Abstract
We tested the opposing views concerning evolution of genes of the innate immune system that (i) being evolutionary ancient, the system may have been highly optimized by natural selection and therefore should be under purifying selection, and (ii) the system may be plastic and continuing to evolve under balancing selection. We have resequenced 12 important innate-immunity genes (CAMP, DEFA4, DEFA5, DEFA6, DEFB1, MBL2, and TLRs 1, 2, 4, 5, 6, and 9) in healthy volunteers (n = 171) recruited from a region of India with high microbial load. We have compared these data with those of European-Americans (EUR) and African-Americans (AFR). We have found that most of the human haplotypes are many mutational steps away from the ancestral (chimpanzee) haplotypes, indicating that humans may have had to adapt to new pathogens. The haplotype structures in India are significantly different from those of EUR and AFR populations, indicating local adaptation to pathogens. In these genes, there is (i) generally an excess of rare variants, (ii) high, but variable, degrees of extended haplotype homozygosity, (iii) low tolerance to nonsynonymous changes, (iv) essentially one or a few high-frequency haplotypes, with star-like phylogenies of other infrequent haplotypes radiating from the modal haplotypes. Purifying selection is the most parsimonious explanation operating on these innate immunity genes. This genetic surveillance system recognizes motifs in pathogens that are perhaps conserved across a broad range of pathogens. Hence, functional constraints are imposed on mutations that diminish the ablility of these proteins to detect pathogens.
Keywords: extended haplotype homozygosity, haplotype networks, neutrality tests, resequencing
The innate immune system developed before the separation of the vertebrates and invertebrates (1). Invertebrates and jawless fish depend solely on the innate immune system. The adaptive immune system depends on the innate immune system because of the need for antigen presentation to the adaptive system, which in turn is critically dependent on recognition of microbial molecules by the innate system (2). The Toll-like receptor (TLR) family of genes is vital to the innate immune system because these genes can recognize the pathogen associated molecular patterns (PAMPs) and are also known to be key regulators of the adaptive immune responses in humans and other mammals (3, 4). Another important family of genes that regulate innate immune responses is the defensins. Epithelial cells in humans secrete antimicrobial peptides that are encoded by the defensin genes (organized in α and β gene clusters). These cationic peptides exhibit a broad spectrum of activity against Gram-positive and Gram-negative bacteria, fungal species and viruses (5), by interacting with negatively charged molecules on the surface of pathogens and permeating their membranes.
There are also other genes that play important roles in the innate immune system. Cathelicidins are small (12–100 aa) cationic peptides that possess broad-spectrum antimicrobial activity, and share features with defensins (6). Although humans and mice each possess a single cathelicidin, other mammalian species, such as cattle and pigs, express many different cathelicidins (7). Mannose-binding lectin (MBL), a member of the collectin family of proteins, binds a broad range of microorganisms and activates the lectin-complement pathway (8). Diversification of the genes of the innate immune system have taken place during evolution possibly in response to the diversification of microbes, especially pathogenic microbes. Gene families, such as TLRs, evolved by gene duplication and individual members of these families have evolved different but related functions, possibly to protect the host against a larger set of diverse pathogens. Individual members of these gene families also exhibit high levels of polymorphism, which is consistent with Haldane's (9) prediction that maintenance of polymorphisms in genes governing host-pathogen interaction is driven by rapid rates of microbial evolution. Indeed, evidence for overdominant selection (heterozygote advantage) has been documented at the major histocompatibility complex class I loci, in which the rate of nonsynonymous (amino acid altering) nucleotide substitutions have been found to be significantly greater than synonymous substitutions in the antigen recognition site (10). With respect to the genes of the innate immune system, however, there is debate whether these genes are continuing to evolve or whether being evolutionarily ancient they have been highly optimized by natural selection (11). Specific polymorphic variants in innate immunity genes have been found to be associated with human diseases (12–15) and some investigations have been carried out on the mode and tempo of evolution of some genes of the innate immune system (16–19).
We have recently documented extensive variation that included the discovery of 259 novel variants, in 12 innate immunity genes (cathelicidin antimicrobial peptide, CAMP; α-defensins, DEFA4, DEFA5 and DEFA6; β-defensin, DEFB1; mannose binding lectin, MBL2; and TLRs 1, 2, 4, 5, 6 and 9) in an Indian population resident in a region with high load of pathogens and have shown that the haplotype structures of these genes differ markedly in this population compared with the HapMap populations (20). Parenthetically, we note that although genes involved in innate immunity continue to be identified, the 12 genes chosen by us are some of the earliest identified innate immunity genes for which there was extensive functional data and no copy number variation in the human genome. The geographic distribution of haplotype frequencies in the TLR4 gene correlates well with the prevalence of various infectious diseases (18). These observations indicate that our innateimmunity genes may have been modified by natural selection through pressures caused by infectious agents. In this study, we have sought to assess the nature and extent of selective forces in shaping the contemporary genetic diversity of these genes. We have used data based on resequencing rather than polymorphism genotyping to detect signatures of natural selection because, as has been correctly emphasized (21), detection of natural selection is primarily based on extraction of properties of the allele frequency spectra of genes; only resequencing permits discovery of the full allele frequency spectrum of a gene, rare alleles included. Further, the standard statistical tests of neutrality (e.g., Tajima's D) are applicable to sequence data, and not to genotype data on a set of preascertained SNPs. In particular, we sought to test whether overdominant selection operates on the TLR genes, because TLR proteins contain leucine rich repeat (LRR) domains (characterized by a segment LxxLxLxxNxL, in which “L” is Leu, Ile, Val, or Phe and “N” is Asn, Thr, Ser, or Cys and “x” is any amino acid) that are responsible for molecular recognition (22). Similar to the antigen recognition site of the MHC locus, individuals who are heterozygous at the variant sites in these LRR coding regions may be capable of expressing various types of microbial recognition peptides and will therefore be expected to enjoy a selective advantage in a population exposed to a diverse array of microbial pathogens.
