Abstract
Aurora kinase-A and -B are key regulators of the cell cycle and tumorigenesis. It has remained a mystery why these 2 Aurora kinases, although highly similar in protein sequence and structure, are distinct in subcellular localization and function. Here, we report the striking finding that a single amino acid residue is responsible for these differences. We replaced the Gly-198 of Aurora-A with the equivalent residue Asn-142 of Aurora-B and found that in HeLa cells, Aurora-AG198N was recruited to the inner centromere in metaphase and the midzone in anaphase, reminiscent of the Aurora-B localization. Moreover, Aurora-AG198N compensated for the loss of Aurora-B in chromosome misalignment and cell premature exit from mitosis. Furthermore, Aurora-AG198N formed a complex with the Aurora-B partners, INCENP and Survivin, and its localization depended on this interaction. Aurora-AG198N phosphorylated the Aurora-B substrates INCENP and Survivin in vitro. Therefore, we propose that the presence of Gly or Asn at a single site assigns Aurora-A and -B to their respective partners and thus to their distinctive subcellular localizations and functions.
Keywords: single amino acid mutation, TPX2, INCENP, Survivin
Aurora kinases have been implicated in several vital events at the spindle pole and the equatorial region during mitosis (reviewed in refs. 1–8). Such diverse roles are accomplished by Aurora-A and -B, which share 70% homology in the catalytic domain and yet are functionally distinct (2–7). In mitotic cells, Aurora-A localizes to the spindle pole and the microtubule, and is mainly involved in centrosome maturation, mitotic entry, and spindle assembly (9–11). On the other hand, Aurora-B persists at the inner centromere from prometaphase to metaphase and relocalizes to the midzone in anaphase, where it participates in chromatin modification, microtubule–kinetochore attachment, the spindle checkpoint, and cytokinesis (12, 13).
To fulfill their routine functions in cell cycle control, both Aurora-A and -B are tightly regulated by their respective partner proteins. TPX2 binding targets Aurora-A to spindle microtubules, enhances its autophosphorylation, and protects the activation site from PP1 dephosphorylation (14–20). Conversely, localization of Aurora-B depends on an intact chromosomal passenger complex containing at least 3 other proteins, INCENP, Survivin, and Borealin (21–26), and the activity of Aurora-B is stimulated mainly by INCENP (27, 28).
To investigate how Aurora-A and -B, despite great similarity in protein sequence and structure, interact with such disparate cellular machinery, we conducted a strategy in which the regulation of Aurora-A or -B by its partner proteins was impaired. When G198 within the human Aurora-A catalytic domain (G205 in Xenopus) is substituted by the equivalent residue N142 in human Aurora-B (N158 in Xenopus), the binding affinity and the activation of Aurora-A by recombinant TPX2 is reduced in vitro (29, 30). Given this, we introduced the Aurora-AG198N mutant into HeLa cells and found that Aurora-AG198N colocalized with Aurora-B at the centromere and the midzone in mitosis, and lost the original Aurora-A localization at the spindle. Moreover, Aurora-AG198N prevented the chromosome misalignment and cell premature exit from mitosis caused by Aurora-B knockdown, indicating functional substitution for Aurora-B in cell cycle regulation.
Results
Aurora-AG198N Colocalizes with Aurora-B at Centromere and Midzone.
We fused the mutants Aurora-AG198N and Aurora-BN142G (Fig. 1A) with GFP and transiently expressed them in HeLa cells [supporting information (SI) Fig. S1A]. Through immunoprecipitation, we found that Aurora-AG198N bound less TPX2 than Aurora-AWT (Fig. 1B), and that Aurora-BN142G bound less INCENP and Survivin than Aurora-BWT (Fig. S2A). Consistent with previous reports (29, 30), the kinase activity of Aurora-AG198N was reduced by 10-fold compared with that of Aurora-AWT (Fig. S3).
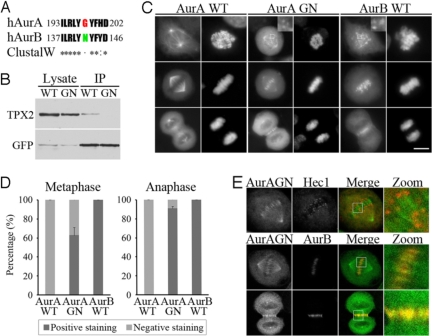
Fig. 1.
