Historical perspectives and the underlying mechanisms of allostery have been recently reviewed (1–4), increasingly highlighting conformation selection and population shift (5). A classical example is transcription factor (TF) catabolite activator protein (CAP) activation by cAMP in response to extracellular stimuli (6). cAMP–CAP–DNA complex was the first TF with 3D structure (7), and it was postulated that cAMP binding causes structural rearrangements allowing CAP to bind DNA (6, 8). Twenty-eight years later, NMR structures have confirmed the allosteric transition paradigm; as reported in this issue of PNAS, Popovych et al. (9) found that without cAMP binding, the CAP apo form loses 3 helical turns and the DNA binding domain rotates, shifting from its DNA binding-ready state. Consistent with theoretical expectation, some mutations shifted the ensemble toward a DNA-compatible active state even without cAMP binding (9). Coiled-coil order-disorder transition coupled with cAMP binding permits efficient allo steric control; yet, it is not necessarily used in other structurally similar cAMP binding domains (10). Protein kinase A undergoes a kink transition sufficient to break the inactive protein kinase A (PKA) tetramer complex on cAMP binding (11). In the CAP/fumarate and nitrate reduction regulator (FNR) superfamily, the CooA (effector: CO) does not involve an order–disorder transition but dissimilative nitrate respiration regulator (DNR) (effector: NO) does (9).
Mechanistically, cAMP concentration in the nucleus can be low and CAP order–disorder transition can amplify allosteric signaling. Allostery can amplify catalysis and signal transduction (12) and intrinsic disorder can maximize (13) allosteric response. Popovych et al. (9) illustrate the order-disorder transition: both cAMP binding to wt-CAP and CAP mutations shift the landscape toward an ordered active state. In disordered states the barriers are low. They are easily overcome by mutational, ligand binding, or posttranslational modification perturbation events affecting the relative residue-residue (or solvent) interaction strengths, shifting a preexisting conformational ensemble (14). All such events are allosteric.
The allosteric order/disorder rationale is consistent with transcription factors and other cell-signaling proteins being disproportionately intrinsically unstructured. A similar mechanism was observed in the related cAMP response element-binding protein (CREB). Although CREB was initially identified as a cAMP pathway target, CREB is now known to be activated by hundreds extracellular stimuli. Many of these transmit through G protein-coupled receptors (GPCRs). CREB can be activated by KID (kinase-inducible domain) Ser-133 phosphorylation by PKA. This allows interaction with the CREB binding protein KIX domain. When unphosphorylated and unbound, KID is disordered. Cooperative folding and binding occur upon pKID–KIX interaction, forming 2 α-helices kinked near the phosphorylated site (15). KIX is an allosteric domain able to bind 2 other proteins cooperatively. Different partners binding at the second site can modulate the conformations and thus the affinity of pKID–KIX interaction, regulating the transactivation complex (16). Indeed, the magnitude of the CREB-dependent transcriptional response is determined by the binding strength between the KID and KIX domains.
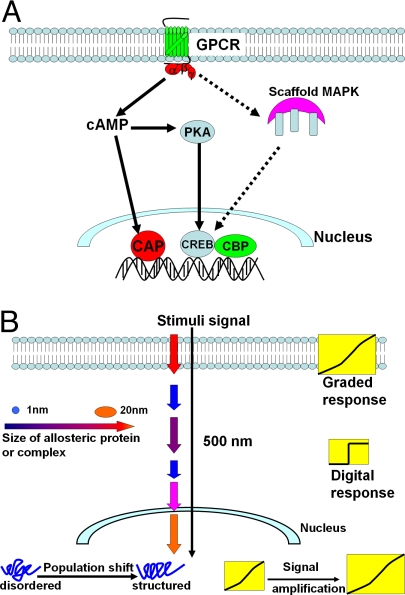
Although allostery is the focus of increasing attention, not much is known about allosteric mechanisms in cellular pathways. Previous studies focused on individual proteins or complexes revealing common features in allosteric regulation and catalysis; yet, allostery is not confined to individual protein systems; rather, allosteric mechanisms are components in the connected systems level signal transduction pathways. Consider allostery within the cellular signal transduction: GPCRs stimulated by hormones and neurotransmitters activate adenylyl cyclase. This membrane-bound enzyme catalyzes cAMP synthesis. cAMP then binds signaling proteins (here, CAP and PKA). Thus, GPCR–cAMP–CAP and GPCR–cAMP–PKA–CREB are 2 “short” cAMP-related signaling pathways (Fig. 1A). Both GPCR and PKA are classical examples of allosteric regulation (11, 17, 18). These pathways span the cell: from membrane-bound GPCR, to cytoplasmic PKA, to CAP or CREB-CBP in the nucleus.