Results
Tests of Selective Neutrality Reveal Purifying Selection Operating on CAMP, TLR2, TLR4, and TLR9 Genes.
Table 1 provides results of tests of selective neutrality, using 4 different statistics, for the 12 innate immunity genes. CAMP showed statistically significant deviation from neutrality by all 4 statistics. Deviation from neutrality was not detected by most of the statistics for the defensin genes or for MBL2. For TLR genes, significant deviations were detected by 3 of the 4 statistics [D*, F*, and FS, but not D; D is known to be statistically less efficient for detecting selection in the presence of background selection (23)] for TLR2, TLR4 and TLR9, but not for TLR1, TLR5 or TLR6. For all of the 4 genes that showed significant deviations from neutrality, the values of the test statistics were negative, indicative of purifying selection. Interestingly, none of these genes show statistically significant evidence of selection in the European-American (EUR) or the African-American (AFR) populations (Table 1). This indicates that local selection may be operating in regions with a high microbial load, although haplotype diversity values are similar for these genes among EUR and AFR and the present study population (Table 1). This scenario is also consistent with local selective sweep, although this is unlikely in view of the results presented later. Contrary to our expectations, the TLR genes do not show signatures of overdominant selection. This issue is examined in greater detail later.
Table 1.
Values of haplotype diversity and statistics for testing departures from neutrality pertaining to innate immunity genes in three populations
| Population | Gene | No. of haplotypes | Haplotype diversity |
Tajima's D |
Fu and Li's D* |
Fu and Li's F* |
Fu's Fs |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Estimate | Variance | Value | P | Value | P | Value | P | Value | P | |||
| Indian (this study) | CAMP | 8 | 0.108 | 0.0005 | −1.79 | < 0.05 | −2.91 | < 0.05 | −3.02 | < 0.02 | −12.28 | < 0.05 |
| DEFA4 | 31 | 0.816 | 0.0001 | −0.07 | > 0.10 | −1.01 | > 0.10 | −0.75 | > 0.10 | −7.40 | < 0.05 | |
| DEFA5 | 21 | 0.699 | 0.0004 | −1.26 | > 0.10 | −1.16 | > 0.10 | −1.45 | > 0.10 | −10.27 | < 0.05 | |
| DEFA6 | 37 | 0.902 | 0.0001 | −0.10 | > 0.10 | −0.92 | > 0.10 | −0.71 | > 0.10 | −16.88 | < 0.05 | |
| DEFB1 | 106 | 0.922 | 0.0001 | 0.00 | > 0.10 | −1.53 | > 0.10 | −0.90 | > 0.10 | −22.54 | < 0.05 | |
| MBL2 | 46 | 0.879 | 0.0001 | 0.40 | > 0.10 | −2.12 | 0.05 < P < 0.10 | −1.13 | > 0.10 | −2.74 | > 0.10 | |
| TLR1 | 45 | 0.757 | 0.0007 | −0.37 | > 0.10 | −1.67 | > 0.10 | −1.29 | > 0.10 | −4.87 | > 0.10 | |
| TLR2 | 34 | 0.659 | 0.0007 | −1.24 | > 0.10 | −2.46 | < 0.05 | −2.35 | < 0.05 | −22.34 | < 0.05 | |
| TLR4 | 58 | 0.78 | 0.0005 | −1.23 | > 0.10 | −3.50 | < 0.02 | −2.98 | < 0.02 | −34.49 | < 0.05 | |
| TLR5 | 35 | 0.848 | 0.0001 | 0.64 | > 0.10 | −0.77 | > 0.10 | −0.16 | > 0.10 | −1.07 | > 0.10 | |
| TLR6 | 34 | 0.833 | 0.0002 | 0.47 | > 0.10 | −0.30 | > 0.10 | 0.07 | > 0.10 | 1.10 | > 0.10 | |