Aurora-AG198N colocalizes with Aurora-B at inner centromere and midzone in mitosis. (A) Scheme depicting the mutation domain of Aurora-A and relative sequence of Aurora-B. G198 (red) in human Aurora-A was altered to the equivalent residue N142 (green) in Aurora-B. Homology of mutation domain is presented by ClustalW. (B) GFP-Aurora-AWT or GFP-Aurora-AG198N was transfected into HeLa cells and immunoprecipitated from cell lysate with GFP antibody. The immunoprecipitated complexes were blotted for TPX2 (Upper) or GFP (Lower). Aurora-AG198N bound less TPX2 compared with Aurora-AWT. (C) Immunofluorescence photomicrographs of HeLa cells transfected with GFP-tagged Aurora-AWT, Aurora-AG198N, or Aurora-BWT (Left). Aurora-AG198N and Aurora-BWT localized to centromere and midzone, whereas Aurora-AWT did not. DNA labeled with DAPI (Right). (Scale bar, 10 μm.) (D) Quantitative characterization of Aurora-AG198N localization in C. Dark-gray bars represent the percentage of cells positively stained with Aurora-AG198N at centromere and midzone, and light-gray bars represent the percentage unstained at these locations. Each data point represents 3 independent experiments, each measuring 250 mitotic cells, and error bars indicate SD (P < 0.01). (E) Confocal images of HeLa cells transfected with GFP-Aurora-AG198N (green) and stained with Hec1 or Aurora-B antibody (red). Aurora-AG198N colocalized with Aurora-B at inner centromere and midzone but not with Hec1 at outer kinetochore.
To evaluate the behaviors of Aurora-AG198N and Aurora-BN142G in the cell, HeLa cells were transfected with GFP-tagged Aurora-AWT, Aurora-AG198N, Aurora-BN142G, and Aurora-BWT and harvested for immunofluorescence microscopy. Whereas Aurora-AWT was concentrated at the centrosome and spindle microtubule as reported (9–11), Aurora-AG198N exhibited a different localization (Fig. 1C). Besides a signal at the centrosome and a diffuse distribution in the cytoplasm, Aurora-AG198N was observed at the centromere in ≈63% of metaphase cells and at the midzone in 91% of anaphase cells (Fig. 1D), reminiscent of Aurora-B localization (12, 13). Similar results were seen with living cell microscopy (Fig. S4). Confocal images further confirmed that Aurora-AG198N colocalized with Aurora-B at the inner centromere and the midzone but not with Hec1 at the outer kinetochore (Fig. 1E). Like Aurora-A, Aurora-BN142G displayed localization at the centrosome and the spindle, in addition to a signal at the centromere and the midzone (Fig. S2 B–D). These results demonstrate that a single amino acid change causes Aurora-A to exhibit a cellular distribution similar to that of Aurora-B apart from remnant at the spindle poles and leads Aurora-B to deviate from its wild-type localization to that of Aurora-A. We next focused on Aurora-AG198N to study the mechanism of the transformation between Aurora-A and -B.
Aurora-AG198N Prevents Chromosome Misalignment and Cell Premature Exit from Mitosis Caused by Aurora-B Knockdown.
We assessed whether Aurora-AG198N was functional by monitoring cell cycle progression after simultaneous Aurora-B knockdown and Aurora-AG198N expression. For this purpose, 2 cell lines were generated using the tet-on system to express Aurora-AWT or Aurora-AG198N in the presence of tetracycline (Fig. S1B). Cells treated with or without tetracycline were cotransfected with Aurora-B RNAi vector and RFP-H2B (ratio 40:1), which is a marker for Aurora-B knockdown and chromosome morphology (Fig. 2A and Fig. S5A). By time-course imaging, we found that Aurora-B knockdown cells could not properly align their chromosomes to the equatorial plate and prematurely exited mitosis from prometaphase without chromosome separation and cytokinesis (Fig. 2B and Movie S1, Movie S2, Movie S3, Movie S4, Movie S5, and Movie S6), indicating compromise of the spindle checkpoint consistent with previous reports (31–34). Expression of Aurora-AWT by tetracycline inducement did not affect progression of the cell cycle (Fig. 2B and Movie S7). In contrast, expression of Aurora-AG198N largely restored the mitotic progression after Aurora-B knockdown (Fig. 2B and Movie S8). RNAi treatment caused a 21% increase in the proportion of prophase to metaphase cells and a corresponding decrease in that of anaphase cells (Fig. 2C), in agreement with previous reports (31, 33). Simultaneous expression of Aurora-AG198N but not Aurora-AWT reduced this increase by 13% (Fig. 2C). Together, these results show that Aurora-AG198N partially compensated for the loss of Aurora-B function in mitosis.
Fig. 2.
Aurora-AG198N prevents chromosome misalignment and cell premature exit from mitosis caused by Aurora-B knockdown. (A) Immunofluorescence photomicrographs of HeLa cells cotransfected with Aurora-B RNAi and RFP-H2B (red) at a ratio of 40:1 and labeled with Aurora-B antibody (green). Note that those cells expressing RFP-H2B showed disperse staining of Aurora-B with decreased fluorescence intensity, as indicated by arrows. Thus, in addition to chromosome morphology, RFP-H2B was a marker for Aurora-B knockdown. DNA labeled with DAPI (blue). (B) HeLa cells induced by tetracycline to express Aurora-AWT or Aurora-AG198N were cotransfected with Aurora-B RNAi and RFP-H2B at a ratio of 40:1, and subjected to time-course fluorescence microscopy. Chromosome morphology, visualized by RFP-H2B staining, was filmed every 5 min. With Aurora-B knockdown, cells could not properly align the chromosomes and exited mitosis without chromosome separation and cytokinesis. Induced expression of Aurora-AG198N, but not Aurora-AWT, prevented this abnormality. Representative cells can be viewed as movies or still images in Movie S1, Movie S2, Movie S3, Movie S4, Movie S5, Movie S6, Movie S7, and Movie S8. (C) Quantitative characterization of mitotic HeLa cells treated as in B. Dark-gray bars represent the percentage of cells from prophase to metaphase, and light-gray bars represent the percentage in anaphase. Cells were classified by chromosome morphology visualized by RFP-H2B. Each data point represents 3 independent experiments, each measuring 250 mitotic cells, and error bars indicate SD (P < 0.01).
