack associations with monocots; however, many basal Curculionidae, particularly Brachycerinae and Dryophthorinae, are associated primarily or solely with them. Monocots were among the most abundant and ecologically successful early angiosperms. Indeed, most early angiosperm fossils, for example Barremian–Aptian, are from Magnoliales or monocots (28–30), not eudicots. Most other Curculionidae, and most extant species of Anthribidae, Attelabidae, and Brentidae, feed on the living, dead, dying, or fungus-infested tissues of core eudicots, the most diverse group of living angiosperms. The large number of monocot-associated taxa near the base of the family Curculionidae is consistent with a common origin of monocot feeding. Some curculionid tribes are intimately associated with gymnosperms; however, they are all nested within clades of angiosperm-feeders. Thus, while some primitive weevils may be primary associates of gymnosperms (e.g., certain Nemonychidae, Belidae, Attelabidae, and Caridae), others, such as gymnosperm-associated Curculionidae and Brentidae, are most likely secondary colonists, an interpretation consistent with other authors (2, 11, 18, 31, 32).†
Basal Curculionidae most likely first colonized core eudicots and certain other land-plant groups that were locally abundant, unoccupied, or underexploited by other herbivores, and occurred in close ecological association with their ancestral monocot hosts. Indeed, other than monocots and core eudicots, the hosts of many relatively basal Curculionidae and Brentidae appear to reflect the kinds of plants that were abundant at the times or in the places (habitats, biogeographic regions) that these groups radiated. Later arising lineages within Curculionidae are generally more specialized in terms of host organ/tissue and taxon associations. For example, increased specialization is apparent in terms of larval host organ/tissue associations in increasingly more derived weevil groups, loosely following the general sequence (in order of increasing specialization): roots and stems (most Brachycerinae, Cyclominae, Dryophthorinae, Entiminae, Ithycerinae, Microcerinae) → wood (most Cossoninae, Molytinae, Scolytinae) → fruits and seeds (most Baridinae, Curculioninae).‡ The apparent nonrandom association between certain plant taxa and certain (especially derived) weevil groups (e.g., among various lineages of higher Curculionidae), is perhaps best explained by a predominance of shifts of weevils onto preexisting closely related plants (and a lesser incidence of shifts onto distant relatives) over parallel speciation (cospeciation) events with plants (33). Not surprisingly, these ecological-evolutionary patterns are similar to those predicted or observed for other herbivorous insects (for example, see ref. 34) and across entire insect herbivore faunas (35). Further exploration of the evolution of ancestral-host associations and larval and adult habits in weevils will require reconstructing their evolutionary histories using phylogenetic comparative methods.
Based on the available data, we propose an initial diversification of weevils into modern families, excepting Brentidae and Curculionidae, on conifers in the Middle to Late Jurassic, when conifers and other gymnospermous plants dominated forest ecosystems.§ Crown-group angiosperms probably first appeared by the end of the Late Jurassic [fossils ≈132–141 Ma (24), molecules ≈140–180 Ma (25)], and were most likely colonized by the ancestors of modern Brentidae and Curculionidae by the mid-Early Cretaceous (ca. Valanginian, 130 Ma). However, the first appearance of crown-group angiosperms evidently did not lead to an immediate and major diversification event in weevils. Massive diversification of the ancestrally angiosperm-associated family Curculionidae started somewhat later, ≈112.0–93.5 Ma (Albian–Cenomanian) (see Figs. 2 and 3) as angiosperms further diversified and for the first time achieved widespread floristic dominance (8, 9).