| TLR9 | 30 | 0.719 | 0.0004 | −1.28 | > 0.10 | −2.66 | < 0.05 | −2.51 | < 0.05 | −15.55 | < 0.05 | |
| European-American | DEFB1 | 17 | 0.875 | 0.0012 | 1.58 | > 0.10 | 0.98 | > 0.10 | 1.43 | > 0.10 | 9.48 | > 0.10 |
| TLR1 | 11 | 0.724 | 0.0027 | −0.50 | > 0.10 | 1.20 | > 0.10 | 0.71 | > 0.10 | 2.60 | > 0.10 | |
| TLR2 | 10 | 0.774 | 0.0021 | −0.44 | > 0.10 | −1.37 | > 0.10 | −1.26 | > 0.10 | −1.85 | > 0.10 | |
| TLR4 | 16 | 0.875 | 0.0009 | −0.26 | > 0.10 | 0.53 | > 0.10 | 0.30 | > 0.10 | −0.17 | > 0.10 | |
| TLR5 | 22 | 0.949 | 0.0002 | 0.60 | > 0.10 | 1.00 | > 0.10 | 1.02 | > 0.10 | 0.90 | > 0.10 | |
| TLR6 | 12 | 0.756 | 0.0026 | 1.58 | > 0.10 | 1.12 | > 0.10 | 1.52 | 0.05 < P < 0.10 | 2.66 | > 0.10 | |
| TLR9 | 11 | 0.762 | 0.0015 | −0.64 | > 0.10 | −2.23 | 0.05 < P < 0.10 | −2.01 | 0.05 < P < 0.10 | −2.65 | > 0.10 | |
| African-American | DEFB1 | 23 | 0.949 | 0.0004 | −0.28 | > 0.10 | 0.38 | > 0.10 | 0.17 | > 0.10 | 1.45 | > 0.10 |
| TLR1 | 14 | 0.878 | 0.0024 | −1.28 | > 0.10 | −2.14 | 0.05 < P < 0.10 | −2.20 | 0.05 < P < 0.10 | −1.55 | > 0.10 | |
| TLR2 | 18 | 0.917 | 0.0003 | −0.96 | > 0.10 | −2.32 | 0.05 < P < 0.10 | −2.20 | 0.05 < P < 0.10 | −8.16 | > 0.10 | |
| TLR4 | 30 | 0.969 | 0.0002 | −0.68 | > 0.10 | −0.37 | > 0.10 | −0.57 | > 0.10 | −6.31 | > 0.10 | |
| TLR5 | 28 | 0.946 | 0.0006 | −0.78 | > 0.10 | −1.35 | > 0.10 | −1.36 | > 0.10 | −0.77 | > 0.10 | |
| TLR6 | 18 | 0.925 | 0.0003 | −0.22 | > 0.10 | 0.21 | > 0.10 | 0.07 | > 0.10 | −0.12 | > 0.10 | |
| TLR9 | 11 | 0.755 | 0.0025 | −0.22 | > 0.10 | −0.13 | > 0.10 | −0.19 | > 0.10 | −1.84 | > 0.10 | |
Many Innate Immunity Genes Show Extended Homozygosity Compared with Neutral Expectations.
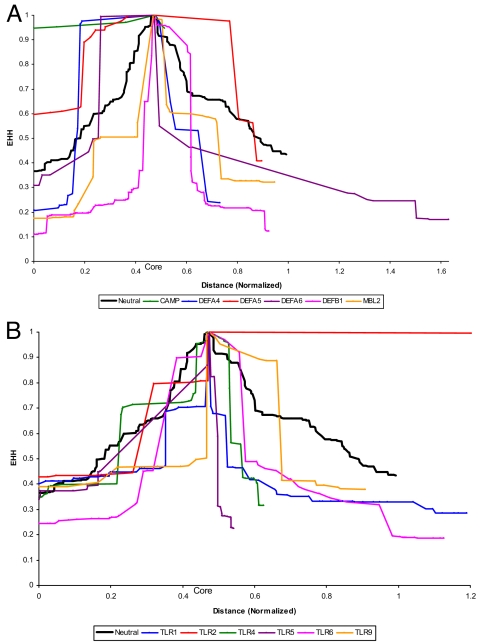
For each gene, haplotypes were reconstructed and their frequencies estimated using PHASE (24) (see Table S1). For CAMP, DEFA5, TLR1, TLR2, TLR4 and TLR9, a small number of haplotypes are in high frequencies; newly arising haplotypes have possibly been eliminated by purifying selection. Therefore, compared with neutral expectations these genes should show higher levels of homozygosity in an extended genomic region around a core region of the dominant (that is, most frequent) haplotype (25). This analysis was performed for each gene. The results are presented in Fig. 1 and compared with neutrality expectations. Fig. 1 shows that for CAMP, DEFA4, DEFA5, DEFA6, TLR2, TLR4 and TLR9 there is evidence of extended homozygosity; which is expected if the dominant haplotypes have risen to high frequencies under the impact of natural selection. The remaining genes do not show any such evidence. It is noteworthy, that except for DEFA4 and DEFA5, the remaining 4 genes showed significant evidence of purifying selection (Table 1). For both DEFA4 and DEFA5, even though 3 of the 4 tests of neutrality were not statistically significant, the values of the statistics were negative for all of the 4 tests, consistent with purifying selection.
Fig. 1.