Fig. 1.
Nanoscale allosteric signal transduction relay from the cell surface to the nucleus. (A) Allosteric proteins and cell signaling. The simplified GPCR-cAMP-CRP and GPCR-cAMP-PKA-CREB pathways represent short routes connecting extracellular stimuli to the nucleus via the cytoplasm. Cytoplasmic proteins often display an on/off transmission switch, like the PKA and scaffold MAPK systems. (B) Allosteric relay in signal transduction can be nanoscale, over hundreds of nanometers.
All dynamic proteins are allosteric (5) and the pure dynamics (19) in one domain can be coupled with conformation change in another domain as in the case of CAP (9). Thus, it is not surprising that catalytic and signal transduction proteins are allosterically regulated. When considering allosteric proteins as modules in signal transduction pathways extending from extracellular stimuli to the responding nuclear events, as in the GPCR to CAP (or CREB) cases, the global allosteric response is not confined to the proteins in their isolated states; rather, allosteric response is system-optimized. Thus, in single proteins an allosteric signal travels from the effector perturbation site to the response ligand binding site over nanoscale distances ranging from 1 to 20 nm; yet, in the cell, signals transmitted from the extracellular surface through the cytoplasm to the nucleus to activate (or repress) transcription are relayed through several proteins over much larger distances (Fig. 1B).
If allosteric regulation is not optimized solely for an individual protein but for a cell-spanning pathway, what are the advantages of allosteric regulation on the cellular scale? First, cross-membrane signaling has to be allosteric; allosteric regulation of a transmembrane receptor allows a graded signal response (Fig. 1B) to fluctuating extracellular stimuli. GPCRs are examples of such a graded response. The ste5 scaffold MAPK system localized near the G protein similarly allows graded signal transduction, with graded response to pheromone level increase (or decrease) (20, 21). Second, the extremely crowded cytoplasmic environment requires an ultrasensitive on/off digital response. cAMP-activated PKA presents such an on/off transition. The ste5 scaffold MAPK system also displays an on/off switch (21). Third, signals reaching the nucleus after traversal over a long distance can be weakened; and optimized transactivation/transcription could require amplification. Intrinsic protein disorder, characteristic of transcription factors, is well-suited for such amplification: the low barriers between the conformers in the ensemble are easily overcome, shifting the equilibrium toward an energetically less-favored, although active binding-ready preexisting state (14). The cAMP-CAP and CREB-CBP constitute such cases. Finally, above we addressed “outside-in” signals traveling from the extramembraneous cell surface to the nucleus; yet, it behooves us to recall that similar considerations apply for “inside-out” signaling. Integrins provide an excellent example; these transmembrane receptors allow both outside-in and inside-out allosteric signaling (22). Their elongated heterodimeric membrane-crossing α- and β-chains (with the extracellular ligand binding domain and cytoplasmic domain ≈20 nm apart in the fully extended active state) present large interdomain, hinge-mediated conformational changes transmitting bidirectional signals (22).
Signal transduction through the plasma membrane allows graded response to stimuli.
In the current era, with accumulating systems and detailed structural information (9), concerted cell-level allosteric regulation can be addressed. Allosteric signal-transmitting proteins span nanoscale sizes and extracellular stimuli are tandemly relayed to the nucleus via the cytoplasm through allosteric proteins over very long distances, reaching tens and even hundreds of nanometers. Allosteric relay is advantageous: signal transduction through the plasma membrane allows graded response to stimuli; in the crowded cytoplasm it allows sensitive digital on/off switch; and in the nucleus—in particular, when intrinsically unstructured proteins are involved—large allosteric population shifts amplify weak signals while retaining graded transcriptional activity.
Acknowledgments.
This was supported by the Intramural Research Program of the Center for Cancer Research, National Cancer Institute, National Institutes of Health Contract N01-CO-12400.