The unusually speciose family Curculionidae most likely first diversified on monocots [stem-group origin 140–150 Ma (37); crown-group origin 112 Ma fossil pollen (28), 130 Ma molecules (10)] and only later colonized and massively diversified in association with core eudicots [stem-group origin 100–120 Ma (37, 38), crown-group origin 93.5–89.3 Ma (39)], thus bypassing a significant diversity of existing early divergent dicots as potential hosts. The observed lag time between the first appearance of crown-group angiosperms and massive diversification of angiosperm-associated Curculionidae is consistent with the hypothesis of sequential evolution (40, 41). Higher-level diversification of Curculionidae continued into the early-to-middle Paleogene as angiosperms, especially core eudicots, continued to diversify and replace conifers and other gymnospermous plants, particularly in lowland “tropical” forests (42), perhaps facilitated by global warming and associated equability, or the K-Pg mass extinction (43, 44). The sequence of first appearances of weevil taxonomic groups in the fossil record, the timing of stem- and crown-group divergences of weevils estimated herein, timing and patterns of diversification in angiosperms, and patterns of angiosperm utilization and association by modern weevils, are consistent with this interpretation.
As for possible mechanisms, the relatively earlier diversification of monocots relative to core eudicots (45) may have facilitated the diversification of early Curculionidae, putting them at an evolutionary advantage over their sister group, the family Brentidae, some of which colonized early eudicots, and Magnoliales, but none of which are known to have successfully colonized the living tissues of monocots. Monocots have less strongly differentiated tissues with fewer numbers and kinds of secondary metabolites and other defences than most eudicots (46), and are predominantly herbaceous, lacking the true woody tissues of eudicots. As a result, they offered rapid growth life histories, Grime's “ruderal growth strategy” (47), and had accommodationist rather than well-defended life-history patterns for deflecting insect herbivores. Thus, monocots may have also offered competition-free and plant-defense-free space relative to other early divergent groups of angiosperms. Regardless of the mechanisms invoked, monocots appear to have played a pivotal and early role in the diversification of Curculionidae.
The unusual diversity of weevils thus appears to be the result of ecological-evolutionary opportunity, combined with morphological, behavioral, and physiological (intrinsic) traits: rostrum, oviposition behavior, larval endophagy, geniculate antennae, among other life-history attributes, fine-tuned, elaborated, and accumulated over the course of a long history of association with living, dead, dying, and decaying plants and plant organs and tissues. The resulting elaborate “trophic repertoire” may have prepared or perhaps even preadapted the most speciose family Curculionidae for feeding on the morphologically, ecologically, developmentally, and biochemically diverse tissues of angiosperms. Fine-tuning and elaboration of the weevil trophic repertoire, for example conveying the ability to metabolize additional- or new-plant secondary metabolites, or to oviposit deep into plant material, not only facilitated colonization and exploitation of diverse living tissues of nearly all other kinds of landplants, but also equipped Curculionidae to adapt to and track (2, 3) the increasing complexity and diversity in chemistry, structure, growth form and habits, habitat associations, and life histories of angiosperms over the course of their evolution. A similar scenario has been proposed for leaf-mining ditrysian Lepidoptera (48), and may be expected for other insect groups (and other elements of the biota) exhibiting close ecological associations with angiosperms.
As a corollary, substantial colonization of core eudicots by Curculionidae during the Late Cretaceous and Paleogene appears only to have occurred once the weevil trophic repertoire had been suitably fine-tuned and elaborated for feeding on their diverse structures and chemistries, and once core eudicots had become sufficiently widespread and abundant to serve as suitable hosts. Other weevil families (e.g., Anthribidae, Belidae, Brentidae, Nemonychidae, and Attelabidae) also colonized angiosperms; however, they lack many of the apparent specializations present in Curculionidae for feeding on (especially living tissues of) angiosperms, and today account for a relatively small fraction of angiosperm-associated weevils. Thus, the extraordinary taxonomic diversity of weevils appears to have been mediated predominantly by the presence of susceptible, abundant, and diverse host resources, and the ability of weevils to use those resources, rather than by the evolution of host taxa themselves. Indeed, the reconstructed evolutionary history of diversification in weevils reveals a deep and complex history of coevolution with angiosperms, including evidence for codiversification (49), resource tracking, and sequential evolution (40, 41).
Materials and Methods
See the SI Materials and Methods for more details.
Taxon Sampling and DNA Sequencing.