Plots of extended haplotype homozygosity for CAMP, Defensin (A) and MBL2 Toll-like Receptor (B) genes.
Haplotype Networks also Reveal Purifying Selection for Many Innate Immunity Genes.
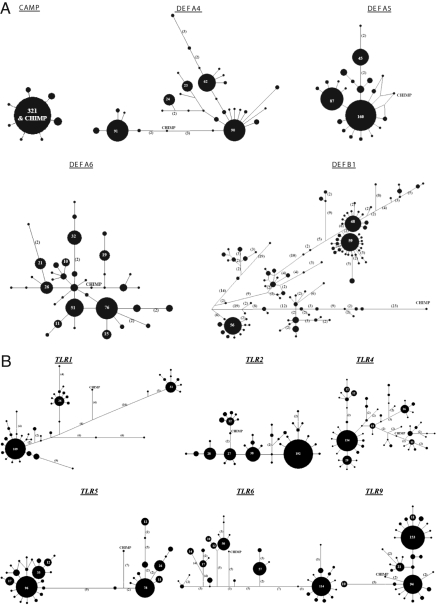
Under a population expansion model, a star-like phylogeny of haplotypes is expected (26, 27). Balancing selection, however, is expected to retain multiple lineages for a long time, resulting in a network in which there are some high-frequency clusters and some small-frequency clusters with long branches (26). Strong purifying selection is expected to retain only a single haplotype, but under weak purifying selection one may expect a small number of haplotypes with moderate frequencies, separated by short branches. We constructed median-joining networks (28) of haplotypes depicting their phylogenetic relationships. For CAMP, essentially there was 1 haplotype (frequency = 94.41%), which was same as the chimpanzee haplotype; the remaining 7 haplotypes were sporadic—observed in only a few individuals—and each was separated by 1 mutation from the dominant haplotype (Fig. 2A). This is indicative of a strong purifying selection operating on CAMP. Haplotype networks for the defensin genes (DEFA4, DEFA5, DEFA6 and DEFB1) essentially show star-like phylogenies (Fig. 2A) consistent with expectations under a population expansion model. However, among these the networks for DEFA4 and DEFB1 reveal some features that are more expected under balancing selection, than under purifying selection. DEFA4 has 2 high frequency (26.47–26.76%) haplotypes that are almost equidistant from the ancestral (chimpanzee) haplotype. DEFB1, however, has 3 high frequency haplotypes (14.04–17.25%) separated by long branches with one another; all these haplotypes are also very distant from the chimpanzee haplotype.
Fig. 2.
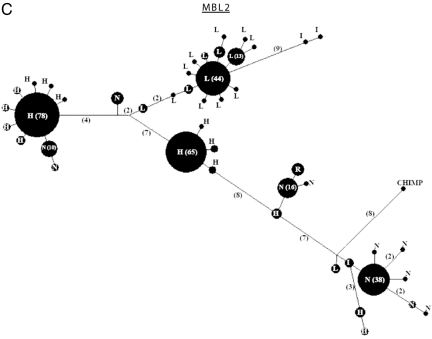
Median-joining networks of haplotypes for CAMP and Defensin genes (A), Toll- like Receptor genes (B), and MBL2 (C). The position of the ancestral (chimpanzee) haplotype is also marked on each network The nodes are proportional to the observed frequencies of the haplotypes; values are marked on nodes that correspond to haplotype frequencies exceeding 10. The numbers on the edges connecting the nodes indicate the number of mutations separating the haplotypes of the corresponding nodes, if the number of mutations exceeds 1.
Haplotype networks of the TLR genes, except TLR5, are consistent with purifying selection, each with one or a few modal haplotypes separated by short branches (Fig. 2B). The chimpanzee haplotype is separated by many mutational steps from the modal human haplotype for most of the TLR genes.
Haplotypes based on some specific SNPs in MBL2 are known to be associated with MBL serum concentration variability (29). MBL serum concentration influences susceptibility to and clinical course of many infectious and chronic diseases (30). We have shown (20) that, although the frequencies of haplotypes in our study population associated with high (53%) and deficient (20%) levels of MBL serum concentration are comparable to those observed worldwide (49% and 22%), the frequencies of intermediate (1.5%) and low (24%) haplotypes are significantly different from the global frequencies (15% and 13%) (17). The significantly higher frequency of the low-secretor haplotype (and, consequently, a lower frequency of intermediate-secretor haplotypes) is noteworthy. The haplotype network for MBL2 (Fig. 2C), the nodes of which have been identified with secretor status, shows an interesting pattern. The ancestral (chimpanzee) haplotype is closer (8 mutational steps away) to the modal haplotype associated with low MBL secretion. There are 2 high-frequency nodes, separated by a long branch, each of which is associated with high MBL secretion. Local star-like phylogenies appear around all of the high-frequency nodes irrespective of MBL secretor status. These features are indicative of balancing selection, although the test for neutrality was not rejected by any of the statistics (Table 1).
Pattern of Nonsynonymous Versus Synonymous Changes Reveal Purifying Selection and Provide No Evidence of Overdominant Selection on TLR Genes.
