Abstract
The fungus Phycomyces blakesleeanus reacts to environmental signals, including light, gravity, touch, and the presence of nearby objects, by changing the speed and direction of growth of its fruiting body (sporangiophore). Phototropism, growth toward light, shares many features in fungi and plants but the molecular mechanisms remain to be fully elucidated. Phycomyces mutants with altered phototropism were isolated ≈40 years ago and found to have mutations in the mad genes. All of the responses to light in Phycomyces require the products of the madA and madB genes. We showed that madA encodes a protein similar to the Neurospora blue-light photoreceptor, zinc-finger protein WC-1. We show here that madB encodes a protein similar to the Neurospora zinc-finger protein WC-2. MADA and MADB interact to form a complex in yeast 2-hybrid assays and when coexpressed in E. coli, providing evidence that phototropism and other responses to light are mediated by a photoresponsive transcription factor complex. The Phycomyces genome contains 3 genes similar to wc-1, and 4 genes similar to wc-2, many of which are regulated by light in a madA or madB dependent manner. We did not detect any interactions between additional WC proteins in yeast 2-hybrid assays, which suggest that MADA and MADB form the major photoreceptor complex in Phycomyces. However, the presence of multiple wc genes in Phycomyces may enable perception across a broad range of light intensities, and may provide specialized photoreceptors for distinct photoresponses.
Keywords: blue light, LOV domain, White Collar protein, zinc finger, gene duplication
Organisms sense and interact with the surrounding environment to increase their probability of survival. Fungi respond to many environmental signals to modify their patterns of growth and behavior (1). Light, particularly blue light, serves as a signal to regulate fungal development and behavior, presumably for the optimization of spore production and dispersal (2). In addition, blue light activates metabolic pathways and directs the growth of fungal structures (3, 4).
The zygomycete fungus Phycomyces blakesleeanus has served as a model organism to investigate the responses of fungi to light (3, 5). Use of Phycomyces in sensory transduction research was promoted by the Nobel laureate Max Delbrück in the 1950s (6). Blue light regulates several aspects of Phycomyces biology: it regulates the development of fruiting bodies (sporangiophores), stimulates the biosynthesis of beta-carotene, and modifies the direction (phototropism) and speed of growth of the sporangiophores (3). In addition, the Phycomyces sporangiophore can change the direction of growth after sensing other environmental signals, like gravity, wind, touch, and the presence of nearby objects, making this unicellular structure a unique experimental object (3). Much of the attention in Phycomyces research has focused on its responses to light. Phycomyces responds to a wide interval of light intensities extending 10 orders of magnitude. This remarkable sensory dexterity approximates that of the human eye and is achieved through the action of 2 photosystems optimized to operate at different light intensities (7).
A genetic screen for phototropic mutants, conducted in Delbrück's lab, allowed the isolation and characterization of mad mutants, and the first outline of the sensory transduction pathway for Phycomyces (8). The discovery of additional mad mutants and detailed genetic characterization led to the identification of 10 unlinked mad genes, madA through madJ (9, 10). Mutants of the madA and madB genes are defective in phototropism and other light responses suggesting that the corresponding gene products play key roles in Phycomyces photobiology (3).
Most of our understanding of fungal photobiology comes from studies with the ascomycete fungus Neurospora crassa. Mutations in the wc-1 or wc-2 genes disrupt all of the responses of Neurospora to blue light (4, 11). The WC-1 protein contains a zinc-finger, a chromophore-binding domain (named LOV), and PAS domains for protein–protein interactions (12). The LOV domain binds the flavin FAD, allowing WC-1 to act as a photoreceptor (13, 14). LOV was initially identified in phototropins, plant blue light photoreceptors for phototropism (15), and the structure of the LOV domain in a small Neurospora photoreceptor, VVD, has been determined (16). The WC-2 protein contains a zinc-finger and 1 PAS domain (17), and interacts with WC-1 to form a complex that binds to the promoters of light-inducible genes, presumably to activate their transcription (13, 18, 19). WC proteins are required for the responses to blue light in the basidiomycete fungi Cryptococcus neoformans (20, 21) and Coprinus cinereus (22), and 3 wc-1 genes have been described in the zygomycetes Rhizopus oryzae and Mucor circinelloides (23, 24). A Mucor WC-1 protein is modified by ubiquitylation, presumably to regulate its activity (25). Red- and blue-light photoreceptors regulate development and secondary metabolism in the ascomycete fungus Aspergillus nidulans (26–28). Protein complexes containing photoreceptors or transcriptional regulators participate in Aspergillus photobiology (27, 29). The existence of proteins similar to WC-1 and WC-2 in ascomycete, basidiomycete, and zygomycete fungi led to the proposal that the White Collar Complex arose early in fungal evolution as a photoreceptive transcription factor (1, 30, 31).