We analyzed up to 8 kb of DNA sequence data from a worldwide sample of 135 weevil genera representing all 7 weevil families, all 26 weevil subfamilies, and 97 genera representing most major tribes in the family Curculionidae (2) (see Tables S1 and S3). Outgroups included 7 subfamilies of basal Chrysomeloidea, the weevil sister group (6, 26), and Ericmodes sylvaticus (Cucujoidea: Protocucujidae). Six genes (2 mitochondrial and 4 nuclear) were used in this study: cytochrome oxidase I (cox I), 16S rDNA, 18S rDNA, 28S rDNA, elongation factor 1-α (EF 1-α), and arginine kinase (AK). For primers used, see Table S4.
Phylogenetic Analyses.
We ran two partitioned BI phylogenetic analyses in the program BEAST 1.4.7 (65–75 million generations, 12 partitions, GTR+I+Γ substitution model, estimated base frequencies) on the maximum- and minimum-age data sets, for a total of 4 analyses. All trees were rooted with Ericmodes sylvaticus (50, 51). Graphical and statistical analyses implemented in the program Tracer 1.4 were used to assess convergence and otherwise check performance and accuracy of the BEAST analyses. Based on these results, we combined the last 5,000 trees from each of the paired minimum- and maximum-age analyses and used them to estimate PPs, to obtain maximum-clade credibility trees, and to estimate divergence times and corresponding 95% confidence intervals (LogCombiner 1.4.7, PAUP* 4.03b10, TreeAnnotator 1.4.7). We implemented a partitioned ML BS analysis (1,000 inferences, 12 partitions, CAT substitution model, individual per partition branch-length optimization) using the program RAxML 7.0.4 on the CIPRES cluster at the San Diego Supercomputing Center.
Hypothesis Testing.
We investigated the degree to which select alternative phylogenetic hypotheses were supported by our data by estimating the posterior probabilities of alternative topologies (under BI), and by comparing the ML trees obtained with and without monophyly constraints on each group of interest using the KH test (52) as implemented in PAUP* 4.03b10.
Divergence-Time Estimates.
Divergence times were coestimated with phylogeny using the Bayesian relaxed molecular clock method (53) (BEAST 1.4.7). We assumed the uncorrelated lognormal prior model of rate change, a Yule prior process to model speciation, and used automatic tuning of operators. We conservatively selected and applied fossil age constraints from 2 recent reviews (2, 5), using only the oldest fossils that could be unequivocally assigned (based on character evidence) to extant weevil subfamilies or families in our analyses (Table S5).
Supplementary Material
Acknowledgments.
We thank R. Anderson, R. Oberprieler, and C. O'Brien for specimens, identifications, and for sharing their observations and insight, W. Farnum for laboratory assistance, and our many other colleagues who generously provided specimens used in this study. This work was supported by grants from the National Science Foundation Assembling the Tree of Life program, and Consejo Nacional de Investigaciones Científicas y Técnicas (PIP 5766).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: DNA sequences reported in this paper have been deposited in the GenBank database (accession nos. FJ859915–FJ859985, FJ867654–FJ867897).
This article contains supporting information online at www.pnas.org/cgi/content/full/0810618106/DCSupplemental.
Computer simulations have shown that Bayesian PP associated with short branch lengths and low values for nonparametric BS support (or other non-Bayesian measures of support) may be excessively liberal, particularly as the size of the data set increases (12, 13). Therefore, relationships for which PP support is elevated as compared to maximum likelihood (ML) BS, should be interpreted cautiously.
Supported by their comparatively early appearance in the fossil record (when known), Gondwanan distributions (at least of the oldest, and often conifer-associated lineages), and phylogenetic position, generally at the base of their respective groups (6).
The larvae of Platypodinae feed on ambrosia fungi cultivated by adults in galleries in wood.)
All living weevil families are known from the fossil record, and all weevil families in the fossil record are extant [Ulyanidae is most likely associated with Nemonychidae (2)], consistent with the contention that reduced extinction has played an important role in the extraordinary diversity of modern insects (36).