For all innate immunity genes, the rate of synonymous change is higher than nonsynonymous change (Table 2), as expected under purifying selection. We have also computed these ratios, using a sliding window approach (31), but did not find any enhanced rate of nonsynonymous, compared with synonymous, substitutions in any specific regions of the genes, including the LRR domains of the TLRs (see Fig. S1). Thus, the expectation that overdominant selection may be operating on the TLR genes is not supported by our data.
Table 2.
Rates of nonsynonymous (dN) and synonymous (dS) substitutions per site and their ratios for innate immunity genes
| Gene* | dN | dS | dN/dS |
|---|---|---|---|
| CAMP | 0.068 | 0.092 | 0.737 |
| DEFA4 | 0.005 | 0.049 | 0.098 |
| DEFA5 | 0.024 | 0.032 | 0.800 |
| DEFA6 | 0.005 | 0.043 | 0.110 |
| DEFB1 | 0.000 | 0.021 | 0.003 |
| MBL2 | 0.004 | 0.012 | 0.341 |
| TLR1 | 0.006 | 0.011 | 0.609 |
| TLR2 | 0.002 | 0.011 | 0.200 |
| TLR4 | 0.002 | 0.006 | 0.409 |
| TLR6 | 0.008 | 0.028 | 0.272 |
| TLR9 | 0.004 | 0.018 | 0.227 |
*Chimpanzee sequence for TLR5 is unavailable.
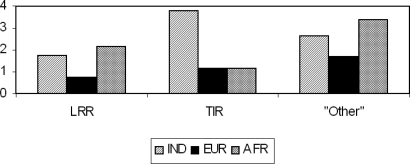
It is noteworthy that the rate of nonsynonymous changes (that is, number of nonsynonymous changes per kilobase) in the TLR genes is the lowest in the EUR population (Fig. 3). In the present study population, this rate is the highest in the TIR domain (the cytoplasmic Toll/Interleukin-1 Receptor (TIR) signaling domain that is instrumental in inducing a signaling cascade upon recognition of specific ligands by TLRs) compared with the remaining regions. The TIR domain in the TLR genes may, therefore, be under stronger impact of natural selection than other regions.
Fig. 3.
Rates per kilobase of nonsynonymous changes in the LRR and TIR domains and in other regions of the TLR genes among Indians, European-Americans, and African-Americans.
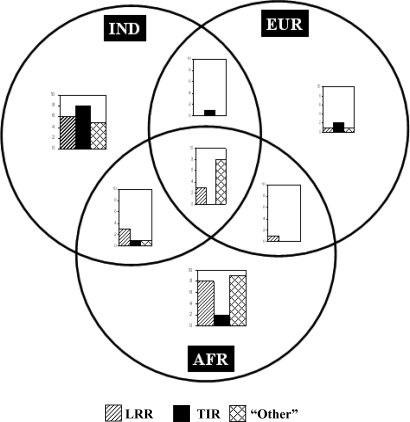
Interestingly, there are no nonsynonymous variants in the TIR domain of the TLR genes that are shared among the Indian, European-American and African-American populations (Fig. 4). The shared nonsynonymous variants are all in the LRR domain or in other regions of these genes. The present study population harbors the maximum number of nonsynonymous variants in the TIR domain of the TLRs; most of these variants are specific to the study population, that is, are unshared with the European-Americans or African-Americans (Fig. 4).
Fig. 4.
Numbers of shared and unshared nonsynonymous variants among Indian, European-American, and African-American populations in different regions of the TLR genes.
Discussion
We have examined the opposing views (11) concerning evolution of genes of the innate immune system that (i) being evolutionary ancient, the system may have been highly optimized by natural selection and therefore should have low tolerance to changes (purifying selection), and (ii) the system may be plastic and continuing to evolve with low selection pressure (balancing selection). We have sampled a large number of residents of a socio-economically depressed region of India, who live in unhygienic conditions and are therefore exposed to a high load of pathogens as evidenced by recurrent outbreaks of typhoid and cholera. In each sampled individual, we have resequenced these 12 genes and have cataloged extensive genomic variation (20). For drawing appropriate inferences, we have compared our data with those available on many of these genes for the European-American and African-American populations. Additionally, for evolutionary calibration of these data on humans, we have used the chimpanzee ancestral sequence.
We find that most of the genes studied by us have been influenced by purifying selection, even though the strengths of the signatures of purifying selection are variable. For most genes, only 1 or 2 haplotypes are present in high frequencies; evolutionary variants that have arisen to create new haplotypes are either eliminated or are present at very low frequencies, being perhaps transient to being eliminated. Nonsynonymous variations in these genes are not tolerated, as indicated by dN/dS ratios being <1, that have been found to be 1.31 and 1.66, respectively, for exons on human chromosomes 21 and 22 (32). Interestingly, most of the modal haplotypes are very dissimilar to the ancestral (chimpanzee) haplotype, perhaps indicating that humans have had to adapt to a different set of pathogens since divergence from their nearest common ancestor ≈5 million years ago. Contrary to our expectation stemming from past studies on MHC genes (10) that overdominant selection would operate, particularly in the LRR domains of the TLR genes that are important for pathogen recognition, we find no compelling evidence to support our expectation. However, it is interesting that compared with the European-American and African-American populations, the Indian population harbors a large number of unique nonsynonymous variants in the LRR and TIR domains of the TLR genes. Perhaps this is a reflection of the greater diversity of pathogens to which the Indian population is exposed, particularly in the region from where individuals were recruited into this study.