In Phycomyces, however, the molecular nature of the photoreceptor or the proteins involved in light-signal transduction remained unknown for many decades. In addition, none of the mad genes had been identified despite their initial isolation in the mid 1960s. Our recent discovery that the Phycomyces madA gene is homologous to Neurospora wc-1 suggested that the MADA protein should act as a light-dependent transcription factor and opened the way to the molecular characterization of Phycomyces photoreception (23).
We report here the identification and characterization of the Phycomyces madB gene, encoding a member of the WC-2 family of zinc-finger proteins. In addition, we describe the complete set of wc genes in the Phycomyces genome and characterize their expression after light exposure. We observe the physical interaction between MADA and MADB, suggesting the presence of a MAD complex that regulates gene transcription by light in Phycomyces. Identification of the madB gene closes a relevant chapter in Phycomyces photobiology by uncovering the molecular identity of a key component of the photoreceptor complex.
Results
Multiple wc-1 and wc-2 Genes in the Phycomyces Genome.
Our discovery that MADA, a protein similar to the Neurospora photoreceptor WC-1, is required for light sensing (23) suggested that Phycomyces might use a protein similar to Neurospora WC-2 to interact with MADA to mediate blue light responses.
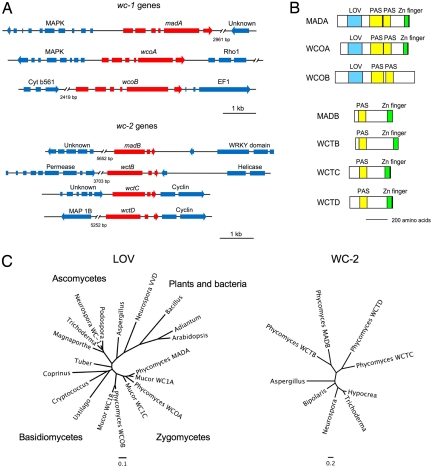
We amplified and cloned a Phycomyces gene similar to wc-2 by PCR. In addition, when the Phycomyces genome sequence became available we analyzed the gene set to identify additional wc genes. The Phycomyces genome contains 3 genes similar to wc-1 and 4 genes similar to wc-2. Two of the wc-1 genes had been identified, madA and wcoA (23), and we named the third gene wcoB (for white collar one gene B). The 4 wc-2 genes were named wctA to wctD (for white collar two gene). The wctA gene was identified initially by PCR, and was later renamed madB based on our genetic and sequencing results. The predicted gene structure for each wc gene was established by PCR amplifying and sequencing the corresponding cDNAs. The only exception was the wctC gene, because we were unable to amplify the cDNA from RNA samples obtained from cultures grown in the dark or after exposure to light. All of the wc-1 genes have 6 exons and 5 introns, and all of the wc-2 genes have 3 exons and 2 introns (Fig. 1A).
Fig. 1.
Phycomyces wc genes and proteins. (A) Genomic structure of Phycomyces wc genes including flanking genes with their putative identities. Exons, introns, and direction of transcription are indicated. (B) WC proteins in the Phycomyces genome and their domains. (C) Phylogenies of the WC proteins. Phylogenetic trees of the LOV domains from fungal WC-1 proteins and photoreceptor proteins from plants and bacteria, and from WC-2 proteins (for accession numbers see SI Text). Bars represent the number of substitutions per site. The wctA gene and the WCTA protein are indicated as madB and MADB, respectively.