Aurora-AG198N Interacts with INCENP and Survivin, and Its Localization Depends on This Interaction.
We asked how Aurora-AG198N was recruited to the inner centromere and the midzone like Aurora-B. Since Aurora-B relies on an intact chromosomal passenger complex for proper localization, we hypothesized that this might also be necessary for Aurora-AG198N. To address this, HeLa cells were synchronized, transfected with GFP-tagged Aurora-AWT, Aurora-AG198N, and Aurora-BWT, and harvested for immunoprecipitation with GFP antibody. Biochemical analysis of the immunoprecipitated complexes revealed that Aurora-AG198N was in a complex with INCENP and Survivin, and this complex did not contain Aurora-B (Fig. 3A). We also tested the interaction between Aurora-AG198N and INCENP in vitro by GST pull-down assay using recombinant proteins. Consistent with the in vivo immunoprecipitation result, Aurora-AG198N bound INCENP826–919, as did Aurora-B, either in the presence or absence of TPX21–43 (Fig. 3E). Surprisingly, we found that Aurora-AWT also bound INCENP826–919 in vitro, but this interaction was remarkably reduced by the addition of TPX21–43 (Fig. 3E), suggesting that TPX2 has higher affinity to Aurora-A than INCENP. Our results show that Aurora-AG198N forms a complex with INCENP and Survivin both in vivo and in vitro.
Fig. 3.
Aurora-AG198N physically interacts with INCENP and Survivin, and its localization depends on them. (A) GFP-tagged Aurora-AWT, Aurora-AG198N, or Aurora-BWT was transfected into HeLa cells and immunoprecipitated from cell lysate with GFP antibody. The immunoprecipitated complexes were blotted for INCENP, endogenous Aurora-B, Survivin, or GFP. Aurora-AG198N bound INCENP and Survivin, not Aurora-B. (B and C) Photomicrographs of tet-on HeLa cells transfected with INCENP RNAi (B) or Survivin RNAi (C) in the presence of tetracycline and labeled with INCENP (B) or Survivin (C) antibody. Knockdown of INCENP or Survivin displaced Aurora-AG198N from centromere and midzone. Arrows indicate lagging chromosomes. (Scale bars, 10 μm.) (D) Quantitative characterization of Aurora-AG198N localization after INCENP or Survivin RNAi. Dark-gray bars represent the percentage of cells positively stained with Aurora-AG198N at centromere and midzone, and light-gray bars represent the percentage unstained at these locations. Each data point represents 3 independent experiments, each measuring 250 mitotic cells, and error bars indicate SD (P < 0.01). (E) GST pull-down assay for recombinant Aurora-AWT, Aurora-AG198N, or Aurora-BWT using GST-INCENP826–919, in the presence or absence of GFP-TPX21–43. Aurora-AG198N bound INCENP826–919 regardless of TPX21–43 addition (Top and Middle). Although Aurora-AWT bound INCENP826–919 (Top), this interaction was remarkably reduced when TPX21–43 was added (Middle). Input proteins were shown in separate lane with corresponding amount (Bottom). Arrowheads indicate Aurora-AWT, Aurora-AG198N, and Aurora-BWT.
We then examined the impact of INCENP and Survivin on Aurora-AG198N localization. In the presence of tetracycline, Aurora-AG198N tet-on cells were depleted of endogenous INCENP or Survivin by RNAi (Fig. S5 B and C) and harvested for immunofluorescence microscopy. Both INCENP and Survivin knockdown completely displaced Aurora-AG198N from the inner centromere and the midzone (Fig. 3 B–D and Fig. S5D). Thus, the localization of Aurora-AG198N to the inner centromere and the midzone is due to its association with INCENP and Survivin as a result of the G198N mutation.
Aurora-AG198N Phosphorylates INCENP and Survivin in Vitro, and Its Kinase Activity Is Enhanced in the Presence of INCENP.
We sought to resolve how Aurora-AG198N compensated for the loss of Aurora-B in mitosis. As a kinase, Aurora-B functions primarily by phosphorylating its substrates. INCENP is phosphorylated by Aurora-B at 3 consecutive residues T893/S894/S895 (28), and Survivin at T117 (35). Thus, we tested if Aurora-AG198N could phosphorylate these proteins. Recombinant Aurora-AG198N was incubated with GST, GST-INCENP826–919, and Survivin in the presence of γ32ATP. Both INCENP826–919 and Survivin were phosphorylated by Aurora-AG198N, Aurora-BWT (Fig. 4A), and Aurora-AWT (36). To test whether Aurora-AG198N could be activated by INCENP as is Aurora-B, kinase assay was performed using MBP as the substrate. The phosphorylation of MBP by Aurora-AG198N was enhanced by the addition of INCENP826–919 (Fig. 4B). These findings suggest that Aurora-AG198N is capable of phosphorylating Aurora-B substrates, and its kinase activity is enhanced by INCENP.