References
- 1.Alonso-Zarazaga MA, Lyal CHC. A World Catalogue of Families and Genera of Curculionoidea (Insecta: Coleoptera) (excepting Scolytidae & Platypodidae) Barcelona: Entomopraxis SCP; 1999. [Google Scholar]
- 2.Oberprieler RG, Marvaldi AE, Anderson RS. Weevils, weevils, weevils everywhere. Zootaxa. 2007;1668:491–520. [Google Scholar]
- 3.Anderson RS. An evolutionary perspective on diversity in Curculionoidea. Mem Ent Soc Wash. 1995;14:103–114. [Google Scholar]
- 4.Crowson RA. The Natural Classification of the Families of Coleoptera. London: Nathan Lloyd; 1955. [Google Scholar]
- 5.Gratshev VG, Zherikhin VV. In: Krzeminska E, Krzeminski W, editors. Acta Zool Cracov; Proceedings of the 2nd Congress on Palaeoentomology; Krakow, Poland. 2003. pp. 129–138. [Google Scholar]
- 6.Farrell BD. “Inordinate Fondness” explained: Why are there so many beetles? Science. 1998;281:555–559. doi: 10.1126/science.281.5376.555. [DOI] [PubMed] [Google Scholar]
- 7.Kuschel G. Past and present of the relict family Nemonychidae (Coleoptera: Curculionoidea) GeoJournal. 1983;7:499–504. [Google Scholar]
- 8.Lidgard S, Crane PR. Quantitative analyses of the early angiosperm radiation. Nature. 1988;331:344–346. [Google Scholar]
- 9.Lidgard S, Crane PR. Angiosperm diversification and Cretaceous floristic trends: A comparison of palynofloras and leaf macrofloras. Paleobiology. 1990;16:77–93. [Google Scholar]
- 10.Bremer K. Early Cretaceous lineages of monocot flowering plants. Proc Natl Acad Sci USA. 2000;97:4707–4711. doi: 10.1073/pnas.080421597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marvaldi AE, Sequeira AS, O'Brien CW, Farrell BD. Molecular and morphological phylogenetics of weevils (Coleoptera, Curculionoidea): Do niche shifts accompany diversification? Syst Biol. 2002;51:761–785. doi: 10.1080/10635150290102465. [DOI] [PubMed] [Google Scholar]
- 12.Lewis PO, Holder MT, Holsinger KE. Polytomies and Bayesian phylogenetic inference. Syst Biol. 2005;54:241–253. doi: 10.1080/10635150590924208. [DOI] [PubMed] [Google Scholar]
- 13.Yang Z. Fair-balance paradox, star-tree paradox, and Bayesian phylogenetics. Mol Biol Evol. 2007;24:1639–1655. doi: 10.1093/molbev/msm081. [DOI] [PubMed] [Google Scholar]
- 14.Kuschel G. A phylogenetic classification of Curculionoidea to families and subfamilies. Mem Ent Soc Wash. 1995;14:5–33. [Google Scholar]
- 15.Marvaldi AE. Higher level phylogeny of Curculionidae (Coleoptera: Curculionoidea) based mainly on larval characters, with special reference to broad-nosed weevils. Cladistics. 1997;13:285–312. doi: 10.1111/j.1096-0031.1997.tb00321.x. [DOI] [PubMed] [Google Scholar]
- 16.Marvaldi AE, Morrone JJ. Phylogenetic systematics of weevils (Coleoptera: Curculionoidea): A reappraisal based on larval and adult morphology. Insect Syst Evol. 2000;31:43–58. [Google Scholar]
- 17.Marvaldi AE. Larval morphology and biology of oxycorynine weevils and the higher phylogeny of the Belidae (Coleoptera, Curculionoidea) Zool Scr. 2005;34:37–48. [Google Scholar]
- 18.Marvaldi AE, Oberprieler RG, Lyal CHC, Bradbury T, Anderson RS. Phylogeny of the Oxycoryninae s.l. (Coleoptera Phytophaga) and evolution of plant-weevil interactions. Invertebr Syst. 2006;20:447–476. [Google Scholar]