The Phycomyces WC proteins are shorter than their Neurospora counterparts and contain a PAS domain for protein interactions, and a zinc finger located at the carboxyl terminus, except for WCOB where we could not identify a standard zinc finger although the presence of conserved cysteine residues suggests WCOB may contain a derivative of the GATA-factor class of zinc fingers (Fig. 1B and Fig. S1). In addition, MADA, WCOA, and WCOB contain an LOV domain that should allow each of these proteins to act as a photoreceptor (Fig. S2). The Phycomyces LOV domains contain a conserved cysteine that would support light-dependent formation of a cysteinyl adduct with the flavin chromophore, as observed in plant LOV domains (15) (Fig. S2). The domains in the Phycomyces WC-1 proteins suggest that these proteins may function as light-regulated transcription factors.
The wc genes were examined by phylogenetic analyses and DNA sequence comparisons to ascertain if these may have arisen through gene duplication, and to assess the extent of duplication in the genome (Fig. 1). The duplication that gave rise to the madA and wcoA gene pair incorporates an upstream gene encoding a MAP kinase. There is no evidence of a MAP kinase upstream of the third homolog, wcoB. The intron positions of the wcoB gene are also conserved, with 4 of the 5 introns sharing the same splice sites with madA and wcoA, which suggests that this gene is related to the other two. Comparison of the zygomycete LOV domains suggests that the event that gave rise to wc-1 gene triplication occurred before the last common ancestor diverged, and that each of the 3 Phycomyces WC-1 homologs is most closely related to its ortholog from the divergent species Mucor circinelloides than to the paralogs within species (Fig. 1C).
Examination of the phylogeny and local gene synteny for the 4 wc-2 homologs reveals that these genes arose through 3 sequential duplication events. All 4 copies of wc-2 contain 2 introns that are conserved in position. The ancestral wc-2 gene duplicated, giving rise to 2 paralogs that both subsequently duplicated, giving rise to 2 pairs of genes, corresponding to wctA/wctB and wctC/wctD. The wctC and wctD genes are each flanked by a cyclin gene that duplicated along with the wct gene. We conclude that the Phycomyces wc genes were derived from limited local duplications within the genome yet all are related to common ancestral wc-1 and wc-2 genes.
wctA Gene Has a Splicing Mutation in the Phototropic Mutant madB.
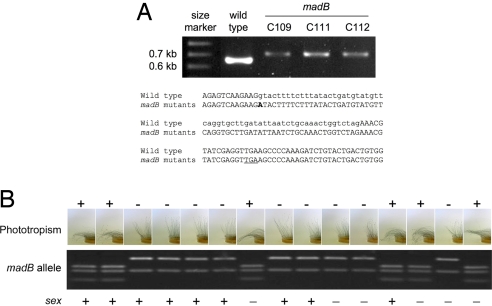
We sequenced all of the 7 wc genes in 50 representative strains carrying all of the mad mutations to identify nucleotide changes that might have been responsible for the deficient phototropic phenotype. We discovered a mutation in the wctA gene in Phycomyces madB mutants. No other mutations were detected in any of the other wc genes in any of the other mad mutants, with the exception of the characterized madA alleles in madA mutants (23). The mutation in madB strains is a G to A transition at nucleotide 907 of the wctA gene (from the initiation ATG) that alters the 5′ splicing site of the first intron. This mutation prevents correct mRNA splicing, resulting in longer mRNA that would yield a truncated protein of 327 aa without the zinc finger. The splicing mutation was confirmed after amplification and sequencing of the corresponding cDNAs in the wild type and several madB mutants (Fig. 2A).
Fig. 2.
The gene madB is similar to Neurospora wc-2. (A) Aberrant cDNA splicing of the wctA gene in the madB mutants. The cDNAs for the wctA gene from the wild-type strain and 3 madB mutants were amplified by PCR and resolved by gel electrophoresis (Upper). Nucleotide sequence of a single transcript cloned from the wild type and madB strains C109, C111, or C112. The G907A mutation is in bold. Coding nucleotides are shown in uppercase, and intron nucleotides are shown in lowercase font. For the mutant transcripts the zinc-finger domain will be deleted by introduction of a premature stop codon (underlined) (Lower). (B) Genetic evidence that the G907A mutation in wctA is linked to reduced phototropism in madB strains. Progeny (15 derived from 15 independent germinated zygospores) from crosses of strains C109 x A56 or C111 x A56 were examined for phototropism, and 50% of the Petri dishes were photographed. The wctA/madB gene was amplified by PCR and cleaved with RsaI to produce 3 (wild type) or 2 (mutant) fragments. The parents madB (phototropism mutant, sex −, 2 fragments) and A56 (phototropism wild type, sex +, 3 fragments) are not shown. The sex locus shows meiotic recombination in the progeny.