Fig. 4.

Aurora-AG198N phosphorylates INCENP and Survivin in vitro, and its activity is enhanced by INCENP. (A) In vitro kinase assay for recombinant Aurora-AG198N using GST, GST-INCENP826–919, and Survivin as substrates. Phosphorylated INCENP and Survivin were detected by autoradiography (Top), and total inputs were visualized by Coomassie blue staining (Middle). Lane 1–3, GST; lane 4–6, GST-INCENP826–919; lane 7–9, Survivin. Lanes 1, 4, and 7, without kinase; lanes 2, 5, and 8, with Aurora-AG198N; lanes 3, 6, and 9, with Aurora-BWT. (B) In vitro kinase assay for recombinant Aurora-AG198N using MBP as substrate, in the presence or absence of GST-INCENP826–919. Phosphorylated MBP was detected by autoradiography. INCENP826–919 stimulated phosphorylation of MBP by Aurora-AG198N.
Both Elongation and Hydrophilicity of the Side Chain at Aurora-AG198 Are Necessary to Convert Aurora-A into Aurora-B-Like Kinase.
We further explored the molecular explanation of how a single amino acid change caused such tremendous transformation of Aurora-A in partner specificity and protein function. According to the crystal structure of human Aurora-A122–403/TPX21–43 (14), the Cα atom of G198 in Aurora-A interacts so closely with TPX2P13 that a longer side chain is intolerable (Fig. 5A i–iv), but the situation is different within the equivalent domain of the Aurora-B complex. According to the structure of Xenopus Aurora-B78–361/INCENP798–840 (37), the side chain of N158 in Aurora-B interacts properly with INCENPY825/I828/D829/S830 in a hydrophilic state; it also forms hydrogen bonds with INCENPY825/I828 to strengthen the interaction (Fig. 5 A v and vi). In our hypothesis, the side chain of N198 in Aurora-AG198N prevents attachment to TPX2, leaving perfect space for INCENP. Moreover, the potential hydrogen bond with human INCENPI872 further stabilizes the complex of Aurora-AG198N:INCENP (Fig. 5A vii and viii).
Fig. 5.
Both elongation and hydrophilicity of the side chain at residue 198 are required to convert Aurora-A into Aurora-B-like kinase. (A) Aurora-A and -B are distinguished by different partner proteins through local structural change at a single amino acid. (i and ii) The complex of human Aurora-A122–403/TPX21–43 (PDB ID code 1OL5 in ref. 14). (v and vi) The complex of Xenopus Aurora-B78–361/INCENP798–840 (PDB ID code 2BFY in ref. 37). (iii, iv, vii, and viii) Predicted by point mutation using PyMol (www.pymol.org). Structurally, G198 of Aurora-A (green) interacts with TPX2P13 (blue) in 3-dimensional space (i and ii). When G198 is replaced by N, the side chain is elongated, and TPX2 is prevented from Aurora-A (iii and iv). In contrast, Aurora-B (yellow) interacts with INCENP (purple) in a looser way within the equivalent domain (v and vi). The side chain of N158 in Xenopus Aurora-B interacts perfectly with INCENPY825/I828/D829/S830 and forms hydrogen bonds with INCENPY825/I828. Thus, Aurora-AG198N can interact with INCENP through its N198, which not only fits the 3-dimensional structure but probably builds hydrogen bonds with human INCENPI872 (vii and viii). (B) Overview of Aurora-A localization after substituting G198 with various amino acids. When the length of side chain reached that of V and L, Aurora-A was prevented from spindle. Hydrophilic side chains N and Q localized Aurora-A to centromere and midzone. (C) HeLa cells were transfected with GFP-tagged Aurora-AWT, Aurora-AG198A, Aurora-AG198V, Aurora-AG198L, Aurora-AG198N, or Aurora-AG198Q (Left, and green in Right), and subjected to immunofluorescence microscopy. DNA labeled with DAPI (Center, and blue in Right). Arrows indicate centromere and midzone. (Scale bar, 10 μm.) (D) GFP-tagged Aurora-AG198A, Aurora-AG198V, Aurora-AG198L, Aurora-AG198N, or Aurora-AG198Q was transfected into HeLa cells and immunoprecipitated from cell lysate with GFP antibody. The immunoprecipitated complexes were blotted for INCENP (Upper) or GFP (Lower). Aurora-AG198N and Aurora-AG198Q bound INCENP, whereas Aurora-AG198A, Aurora-AG198V, and Aurora-AG198L did not.