- 19.Thompson RT. Observations on the morphology and classification of weevils (Coleoptera: Curculionoidea) with a key to the major groups. J Nat Hist. 1992;26:835–891. [Google Scholar]
- 20.Kuschel G, Leschen RAB, Zimmerman EC. Platypodidae under scrutiny. Invertebr Taxon. 2000;14:771–806. [Google Scholar]
- 21.Kuschel G. A cossonine genus with bark-beetle habits, with remarks on relationships and biogeography (Coleoptera, Curculionidae) N Z J Sci. 1966;9:3–29. [Google Scholar]
- 22.Kuschel G. The subfamily Molytinae (Coleoptera: Curculionidae): General notes and descriptions of new taxa from New Zealand and Chile. New Zealand Ent. 1987;9:11–29. [Google Scholar]
- 23.Oberprieler RG. Systematics and evolution of the cycad-associated weevil genus Apinotropis Jordan (Coleoptera: Anthribidae) Afr Entomol. 1999;7:1–33. [Google Scholar]
- 24.Brenner G. In: Flowering Plant Origin, Evolution, and Phylogeny. Taylor DW, Hickey LJ, editors. New York: Chapman & Hall; 1996. pp. 91–115. [Google Scholar]
- 25.Bell C, Soltis DE, Soltis PS. The age of the angiosperms: A molecular timescale without a clock. Evolution. 2005;59:1245–1258. [PubMed] [Google Scholar]
- 26.Marvaldi AE, Duckett CN, Kjer KM, Gillespie JJ. Structural alignment of 18S and 28S rDNA sequences provides insights in to phylogeny of Phytophaga (Coleoptera: Curculionoidea and Chrysomeloidea) Zool Scr. 2009;38:63–77. [Google Scholar]
- 27.Kuschel G, May BM. Palophaginae, a new subfamily for leaf-beetles, feeding as adults and larva on araucarian pollen in Australia (Coleoptera: Megalopodidae) Invertebr Taxon. 1990;3:697–719. [Google Scholar]
- 28.Friis EM, Pedersen KR, Crane PR. Araceae from the Early Cretaceous of Portugal: Evidence on the emergence of monocotyledons. Proc Natl Acad Sci USA. 2004;101:16565–16570. doi: 10.1073/pnas.0407174101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crane PR, Friis EM, Pedersen KJ. The origin and early diversification of angiosperms. Nature. 1995;374:27–33. [Google Scholar]
- 30.Friis EM, Pedersen KR, Crane PR. Fossil evidence of water lilies (Nymphaeales) in the Early Cretaceous. Nature. 2001;410:357–360. doi: 10.1038/35066557. [DOI] [PubMed] [Google Scholar]
- 31.Sequeira AS, Normark BB, Farrell BD. Evolutionary assembly of the conifer fauna: Distinguishing ancient from recent associations in bark beetles. Proc R Soc Lond B Biol Sci. 2000;267:2359–2366. doi: 10.1098/rspb.2000.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sequeira AS, Farrell BD. Evolutionary origins of Gondwanan interactions: How old are Araucaria beetle herbivores? Biol J Linn Soc Lond. 2001;74:459–474. [Google Scholar]
- 33.Farrell BD, et al. The evolution of agriculture in beetles (Curculionidae: Scolytinae and Platypodinae) Evolution. 2001;55:2011–2027. doi: 10.1111/j.0014-3820.2001.tb01318.x. [DOI] [PubMed] [Google Scholar]
- 34.Berenbaum M. Coumarins and caterpillars: A case for coevolution. Evolution. 1983;37:163–179. doi: 10.1111/j.1558-5646.1983.tb05524.x. [DOI] [PubMed] [Google Scholar]