Mutation in wctA Cosegregates with Impaired Phototropism in madB Mutants in a Genetic Analysis.
To test the hypothesis that wctA corresponds to the madB locus, genetic crosses between madB strains (C109 or C111) and an isogenic strain (A56) were performed. The wctA gene was amplified by PCR from each progeny DNA, and cleaved with the restriction enzyme RsaI. The G907A mutation identified in madB strains mutates an RsaI recognition site, resulting in 2 fragments for this wctA allele after enzymatic digestion, compared with 3 fragments for the wild type allele. From the C109 x A56 cross (24 progeny from 7 zygospores) the G907A mutation cosegregated with the 16 progeny exhibiting reduced phototropism whereas the remaining 8 progeny with wild type phototropism contained the wild type wctA allele. After the C111 x A56 cross (38 progeny from 14 independent zygospores) 15 progeny with reduced light sensitivity had the mutated wctA allele with the 23 other wild type progeny containing the wild type allele. A subset of this analysis is illustrated in Fig. 2B. Sex segregated independently of the madB mutation (Fig. 2B). In addition, we confirmed the presence of the G907A mutation in the madB progeny (A820 and A821) but not in the wild type progeny (A818 and A819) obtained after a cross between a madB strain (A520) and a phototropic wild type (C169).
As all of the madB strains have a mutation in the wctA gene, and the mutation in wctA cosegregates with the madB phenotype in genetic crosses, we conclude that the madB phenotype is caused by a mutation in the wctA gene that we will henceforth call madB.
Transcriptional Regulation of the wc Genes by Blue Light.
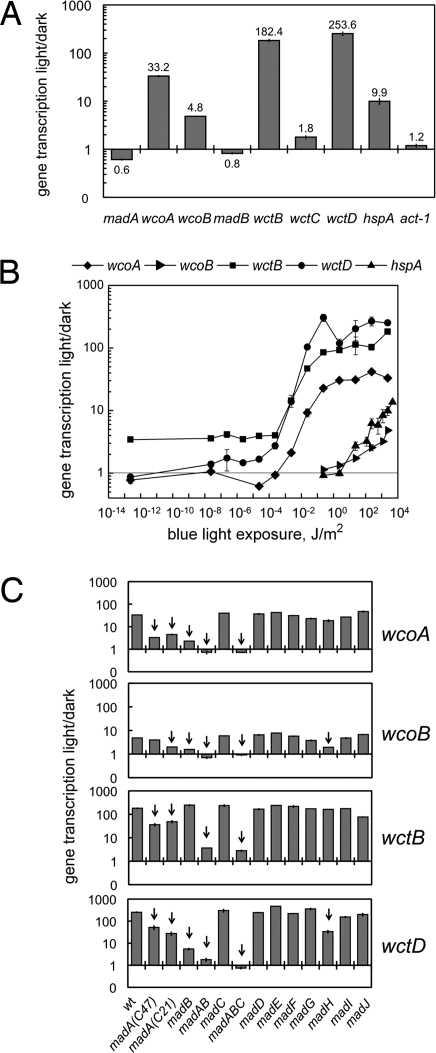
Blue light induced the expression of several Phycomyces wc genes to different levels. After exposing Phycomyces mycelia to 30 min of blue light we observed a 5-fold increase in wcoB mRNA, and a thirtyfold increase in wcoA mRNA. The induction by blue light was more pronounced for the wctB and wctD genes, 180- to 250-fold (Fig. 3A). In comparison, we observed a 10-fold photoactivation for the heat-shock gene hspA (32). On the contrary, expression of madA, madB, and wctC genes was not induced by light, and a slight photorepression was detected for madA and madB (Fig. 3A).
Fig. 3.