To test this speculation, we generated a series of Aurora-A mutants by replacing G198 with various amino acids (Fig. 5B). Aurora-AG198A localized to the spindle, as did Aurora-AWT (Fig. 5C). However, when the side chain of this residue was elongated to the length of that of V or L, Aurora-A did not localize to the spindle (Fig. 5C). Furthermore, when G198 was replaced by N or Q, similar in length to V and L but hydrophilic, Aurora-A localized to the centromere and the midzone (Fig. 5C). To quantitatively analyze the INCENP-binding activity of all mutants, HeLa cells were synchronized, transfected with GFP-tagged Aurora-AG198A, Aurora-AG198V, Aurora-AG198L, Aurora-AG198N, and Aurora-AG198Q, and harvested for immunoprecipitation with GFP antibody. Biochemical analysis revealed that Aurora-AG198N and Aurora-AG198Q formed complexes with INCENP, whereas Aurora-AG198A, Aurora-AG198V, and Aurora-AG198L did not (Fig. 5D). Together, these results show that the elongation and the hydrophilicity of the side chain at Aurora-AG198 determine its interaction with INCENP instead of TPX2 and, therefore, are required for its conversion into Aurora-B-like kinase.
Discussion
Our study reveals that G198 of Aurora-A is a key residue in terms of discriminating the behaviors of Aurora-A and -B in the cell. Because the protein sequences of their catalytic domains are very similar, we are interested in understanding how distinct localization and function are determined. Although it has been reported that G198 in human Aurora-A (29) [G205 in Xenopus (30)] is a determinant for its regulation by TPX2 in vitro, the molecular mechanism and the biological effects of the G–N mutation were unknown. Here, we propose that the G–N mutation converts Aurora-A into Aurora-B-like kinase in cell (Fig. 6). It is achieved through the alteration of partner specificity, which in turn, defines subcellular localization and substrate phosphorylation. First, the G–N mutation enables Aurora-A to interact with INCENP instead of TPX2. Because INCENP, Survivin, and Borealin associate with each other through a tight 3-helical bundle (38), once Aurora-A is connected to INCENP, it is brought into the chromosomal passenger complex and is localized where this complex settles. Finally, Aurora-AG198N phosphorylates the Aurora-B substrates (Fig. 4A); thus, it probably works as an enzymatic component in the complex substituting for Aurora-B. Accordingly, the cell cycle proceeds successfully even when Aurora-B is absent (Fig. 6). To our surprise, recombinant Aurora-A, purified from either Escherichia coli or Sf21 insect cells, binds INCENP826–919 in a GST pull-down assay (Fig. 3E), and phosphorylates INCENP826–919 and Survivin in vitro (36), indicating the potential of Aurora-A to act like Aurora-B. Addition of TPX21–43 eliminates the interaction between Aurora-A and INCENP in the GST pull-down assay (Fig. 3E), suggesting that in vivo, this potential of Aurora-A is probably restrained due to the anchoring effect by TPX2 that keeps Aurora-A to the spindle and away from the centromere and the midzone.
Fig. 6.
Model depicting the relationship between Aurora-A and -B: Aurora-A substitutes for Aurora-B in mitosis when it is displaced to the location of Aurora-B. (A) Under conventional condition, Aurora-A (green) binds TPX21–43 (blue) and localizes to spindle microtubule, and Aurora-B (yellow) binds INCENP826–919 (purple) and localizes to centromere and midzone. (B) When Aurora-A is altered at G198 by N, it binds INCENP instead of TPX2. In this complex containing at least Aurora-AG198N, INCENP, and Survivin, Aurora-AG198N is activated and phosphorylates Aurora-B substrates. Hence, when Aurora-B is absent from the cell, Aurora-AG198N maintains normal progression of the cell cycle.
We also observed in our experiments that when the equivalent residue N142 of human Aurora-B is replaced with G to make an Aurora-A-like mutant, it exhibits additional localization to the centrosome and the spindle, deviating from the characteristics of Aurora-B to those of Aurora-A. The affinity of this mutant to INCENP and Survivin is lower than that of Aurora-BWT, although it still maintains a high level. This result suggests that there might be other residues responsible for the interaction between Aurora-B and its partners. Because Aurora-BN142G has a strong signal at the centromere and the midzone like Aurora-BWT, it is difficult to further analyze the different protein interactions between them by biochemical approaches. Thus, our experiments concentrated on understanding how Aurora-A is converted into Aurora-B-like kinase. In vertebrates, there is a distinction between Aurora-A and -B within this particular site, with Gs in Aurora-A and Ns in Aurora-B. However, in those organisms with only 1 Aurora, such as Saccharomyces cerevisiae and Saccharomyces pombe, whose Aurora works like Aurora-B (39–40), or Dictyostelium, whose Aurora has the property of both Aurora-A and -B (41), there are Gs instead of Ns, suggesting broader application of G in this site during evolution. Because a single amino acid change imparts Aurora-B-like behavior on Aurora-A and confers some Aurora-A-like properties on Aurora-B, we propose that our observations might be of evolutionary significance, suggesting that a new protein might be generated from its precursor by point mutation and evolutionary selection.
In conclusion, we demonstrate that a single amino acid change within the Aurora-A catalytic domain is sufficient to convert it into an Aurora-B-like kinase in terms of partner specificity and function in the cell. Through delicate differentiation in the local structure, generated by the presence of either Gly or Asn, Aurora-A and -B are matched with different sets of partner proteins, brought to different subcellular locations, and assigned distinct functions.
Materials and Methods
For additional information on materials and methods used, see SI Text.