A recent study (19) claims that balancing selection is the main force operating on the innate immune system. We note that only 4 genes—TLR1, TLR4, TLR6 and TLR9—are common between our study and that of Ferrer-Admetlla et al. (19). As stated by the authors themselves, Ferrer-Admetlla et al. (19) did not find a conclusive statistical evidence of excess of intermediate variants in TLR1, TLR4 and TLR6, as expected under balancing selection. In fact, for TLR9 they have found an excess of rare variants, which is what is expected under purifying selection. Similarly, another study (17) has inferred that globally the variation in MBL2 conforms to neutrality, whereas our data indicate that balancing selection is the most parsimonious explanation. Although these inferences may seem apparently contradictory, geographical variation in selection regimes is possible. Although low MBL concentration is known to be clinically disadvantageous (30), the high frequency of the haplotypes associated with low MBL concentration in our study population resident in an area with a high load of pathogens may be indicative of selective advantage being conferred on the carriers of these haplotypes (33), resulting in balancing selection acting locally.
Our major result that purifying selection operates on genes of the innate immune system is in striking contrast with previous findings on genes of the adaptive immune system, such as genes of the major histocompatibility complex or Ig genes. The adaptive immunity genes are under either balancing or positive selection (10, 40). A simple explanation of our finding of purifying selection operating on the genes of the innate immune system is that this genetic surveillance system recognizes motifs in pathogens that are conserved across a broad range of pathogens. Therefore, functional constraints are imposed on mutations that diminish the ability of innate immunity proteins to detect pathogens. Thus, this system in the host does not coevolve with the mutations in genomes of pathogens. We note that purifying selection has also been detected in innate immunity genes in Drosophila melanogaster (41) and Daphnia (42) and in some genes of the TLR family in humans (43, 44).
In summary, our study has revealed that (i) although there is considerable genetic and haplotypic diversity in the innate immune system, one or a few haplotypes are modal, indicating a high degree of optimization of the innate immune system (11); (ii) the signatures of selection on these genes are variable, but purifying selection is the most dominant mode of selection, indicating that there is low tolerance to nonsynonymous changes; and (iii) there may be local variations in the nature of selection that is perhaps modulated by local differences in pathogen diversities and loads, implying that the extent of optimization of the innate immune system can be local and not necessarily global.
Materials and Methods
Study Populations and Participants.
Unrelated, healthy (ascertained to have not suffered from any major infection during the last 6 months or were not suffering from any chronic disease) individuals (n = 171), of both genders and of ages 12 years or older, were recruited into this study. All were residents of an economically depressed area of Kolkata (India), with poor hygienic conditions. Blood samples were collected from them by venipuncture with voluntary, informed and written consent, after obtaining institutional ethical approval.
Collation of Data from Public-Domain Databases.
For the purpose of comparison, we have downloaded sequence data available on 7 (DEFB1, TLR1, TLR2, TLR4, TLR5, TLR6 and TLR9) of the 12 genes for European-American (n = 23) and African-American (n = 24) populations from http://innateimmunity.net (34). For evolutionary calibration, the chimpanzee reference sequence for each gene was obtained from the National Center for Biotechnology Information database (www.ncbi.nlm.nih.gov/).
DNA Analysis.
From each blood sample collected, DNA was isolated using Qiagen columns, using the manufacturer's protocol. Double-pass DNA resequencing of these genes was carried out. (See Ref. 20 for details.) Analyses of sequence chromatograms and genotype calls were carried out using SeqScape v2.5 (Applied Biosystems) and PolyPhred (http://droog.mbt.washington.edu/PolyPhred.html) software packages. Coding regions were translated using DNASTAR and BioEdit packages.
Statistical Analysis.