Regulation of gene transcription by blue light. Total RNAs were isolated from mycelia exposed to blue light, or kept in the dark. The amount of mRNA for each gene was assayed by quantitative RT-PCR. Each fluorescent signal was first normalized to the corresponding actin signal to correct for loading errors and then was normalized to the signal obtained in the dark. The plots show the relative photoactivation in 2–15 independent experiments (average ± SEM). (A) Photoactivation of gene expression in the wild type after 30 min of blue light (2.3 × 103 J/m2). (B) Threshold determination for the photoactivation of gene expression in the wild type. Mycelia were exposed to blue light of different intensities during 30 min before mRNA extraction. 1 J/m2 of blue light corresponds to 4 μmol/m2 of 450 nm. (C) Photoactivation of gene expression in the wild type and mad mutants after 30 min of blue light (2.3 × 103 J/m2). The madA madB double mutant is shown as madAB, and the madA madB madC triple mutant is shown as madABC. For strain numbers see SI Text.
Blue-light exposures of different duration showed that maximum mRNA accumulation for wcoA, wcoB, wctB, and wctD genes occurred after 15–30 min of light, but photoactivation was observed after 5-min exposures (Fig. S3). Longer light exposures reduced light-dependent mRNA accumulation, but gene photoactivation was still observed in mycelia exposed to light for 2 hours. On the contrary, blue light caused a reduction in the amount of madA mRNA (Fig. S3).
We observed different thresholds for the photoactivation of the wc genes. A low light exposure, 10−5-10−4 J/m2 (4 × 10−5-10−4 μmol/m2 for 450 nm), was sufficient to induce the expression of wctB, wctD, and wcoA. We observed a small but detectable photoinduction for wctB at <10−4 J/m2 that we did not explore further. However, the threshold for the photoactivation of wcoB, 1 J/m2 (4 μmol/m2 for 450 nm), was similar to that of hspA but markedly higher than that for the other wc genes (Fig. 3B).
The photoactivation of the wc genes required the MADA and MADB proteins. A reduced photoactivation was observed in Phycomyces strains carrying madA alleles that either resulted in an amino acid change in the LOV domain (strain C47), or produced aberrant mRNAs because of a splicing mutation (strain C21) (23). Similarly, the splicing mutation in the madB strain reduced the photoactivation of wcoA, wcoB, and wctD, but not wctB (Fig. 3C). However, MADB was required for wctB photoactivation as shown by the differences in photoactivation observed in madA and madA madB double mutants (Fig. 3C). Mutations in other mad genes did not change the relative accumulation of wc mRNAs in light-exposed mycelia. A notable exception was a madH strain that showed a reduced photoactivation for wcoB and wctD, similar to the phenotype observed in madA strains (Fig. 3C).
MADA and MADB Proteins Interact to Form a MAD Complex.
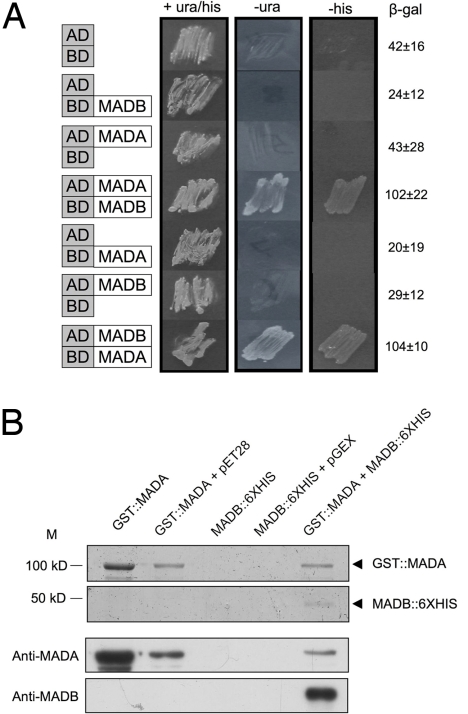
To test protein–protein interactions, the madA, wcoA, wcoB, madB, wctB and wctD cDNAs were fused in frame to the activator (AD) or DNA binding (BD) domains of S. cerevisiae Gal4 in vectors for yeast 2-hybrid analysis. The recipient yeast strain carries URA3, HIS3 and lacZ reporter genes controlled by the GAL4 promoter region to detect the formation of a complex by their growth on minimal agar and the detection of β-galactosidase activity. When S. cerevisiae cells expressed MADA and MADB fused to the AD or BD domains, the HIS3 and URA3 reporter genes were both induced and cells grew in the absence of uracil or histidine. In addition, the lacZ reporter gene was induced, further validating that MADA and MADB form a complex in yeast cells (Fig. 4A). In contrast, S. cerevisiae cells expressing only MADA or MADB fused either to AD or BD domains did not grow on selective media. We did not observe any effect of light on the reporter gene-dependent growth of the strains. No other positive interactions were observed in yeast 2-hybrid assays between any of the other WC proteins: WCOA, WCOB, WCTB, and WCTD (Fig. S4).