Molecular Cloning, Protein Purification, and Antibody Preparation.
Wild-type Aurora-A, -B, Survivin, TPX2, and PRC1 were cloned by RT-PCR from HeLa cell lysate. Mutants of Aurora-A and -B were generated by point mutation PCR. GFP was cloned by PCR from pEGFP vector. For overexpression in HeLa cells, wild type and mutants of Aurora-A and -B were inserted into pEGFP-C. For expression of recombinant proteins in BL21 E. coli, Aurora-AWT, Aurora-AG198N, Aurora-BWT, Survivin, TPX2, and PRC1 were inserted into pET28a. Proteins were affinity purified using nickel beads (Qiagen). Recombinant Aurora-A (active, from Sf21 cells) and GST-INCENP826–919 were from Upstate Biotechnology. Antibodies to Aurora-A, TPX2, GFP, and PRC1 were raised in rabbits or mice. Antibodies to INCENP and Hec1 were from Abcam, antibody to Aurora-B from BD Biosciences, antibodies to Survivin and actin from Santa Cruz Biotechnology, and antibody to phospho-histone H3 (Ser-10) from Upstate Biotechnology.
Cell Line Generation and Cell Culture.
Tet-on cell lines were generated according to the supplier's instructions (plasmids and drugs from Invitrogen). HeLa cells were transfected with pcDNA6/TR, and cultured in the presence of 10 μg/mL blasticidin for 2 weeks. Positive clones were selected for further transfection of pcDNA4/TO-GFP-tagged Aurora-AWT, Aurora-AG198N, Aurora-BWT, or Aurora-BN142G and cultured in the presence of 200 μg/mL Zeocin for 2 more weeks. Again, positive clones were selected and cultured for use. GFP-tagged Aurora-AWT, Aurora-AG198N, Aurora-BWT, or Aurora-BN142G was expressed when 1 μg/mL tetracycline was added.
For immunofluorescence, cells were plated on coverslips 1 day before transfection by using a standard calcium phosphate transfection protocol and subjected to immunofluorescence microscopy 48 h later. For living-cell imaging, cells were plated in glass-bottomed culture dishes and observed 48 h after transfection. DNA was labeled with 100 ng/mL Hoechst 33342 for 30 min at 37 °C before observation. In time-lapse experiments, images were acquired every 5 min by using a Zeiss Axiovert 200M microscope and AxioVision image acquisition software.
Immunoprecipitation.
Fifteen microliters of Protein-A Sepharose beads (GE Healthcare) were washed 3 times with PBS, diluted into 500 μL and incubated with 5 μg of rabbit-anti-GFP antibody for 1 h at 4 °C on a rotator. Beads were then washed 3 times with lysis buffer, consisting of 0.5% Nonidet P-40, 20 mM Tris·HCl (pH 7.4), 500 mM NaCl, 0.5 mM EGTA, 10 mM NaF, 2 mM sodium vanadate, 10 mM β-glycerophosphate, 10 μg/mL leupeptin, 0.7 μg/mL pepstatin A, 1.5 μg/mL aprotinin, and 1 mM PMSF. Mitotic HeLa cells transfected with GFP-tagged Aurora-AWT, Aurora-AG198N, Aurora-BWT, or Aurora-BN142G were shaken off the culture dish 11 h after release from double thymidine block. For immunoprecipitation, the cells were lysed on ice for 30 min with lysis buffer. Lysates were centrifuged at 16,000 × g for 15 min, and supernatants were incubated with beads for 1 h at 4 °C on the rotator. After 6 washes with lysis buffer, the bound proteins were analyzed by Western blotting.
Immunofluorescence Microscopy.
Cells grown on coverslips were fixed with precooled methanol for 6 min on ice. After rehydration in TBS, the cells were incubated with the primary antibody overnight at 4 °C. Then they were washed 3 times in TBS and incubated with the secondary antibody for 45 min at room temperature. Coverslips were mounted onto slides by using Mowiol containing 1 μg/mL DAPI and analyzed under a Zeiss Axiovert 200M microscope. Images were captured with a Zeiss MRM CCD camera and Axiovision image acquisition software. The antibodies were diluted in 3% BSA-TBS before use: Hec1 (1:100; Abcam), Aurora-B (1:100; BD Biosciences), INCENP (1:1,000; Abcam), Survivin (1:100; Santa Cruz), PRC1 (1:100), Alexa Fluor 488 donkey anti-mouse (1:100; Invitrogen), Alexa Fluor 549 donkey anti-mouse (1:100; Invitrogen), and TRITC-conjugated anti-rabbit (1:100; Jackson ImmunoResearch).
RNA Interference.
pSUPER mammalian expression vector was used to direct the synthesis of siRNA-like transcripts. Oligonucleotides for suppressing INCENP, Aurora-B, and Survivin were synthesized as previously reported (42–44): INCENP, 5′-gatccccgaagagacggatttcttatttcaagagaataagaaatccgtctcttcttttta-3′ and 5′-agcttaaaaagaagagacggatttcttattctcttgaaataagaaatccgtctcttcggg-3′; Aurora-B, 5′-gatccccgagcctgtcaccccatctgttcaagagacagatggggtgacaggctctttttggaaa-3′ and 5′-agcttttccaaaaagagcctgtcaccccatctgtctcttgaacagatggggtgacaggctcggg-3′; and Survivin, 5′-gatccccgaggctggcttcatccactttcaagagaagtggatgaagccagcctcttttta-3′ and 5′-agcttaaaaagaggctggcttcatccacttctcttgaaagtggatgaagccagcctcggg-3′.