- 35.Novotny V, Basset Y. Host specificity of insect herbivores in tropical forests. Proc R Soc Lond B Biol Sci. 2005;272:1083–1090. doi: 10.1098/rspb.2004.3023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Labandeira CC, Sepkoski JJ., Jr Insect diversity in the fossil record. Science. 1993;261:310–315. doi: 10.1126/science.11536548. [DOI] [PubMed] [Google Scholar]
- 37.Chaw SM, Chang CC, Chen HL, Li WH. Dating the monocot–dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol. 2004;58:424–441. doi: 10.1007/s00239-003-2564-9. [DOI] [PubMed] [Google Scholar]
- 38.Anderson CL, Bremer K, Friis EM. Dating phylogenetically basal eudicots using rbcL sequences and multiple fossil reference points. Am J Bot. 2005;92:1737–1748. doi: 10.3732/ajb.92.10.1737. [DOI] [PubMed] [Google Scholar]
- 39.Friis EM, Pedersen KR, Crane PR. When Earth started blooming: Insights from the fossil record. Curr Opinion Plant Biol. 2005;8:5–12. doi: 10.1016/j.pbi.2004.11.006. [DOI] [PubMed] [Google Scholar]
- 40.Jermy T. Insect-host-plant relationship—Coevolution or sequential evolution? Symp Biol Hung. 1976;16:109–113. [Google Scholar]
- 41.Jermy T. Evolution of insect/hostplant relationships. Am Nat. 1984;124:609–630. [Google Scholar]
- 42.Jaramillo CA. Cenozoic plant diversity in the Neotropics. Science. 2006;311:1893–1896. doi: 10.1126/science.1121380. [DOI] [PubMed] [Google Scholar]
- 43.Zachos J, Pagani M, Sloan L, Thomas E, Billups K. Trends, rhythms, and aberrations in global climate 65 Ma to present. Science. 2001;292:686–693. doi: 10.1126/science.1059412. [DOI] [PubMed] [Google Scholar]
- 44.Labandeira CC, Johnson KR, Wilf P. Impact of the terminal Cretaceous event on plant-insect associations. Proc Natl Acad Sci USA. 2002;99:2061–2066. doi: 10.1073/pnas.042492999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Magallon S, Crane PR, Herendeen PS. Phylogenetic pattern, diversity, and diversification of eudicots. Ann MO Bot Gard. 1999;86:297–372. [Google Scholar]
- 46.Coley PD, Barone JA. Herbivory and plant defenses in tropical forests. Annu Rev Ecol Syst. 1996;27:305–335. [Google Scholar]
- 47.Grime JP. Evidence for the existence of three primary strategies in plants and its relevance to ecological and evolutionary theory. Amer Nat. 1977;111:1169–1195. [Google Scholar]
- 48.Lopez-Vaamonde C, et al. Fossil calibrated molecular phylogenies reveal that leaf-mining moths radiated millions of years after their host plants. J Evolution Biol. 2006;19:1314–1326. doi: 10.1111/j.1420-9101.2005.01070.x. [DOI] [PubMed] [Google Scholar]
- 49.Ehrlich PR, Raven PH. Butterflies and plants: A case for coevolution. Evolution. 1964;18:586–608. [Google Scholar]
- 50.McKenna DD, Farrell BD. In: Timetrees of Life. Hedges B, Kumar S, editors. Oxford: Oxford Univ Press; 2009. pp. 278–289. [Google Scholar]
- 51.Hunt T, et al. A comprehensive phylogeny of beetles reveals the evolutionary origins of a superradiation. Science. 2007;318:1913–1916. doi: 10.1126/science.1146954. [DOI] [PubMed] [Google Scholar]
- 52.Kishino H, Hasegawa M. Evaluation of the maximum likelihood estimation of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J Mol Evol. 1989;29:170–179. doi: 10.1007/BF02100115. [DOI] [PubMed] [Google Scholar]
- 53.Drummond AJ, Ho SYW, Philipps MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PLoS Biol. 2006;4:e88. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.