Fig. 4.
MADA and MADB interact physically. (A) Yeast 2-hybrid assays. The coding regions of the madA and madB genes were fused adjacent to the AD and BD segments of S. cerevisiae GAL4. Plasmids were cotransformed into a S. cerevisiae strain in which the GAL4 UAS regulates URA3, HIS3, and lacZ genes. Growth of strains in the absence of uracil (-ura) or histidine (-his) and increased β-galactosidase activitiy (β-gal ± SE, Miller units) indicate protein–protein interactions. (B) MADA and MADB form a complex when coexpressed in E. coli. SDS/PAGE stained with coomassie to detect proteins purified with a GST resin. Arrowheads indicate the MADA and MADB recombinant proteins (Upper). Western hybridisations with anti-MADA and anti-MADB after purification with a GST resin (Lower).
As confirmation of the MADA/MADB interaction we expressed the cDNAs for these 2 genes in E. coli and carried out copurification assays (Fig. 4B). The madA cDNA was fused in frame to a GST domain, and the madB cDNA was fused in frame to a 6-His tag. The E. coli protein fraction that bound the GST resin was eluted, and resolved by electrophoresis. The molecular nature of each protein band was revealed after hybridization with specific polyclonal antisera against MADA or MADB. Because we observed the MADB-His tagged protein after GST purification we concluded that the GST resin purified a MADA-MADB protein complex, corroborating the yeast 2-hybrid results (Fig. 4B).
Discussion
Research on the molecular biology of Phycomyces sensory transduction has been limited despite the experimental advantages of a large unicellular structure capable of sensing many environmental signals. However, the isolation and characterization of phototropic mad mutants, initially in Delbrück's lab and later in other laboratories, allowed the identification of genetic loci that are required for reception and processing of the light signal. Among them, the madA and madB gene products are key components of the Phycomyces light-transduction pathway (3). The identification of the nature of madB as a wc-2 gene, together with our previous discovery that madA is a wc-1 gene (23), provide 2 essential elements of Phycomyces photoreception.
The MADA and MADB interaction suggests that this photoresponsive complex acts as its Neurospora counterpart by binding to the promoters of light-regulated genes. However, the role of the MAD complex in short-term phototropic photoresponses is less clear. The Phycomyces sporangiophore is a single cell cylinder, and phototropic bending is observed within 5 minutes after applying unilateral light (33), a short time for a light-dependent transcription factor to operate efficiently through gene transcription and protein translation. It is possible that, in addition, the MAD complex may operate through the light-dependent postranslational modification of other regulatory proteins or by other mechanisms not yet described for WC-1 and WC-2 in Neurospora.
The Phycomyces genome contains 3 wc-1 and 4 wc-2 genes that originated via gene duplication events that occurred early in the zygomycete evolutionary lineage, and have not been observed in other fungal groups. The absence of any additional interactions between WC proteins in yeast 2-hybrid assays suggests that either these proteins participate in weak interactions that are not sufficient for detection in yeast, that species-specific modifications are required, or they do not interact at all, a surprising result considering the presence of PAS domains in all of the Phycomyces WC proteins. It is possible that additional proteins, perhaps the MAD complex, serve as scaffolds for additional interactions between the remaining WC proteins.