Footnotes
The authors declare no conflict of interest.
See companion article on page 6927.
References
- 1.Changeux JP, Edelstein SJ. Allosteric mechanisms of signal transduction. Science. 2005;308:1424–1428. doi: 10.1126/science.1108595. [DOI] [PubMed] [Google Scholar]
- 2.Cui Q, Karplus M. Allostery and cooperativity revisited. Protein Sci. 2008;17:1295–1307. doi: 10.1110/ps.03259908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goodey NM, Benkovic SJ. Allosteric regulation and catalysis emerge via a common route. Nat Chem Biol. 2008;4:474–482. doi: 10.1038/nchembio.98. [DOI] [PubMed] [Google Scholar]
- 4.Tsai CJ, Del Sol A, Nussinov R. Protein allostery, signal transmission and dynamics: A classification scheme of allosteric mechanisms. Mol Biosyst. 2009;5:207–216. doi: 10.1039/b819720b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunasekaran K, Ma B, Nussinov R. Is allostery an intrinsic property of all dynamic proteins? Proteins. 2004;57:433–443. doi: 10.1002/prot.20232. [DOI] [PubMed] [Google Scholar]
- 6.Passner JM, Schultz SC, Steitz TA. Modeling the cAMP-induced allosteric transition using the crystal structure of CAP-cAMP at 2.1 A resolution. J Mol Biol. 2000;304:847–859. doi: 10.1006/jmbi.2000.4231. [DOI] [PubMed] [Google Scholar]
- 7.McKay DB, Steitz TA. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981;290:744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- 8.Li L, et al. A computational investigation of allostery in the catabolite activator protein. J Am Chem Soc. 2007;129:15668–15676. doi: 10.1021/ja076046a. [DOI] [PubMed] [Google Scholar]
- 9.Popovych N, et al. Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc Natl Acad Sci USA. 2009;106:6927–6932. doi: 10.1073/pnas.0900595106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman HM, et al. The cAMP binding domain: An ancient signaling module. Proc Natl Acad Sci USA. 2005;102:45–50. doi: 10.1073/pnas.0408579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taylor SS, et al. Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design. Biochim Biophys Acta. 2008;1784:16–26. doi: 10.1016/j.bbapap.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu L, Anslyn EV. Signal amplification by allosteric catalysis. Angew Chem Int Ed Engl. 2006;45:1190–1196. doi: 10.1002/anie.200501476. [DOI] [PubMed] [Google Scholar]
- 13.Hilser VJ, Thompson E.B. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci USA. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma B, Nussinov R. Regulating highly dynamic unstructured proteins and their coding mRNAs. Genome Biol. 2009;10:204. doi: 10.1186/gb-2009-10-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sugase K, Dyson HJ, Wright PE. Mechanism of coupled folding and binding of an intrinsically disordered protein. Nature. 2007;447:1021–1025. doi: 10.1038/nature05858. [DOI] [PubMed] [Google Scholar]
- 16.Goto NK, et al. Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP). The mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J Biol Chem. 2002;277:43168–43174. doi: 10.1074/jbc.M207660200. [DOI] [PubMed] [Google Scholar]
- 17.Kobilka BK, Deupi X. Conformational complexity of G-protein-coupled receptors. Trends Pharmacol Sci. 2007;28:397–406. doi: 10.1016/j.tips.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 18.Suel GM, et al. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- 19.Tsai CJ, del Sol A, Nussiniv R. Allostery: Absence of a change in shape does not imply that allostery is not at play. J Mol Biol. 2008;378:1–10. doi: 10.1016/j.jmb.2008.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi S, Pryciak PM. Membrane localization of scaffold proteins promotes graded signaling in the yeast MAP kinase cascade. Curr Biol. 2008;18:1184–1191. doi: 10.1016/j.cub.2008.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dohlman HG. A scaffold makes the switch. Sci Signal. 2008;1:pe46. doi: 10.1126/scisignal.142pe46. [DOI] [PubMed] [Google Scholar]
- 22.Hynes RO. Integrins: Bidirectional, allosteric signaling machines. Cell. 2002;110:673–687. doi: 10.1016/s0092-8674(02)00971-6. [DOI] [PubMed] [Google Scholar]