Estimation of allele frequencies and tests for Hardy-Weinberg equilibrium were carried out using MAXLIK (35). Haplotype identification and estimation of haplotype frequencies were done using PHASE for Windows, version 2.1 (www.stat.washington.edu/stephens) (24). Sequences were aligned using CLUSTALW (www.ebi.ac.uk/clustalw). Statistics for evaluating departures of allele frequency spectra from neutrality (Tajima's D, Fu and Li's D* and F*, and Fu's F values) were computed using DnaSP, version 4.10 (36). Coalescent simulations, using DnaSP were carried out to test the statistical significance of Fu's FS. DnaSP was also used for calculation of haplotype diversity. For each gene, extended homozygosity of the most frequent haplotype was calculated around a “core” using the method suggested by Sabeti et al. (25) The “core” was taken to be a centrally-located SNP within the gene and with low heterozygosity. A random sample of 5000 of 25,000 simulated haplotypes generated under the neutral model, using the computer program ms (37), with θ = 1, were analyzed for extended haplotype homozygosity. (θ = 0.5 did not produce any strikingly different result.) Haplotype networks were drawn using Phylogenetic Network Software NETWORK 4.2.0.1, website: fluxus-engineering.com (28). To assess the impact of natural selection on these genes, we estimated the rates of nonsynonymous (dN) and synonymous (dS) substitutions by the Nei–Gojobori (NG) (38) method, using PAML4 (39). Because for each gene different haplotypes were represented in varying numbers of individuals, we took care to represent each distinct haplotype by its observed frequency in the input file of PAML4 used to obtain the estimates of dN and dS. To identify specific regions of a gene with possible differential impact of natural selection, we estimated the dN:dS ratio in various segments of a gene by performing a sliding window analysis (31), with window-width of 20 codons and a slide parameter of 1 codon (http://ibio.jp/∼tendo/etools/wina). Weighted averages of resulting dN:dS estimates were taken.
Supplementary Material
Acknowledgments.
We thank all members of The Chatterjee Group—Indian Statistical Institute Centre for Population Genomics, Kolkata, and The Centre for Genomic Application, New Delhi, India, for logistical and infrastructural support. This work was financially supported by U.S. National Institute of Allergy and Infectious Diseases, National Institutes of Health Contract HHSN200400067C.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811357106/DCSupplemental.
References
- 1.Kimbrell DA, Beutler B. The evolution and genetics of innate immunity. Nat Rev Genet. 2001;2:256–267. doi: 10.1038/35066006. [DOI] [PubMed] [Google Scholar]
- 2.Hoebe K, Janssen E, Beutler B. The interface between innate and adaptive immunity. Nature Immnol. 2004;5:971–974. doi: 10.1038/ni1004-971. [DOI] [PubMed] [Google Scholar]
- 3.Cook DN, Pisetsky DS, Schwartz DA. Toll-like receptors in the pathogenesis of human disease. Nat Immunol. 2004;5:975–979. doi: 10.1038/ni1116. [DOI] [PubMed] [Google Scholar]
- 4.Iwasaki A, Medzhitov R. Toll-like receptor control of the adaptive immune responses. Nat Rev Genet. 2004;5:987–995. doi: 10.1038/ni1112. [DOI] [PubMed] [Google Scholar]
- 5.Ganz T, Lehrer RI. Antimicrobial peptides of vertebrates. Curr Opin Immunol. 1998;10:41–44. doi: 10.1016/s0952-7915(98)80029-0. [DOI] [PubMed] [Google Scholar]
- 6.Nizet V, Gallo RL. Cathelicidins and innate defense against invasive bacterial infection. Scand J Infect Dis. 2003;35:670–676. doi: 10.1080/00365540310015629. [DOI] [PubMed] [Google Scholar]
- 7.Ramanathan B, Davis EG, Ross CR, Blecha F. Cathelicidins: Microbicidal activity mechanisms of action and roles in innate immunity. Microbes Infect. 2002;4:361–372. doi: 10.1016/s1286-4579(02)01549-6. [DOI] [PubMed] [Google Scholar]
- 8.Fujita T, Matsushita M, Endo Y. The lectin-complement pathway—its role in innate immunity and evolution. Immunol Rev. 2004;198:185–202. doi: 10.1111/j.0105-2896.2004.0123.x. [DOI] [PubMed] [Google Scholar]
- 9.Haldane JBS. Disease and evolution. Ric Sci Suppl A. 1949;19:68–76. [Google Scholar]
- 10.Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 11.Parham P. The unsung heroes. Nature. 2003;423:20. doi: 10.1038/423020a. [DOI] [PubMed] [Google Scholar]
- 12.Hawn TR, et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires' disease. J Exp Med. 2003;198:1563–1572. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hawn TR, et al. A stop codon polymorphism of Toll-like receptor 5 is associated with resistance to systemic lupus erythematosus. Proc Natl Acad Sci USA. 2005;102:10593–10597. doi: 10.1073/pnas.0501165102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lorenz E, Mira JP, Cornish KL, Arbour NC, Schwartz DA. A novel polymorphism in the toll-like receptor 2 gene and its potential association with staphylococcal infection. Infect Immun. 2000;68:6398–6401. doi: 10.1128/iai.68.11.6398-6401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yim JJ, et al. The association between microsatellite polymorphisms in intron II of the human Toll-like receptor 2 gene and tuberculosis among Koreans. Genes Immun. 2006;7:150–155. doi: 10.1038/sj.gene.6364274. [DOI] [PubMed] [Google Scholar]