The observation that madA and madB are not induced by light suggests that the proteins are present in the dark poised to activate gene transcription and phototropism upon light exposure. The activation by light of wcoA, wcoB, wctB, and wctD suggests that the corresponding gene products may be required after initial exposure to light, or to respond to higher light intensities. These genes may be required for a particular light response in Phycomyces, as has been shown for some of the wc-1 genes in Mucor (24). We can propose that in the dark the MAD complex will be available for light perception. After low light exposure, the activation of wcoA, wctB, and wctD genes would provide additional photoreceptors that could be devoted to low light reception. Higher light intensities would activate wcoB, perhaps to perceive high-intensity photons. However, the differences in the thresholds for wc gene activation suggest that additional photoreceptor proteins, most likely WCOA or WCOB, collaborate in the dark with the MAD complex to activate gene transcription at different intensity ranges. Our observation that madA and madB are required for the photoactivation of these genes supports this hypothesis. Further characterization of the Phycomyces WC proteins will allow us to establish the detailed role these proteins play in photoreception.
It is remarkable that of the remaining 8 mad photoresponse mutants (madC-madJ), none result from mutations in the remaining 5 wc genes. The identity of the gene products encoded by madC-madJ will be uncovered by meiotic mapping studies and may reveal light sensing machinery, or components that lie downstream of the primary MAD photoreceptor complex in the sensory transduction cascade. Our discovery that phototropism and other responses to light in Phycomyces require the activity of a complex composed of MADA and MADB proteins will help to initiate the molecular characterization of Phycomyces photoreception, but it is clear that much remains to be learned from this model fungal system that will advance our understanding of how organisms perceive their environment.
Materials and Methods
Strains.
We used the standard wild type strain of P. blakesleeanus (NRRL1555). Other wild type and mad mutant strains are described in Table S1. Escherichia coli DH5-alpha was used for cloning plasmids.
Cloning and Characterization of Phycomyces wc Genes in the Wild Type and mad Mutant Strains.
A fragment of the Phycomyces wc-2 gene was amplified by PCR from genomic DNA using primers for conserved areas of the gene (Table S2). The 1 kb PCR product was cloned into plasmid pGEM-T (Promega) and sequenced. Other wc-1 and wc-2 genes were identified in the subsequently released Phycomyces genome sequence (Joint Genome Institute http://www.jgi.doe.gov). The predicted ORFs were confirmed or corrected after amplification and sequencing the cDNAs (see primers in Table S2), except the cDNA for wcoB that was obtained in 2 steps (see SI Text). Phycomyces genomic DNA was used to amplify and sequence the wc genes in mad mutant strains (see the list of strains in Table S1 and the list of primers in Table S2). DNA manipulations followed standard methods (34). For domain predictions, protein comparisons, and phylogenetic analysis see SI Text.
Mating Assays and Genetic Segregation.
Crosses were conducted between madB mutant strains C109 and C111 (−) and isogenic strain A56 (+) following methods described in ref. 23. A 478-bp fragment of the madB gene was amplified with primers (Table S2), and cut with RsaI restriction enzyme. The wild type allele results in 3 fragments (217, 169 and 92 bp) and the mutant allele in 2 fragments (309 and 169 bp). Mating type for all progeny was assessed by crossing to (+) and (−) parental strains to confirm meiotic segregation.
Light Exposures, RNA Isolation and cDNA Synthesis, Quantitative RT-PCR.
The Phycomyces dark-grown mycelia were exposed to blue light at the age of 48 h during 30 min unless otherwise stated, and used for RNA extraction as reported (32). Total RNA was used for cDNA synthesis and amplification (see primers in Table S2). Quantitative PCRs were performed to determine relative mRNA abundance using 1-step RT-PCR in a 7500 Real Time PCR System (Applied Biosystems) as described in ref. 35 (see primers in Table S2). The results for each gene were normalized to the corresponding results obtained with the actin gene (act-1) to correct for sampling errors. Then, the results obtained with each sample were normalized to the RNA sample from the corresponding mycelia grown in the dark. For phototropism measurements, strains were illuminated with white light of an intensity below that required for madB response, as determined empirically.
Yeast 2-Hybrid Experiments.
cDNAs of madA, wcoA, wcoB, madB, wctB and wctD were amplified by RT-PCR, and cloned into plasmids pDESTTM32 and pDESTTM22 in the S. cerevisiae reporter strain MaV203 using the ProQuestTM Two-Hybrid System (Invitrogen). Double transformants were selected on media lacking leucine and tryptophan. Interactions were assessed by growth in the absence of uracil or histidine with 10 mM 3-aminotriazole, and by β-galactosidase assays.
Expression and Detection of the MADA/MADB Complex in E. coli.