Oligonucleotides were annealed and ligated into the BglII/HindIII restriction sites of digested pSUPER vector (45).
Kinase Assays.
To measure the kinase activity of Aurora-AG198N, 1 μg of recombinant Aurora-AG198N was incubated with 1 μg of GST, GST-INCENP826–919 or Survivin for 20 min at 30 °C. Kinase assays were performed in 50 mM Tris·HCl (pH 7.4), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, and 100 μM ATP containing 3,000 Ci/mmol [γ-32P]ATP. Reactions were quenched with SDS sample buffer and analyzed by SDS/PAGE and autoradiography.
Supplementary Material
Acknowledgments.
We thank other members in our laboratory for helpful comments and Eleanor Erikson and Christine Tran for critical reading of this manuscript. This work was supported by National Natural Science Foundation of China Grants 30721064, 30225016, and 30330200, National Basic Research Program of China Grants 2004CB720003 and 2006CB0D0100, and National Key Scientific Program of China Grant 2007CB914502.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0900833106/DCSupplemental.
References
- 1.Taylor S, Peters JM. Polo and Aurora kinases: Lessons derived from chemical biology. Curr Opin Cell Biol. 2008;20:77–84. doi: 10.1016/j.ceb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- 2.Fu J, Bian M, Jiang Q, Zhang C. Roles of Aurora kinases in mitosis and tumorigenesis. Mol Cancer Res. 2007;5:1–10. doi: 10.1158/1541-7786.MCR-06-0208. [DOI] [PubMed] [Google Scholar]
- 3.Giet R, Petretti C, Prigent C. Aurora kinases, aneuploidy and cancer, a coincidence or a real link? Trends Cell Biol. 2005;15:241–250. doi: 10.1016/j.tcb.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Meraldi P, Honda R, Nigg EA. Aurora kinases link chromosome segregation and cell division to cancer susceptibility. Curr Opin Genet Dev. 2004;14:29–36. doi: 10.1016/j.gde.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 5.Keen N, Taylor S. Aurora-kinase inhibitors as anticancer agents. Nat Rev Cancer. 2004;4:927–936. doi: 10.1038/nrc1502. [DOI] [PubMed] [Google Scholar]
- 6.Carmena M, Earnshaw WC. The cellular geography of aurora kinases. Nat Rev Mol Cell Biol. 2003;4:842–854. doi: 10.1038/nrm1245. [DOI] [PubMed] [Google Scholar]
- 7.Andrews PD, Knatko E, Moore WJ, Swedlow JR. Mitotic mechanics: The auroras come into view. Curr Opin Cell Biol. 2003;15:672–683. doi: 10.1016/j.ceb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff JR, Plowman GD. The Aurora/Ipl1p kinase family: Regulators of chromosome segregation and cytokinesis. Trends Cell Biol. 1999;9:454–459. doi: 10.1016/s0962-8924(99)01658-x. [DOI] [PubMed] [Google Scholar]
- 9.Barr AR, Gergely F. Aurora-A: The maker and breaker of spindle poles. J Cell Sci. 2007;120:2987–2996. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- 10.Marumoto T, Zhang D, Saya H. Aurora-A—A guardian of poles. Nat Rev Cancer. 2005;5:42–50. doi: 10.1038/nrc1526. [DOI] [PubMed] [Google Scholar]
- 11.Dutertre S, Descamps S, Prigent C. On the role of aurora-A in centrosome function. Oncogene. 2002;21:6175–6183. doi: 10.1038/sj.onc.1205775. [DOI] [PubMed] [Google Scholar]
- 12.Vader G, Medema RH, Lens SM. The chromosomal passenger complex: Guiding Aurora-B through mitosis. J Cell Biol. 2006;173:833–837. doi: 10.1083/jcb.200604032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: Conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 14.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 15.Eyers PA, Erikson E, Chen LG, Maller JL. A novel mechanism for activation of the protein kinase Aurora A. Curr Biol. 2003;13:691–697. doi: 10.1016/s0960-9822(03)00166-0. [DOI] [PubMed] [Google Scholar]
- 16.Eyers PA, Maller JL. Regulation of Xenopus Aurora A activation by TPX2. J Biol Chem. 2004;279:9008–9015. doi: 10.1074/jbc.M312424200. [DOI] [PubMed] [Google Scholar]
- 17.Kufer TA, et al. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ozlü N, et al. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev Cell. 2005;9:237–248. doi: 10.1016/j.devcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Trieselmann N, Armstrong S, Rauw J, Wilde A. Ran modulates spindle assembly by regulating a subset of TPX2 and Kid activities including Aurora A activation. J Cell Sci. 2003;116:4791–4798. doi: 10.1242/jcs.00798. [DOI] [PubMed] [Google Scholar]
- 20.Tsai MY, et al. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat Cell Biol. 2003;5:242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- 21.Adams RR, et al. INCENP binds the Aurora-related kinase AIRK2 and is required to target it to chromosomes, the central spindle and cleavage furrow. Curr Biol. 2000;10:1075–1078. doi: 10.1016/s0960-9822(00)00673-4. [DOI] [PubMed] [Google Scholar]