- 16.Tomasinsig L, Zanetti M. The cathelicidins—structure function and evolution. Curr Prot Pep Sci. 2005;6:23–34. doi: 10.2174/1389203053027520. [DOI] [PubMed] [Google Scholar]
- 17.Verdu P, et al. Evolutionary insights into the high worldwide prevalence of MBL2 deficiency alleles. Hum Mol Genet. 2006;15:2650–2658. doi: 10.1093/hmg/ddl193. [DOI] [PubMed] [Google Scholar]
- 18.Ferwerda B, et al. TLR4 polymorphisms infectious diseases and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci USA. 2007;104:16645–16650. doi: 10.1073/pnas.0704828104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ferrer-Admetlla A, et al. Balancing selection is the main force shaping the evolution of innate immunity genes. J Immunol. 2008;181:1315–1322. doi: 10.4049/jimmunol.181.2.1315. [DOI] [PubMed] [Google Scholar]
- 20.Bairagya B, et al. Genetic variation and haplotype stuctures of innate immunity genes in eastern India. Infect Gen Evol. 2008;8:360–366. doi: 10.1016/j.meegid.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kreitman M, di Rienzo A. Balancing claims for balancing selection. Trends Genet. 2004;20:300–304. doi: 10.1016/j.tig.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 22.Matsushima N, et al. Comparative sequence analysis of leucine-rich repeats (LRRs) within vertebrate toll-like receptors. BMC Genomics. 2007;8:124. doi: 10.1186/1471-2164-8-124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charlesworth D, Charles B, Morgan MT. The pattern of neutral molecular variation under the background selection model. Genetics. 1995;141:1605–1617. doi: 10.1093/genetics/141.4.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Amer J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabeti PC, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- 26.Takahata N, Nei M. Allelic genealogy under overdominant and frequency-dependent selection and polymorphism of major histocompatibility complex loci. Genetics. 1990;124:967–978. doi: 10.1093/genetics/124.4.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rogers AR, Harpending H. Population growth makes waves in the distribution of pairwise genetic differences. Mol Biol Evol. 1992;9:552–569. doi: 10.1093/oxfordjournals.molbev.a040727. [DOI] [PubMed] [Google Scholar]
- 28.Bandelt H-J, Forster P, Röhl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]
- 29.Garred P, Larsen F, Seyfarth J, Fujita R, Madsen HO. Mannose-binding lectin and its genetic variants. Genes Immun. 2006;7:85–94. doi: 10.1038/sj.gene.6364283. [DOI] [PubMed] [Google Scholar]
- 30.Nuytinck L, Shapiro F. Mannose-binding lectin: Laying the stepping stones from clinical research to personalized medicine. Personalized Med. 2004;1:35–52. doi: 10.1517/17410541.1.1.35. [DOI] [PubMed] [Google Scholar]
- 31.EndoT, IkeoK, Gojobori T. Large-scale search for genes on which positive selection may operate. Mol Biol Evol. 1996;13:685–690. doi: 10.1093/oxfordjournals.molbev.a025629. [DOI] [PubMed] [Google Scholar]
- 32.Balasubramanian S, et al. SNPs on human chromosomes 21 and 22—analysis in terms of protein features and pseudogenes. Pharmacogenomics. 2002;3:393–402. doi: 10.1517/14622416.3.3.393. [DOI] [PubMed] [Google Scholar]
- 33.Seyfarth J, Garred P, Madsen HO. The “involution” of mannose-binding lectin. Hum Mol Genet. 2005;14:2859–2869. doi: 10.1093/hmg/ddi318. [DOI] [PubMed] [Google Scholar]
- 34.Lazarus R, et al. Single nucleotide polymorphisms in innate immunity genes: Abundant variation and potential role in complex human disease. Immunol Rev. 2002;190:9–25. doi: 10.1034/j.1600-065x.2002.19002.x. [DOI] [PubMed] [Google Scholar]
- 35.Reed TE, Schull WJ. A general maximum likelihood estimation program. Amer J Hum Genet. 1968;20:579–580. [PMC free article] [PubMed] [Google Scholar]
- 36.Rozas J, Sánchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 37.Hudson RR. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 38.Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- 39.Yang Z. PAML 4: A program package for phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 40.Tanaka T, Nei M. Positive Darwinian selection observed at the variable-region genes of immunoglobulins. Mol Biol Evol. 1989;6:447–459. doi: 10.1093/oxfordjournals.molbev.a040569. [DOI] [PubMed] [Google Scholar]
- 41.Jiggins FM, Hurst GDD. The evolution of parasite recognition genes in the innate immune system: Purifying selection on Drosophila melanogaster peptidoglycan recognition proteins. J Mol Evol. 2003;57:598–605. doi: 10.1007/s00239-003-2506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Little T, Colbourne J, Crease T. Molecular evolution of Daphnia immunity genes: Polymorphism in a Gram-negative binding protein gene and an α-2-macroglobulin gene. J Mol Evol. 2004;59:498–506. doi: 10.1007/s00239-004-2641-8. [DOI] [PubMed] [Google Scholar]
- 43.Smirnova I, Hamblin MT, McBride C, Beutler B, Di Rienzo A. Excess of rare amino acid polymorphisms in the Toll-like receptor 4 in humans. Genetics. 2001;158:1657–1664. doi: 10.1093/genetics/158.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tapping RI, Omueti KO, Johnson CM. Genetic polymorphisms within the human Toll-like receptor 2 subfamily. Biochem Soc Trans. 2007;35:1445–1448. doi: 10.1042/BST0351445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.