- 22.Kaitna S, Mendoza M, Jantsch-Plunger V, Glotzer M. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- 23.Wheatley SP, Carvalho A, Vagnarelli P, Earnshaw WC. INCENP is required for proper targeting of Survivin to the centromeres and the anaphase spindle during mitosis. Curr Biol. 2001;11:886–890. doi: 10.1016/s0960-9822(01)00238-x. [DOI] [PubMed] [Google Scholar]
- 24.Bolton MA, et al. Aurora B kinase exists in a complex with survivin and INCENP and its kinase activity is stimulated by survivin binding and phosphorylation. Mol Biol Cell. 2002;13:3064–3077. doi: 10.1091/mbc.E02-02-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gassmann R, et al. Borealin: A novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol. 2004;166:179–191. doi: 10.1083/jcb.200404001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sampath SC, et al. The chromosomal passenger complex is required for chromatin-induced microtubule stabilization and spindle assembly. Cell. 2004;118:187–202. doi: 10.1016/j.cell.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Bishop JD, Schumacher JM. Phosphorylation of the carboxyl terminus of inner centromere protein (INCENP) by the Aurora B kinase stimulates Aurora B kinase activity. J Biol Chem. 2002;277:27577–27580. doi: 10.1074/jbc.C200307200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Honda R, Korner R, Nigg EA. Exploring the functional interactions between Aurora B, INCENP, and survivin in mitosis. Mol Biol Cell. 2003;14:3325–3341. doi: 10.1091/mbc.E02-11-0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayliss R, et al. Determinants for Aurora-A activation and Aurora-B discrimination by TPX2. Cell Cycle. 2004;3:404–407. [PubMed] [Google Scholar]
- 30.Eyers PA, Churchill ME, Maller JL. The Aurora A and Aurora B protein kinases: A single amino acid difference controls intrinsic activity and activation by TPX2. Cell Cycle. 2005;4:784–789. doi: 10.4161/cc.4.6.1693. [DOI] [PubMed] [Google Scholar]
- 31.Adams RR, Maiato H, Earnshaw WC, Carmena M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J Cell Biol. 2001;153:865–880. doi: 10.1083/jcb.153.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol. 2002;12:900–905. doi: 10.1016/s0960-9822(02)00887-4. [DOI] [PubMed] [Google Scholar]
- 33.Ditchfield C, et al. Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol. 2003;161:267–280. doi: 10.1083/jcb.200208091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hauf S, et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J Cell Biol. 2003;161:281–294. doi: 10.1083/jcb.200208092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wheatley SP, et al. Aurora-B phosphorylation in vitro identifies a residue of survivin that is essential for its localization and binding to inner centromere protein (INCENP) in vivo. J Biol Chem. 2004;279:5655–5660. doi: 10.1074/jbc.M311299200. [DOI] [PubMed] [Google Scholar]
- 36.Katayama H, et al. Aurora kinase-A regulates kinetochore/chromatin associated microtubule assembly in human cells. Cell Cycle. 2008;7:2691–2704. doi: 10.4161/cc.7.17.6460. [DOI] [PubMed] [Google Scholar]
- 37.Sessa F, et al. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol Cell. 2005;18:379–391. doi: 10.1016/j.molcel.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 38.Jeyaprakash AA, et al. Structure of a Survivin-Borealin-INCENP core complex reveals how chromosomal passengers travel together. Cell. 2007;131:271–285. doi: 10.1016/j.cell.2007.07.045. [DOI] [PubMed] [Google Scholar]
- 39.Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petersen J, et al. The S. pombe aurora-related kinase Ark1 associates with mitotic structures in a stage dependent manner and is required for chromosome segregation. J Cell Sci. 2001;114:4371–4384. doi: 10.1242/jcs.114.24.4371. [DOI] [PubMed] [Google Scholar]
- 41.Li H, et al. Dictyostelium Aurora kinase has properties of both Aurora A and Aurora B kinases. Eukaryot Cell. 2008;7:894–905. doi: 10.1128/EC.00422-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi W, Tang Z, Yu H. Phosphorylation- and polo-box-dependent binding of Plk1 to Bub1 is required for the kinetochore localization of Plk1. Mol Biol Cell. 2006;17:3705–3716. doi: 10.1091/mbc.E06-03-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lens SM, et al. Survivin is required for a sustained spindle checkpoint arrest in response to lack of tension. EMBO J. 2003;22:2934–2947. doi: 10.1093/emboj/cdg307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scrittori L, et al. A small C-terminal sequence of Aurora B is responsible for localization and function. Mol Biol Cell. 2005;16:292–305. doi: 10.1091/mbc.E04-06-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.