Phycomyces cDNAs for MADA and MADB were amplified and cloned into the expression vectors pGEX4T1 (GST-tag) (GE Healthcare) and pET28a (6XHis) (Novagen). The fusion proteins were expressed in E. coli strain Rosetta2 (DE3) pLysS (Novagen) transformed with each or both recombinant plasmids, and the GST proteins purified with a GST-bind resin (Novagen). Purified proteins were used for SDS/PAGE and western hybridization using an anti-MADA polyclonal antibody and an anti-MADB polyclonal antibody (Pacific Inmunology) (see SI Text for details).
Supplementary Material
Acknowledgments.
We thank E. Cerdá-Olmedo for scientific advice, D. Perez del Camino for technical help, and G. Gutiérrez for phylogenies. This work was supported by European funds (ERDF), Spanish Grants BIO2006–14897, AGL2005–08081, P06-CVI-01650 (J. Andalucía), and GR64 (to J. Castilla-León); and National Institutes of Health Grants AI039115 and AI073917. J.R.R. held an European Molecular Biology Organization short-term fellowship in the laboratory of J.M.C. We acknowledge access to the Phycomyces genome sequence (US Department of Energy, Joint Genome Institute).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the EMBL Nucleotide Sequence Database (FM178798 wcoB, FM178799 madB, FM178800 wctB, FM179475 wctC, FM178801 wctD).
This article contains supporting information online at www.pnas.org/cgi/content/full/0900879106/DCSupplemental.
References
- 1.Bahn YS, et al. Sensing the environment: Lessons from fungi. Nat Rev Microbiol. 2007;5:57–69. doi: 10.1038/nrmicro1578. [DOI] [PubMed] [Google Scholar]
- 2.Corrochano LM, Galland P. In: The Mycota I Growth, Differentiation and Sexuality, The Mycota. 2nd Ed. Kües U, Fischer R, editors. Vol I. Berlin: Springer-Verlag; 2006. pp. 233–259. [Google Scholar]
- 3.Cerdá-Olmedo E. Phycomyces and the biology of light and color. FEMS Microbiol Rev. 2001;25:503–512. doi: 10.1111/j.1574-6976.2001.tb00588.x. [DOI] [PubMed] [Google Scholar]
- 4.Linden H, Ballario P, Macino G. Blue light regulation in Neurospora crassa. Fungal Genet Biol. 1997;22:141–150. doi: 10.1006/fgbi.1997.1013. [DOI] [PubMed] [Google Scholar]
- 5.Cerdá-Olmedo E, Lipson ED, editors. Phycomyces. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1987. [Google Scholar]
- 6.Bergman K, et al. Phycomyces. Bacteriol Rev. 1969;33:99–157. doi: 10.1128/br.33.1.99-157.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Galland P, Lipson ED. Blue-light reception in Phycomyces phototropism: Evidence for two photosystems operating in low- and high-intensity ranges. Proc Natl Acad Sci USA. 1987;84:104–108. doi: 10.1073/pnas.84.1.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergman K, Eslava AP, Cerdá-Olmedo E. Mutants of Phycomyces with abnormal phototropism. Mol Gen Genet. 1973;123:1–16. doi: 10.1007/BF00282984. [DOI] [PubMed] [Google Scholar]
- 9.Orejas M, Peláez MI, Alvarez MI, Eslava AP. A genetic map of Phycomyces blakesleeanus. Mol Gen Genet. 1987;210:69–76. doi: 10.1007/BF00337760. [DOI] [PubMed] [Google Scholar]
- 10.Campuzano V, Galland P, Eslava AP, Alvarez MI. Genetic characterization of two phototropism mutants of Phycomyces with defects in the genes madI and madJ. Curr Genet. 1995;27:524–527. doi: 10.1007/BF00314442. [DOI] [PubMed] [Google Scholar]
- 11.Liu Y, He Q, Cheng P. Photoreception in Neurospora: A tale of two White Collar proteins. Cell Mol Life Sci. 2003;60:2131–2138. doi: 10.1007/s00018-003-3109-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ballario P, et al. White collar-1, a central regulator of blue light responses in Neurospora, is a zinc finger protein. EMBO J. 1996;15:1650–1657. [PMC free article] [PubMed] [Google Scholar]
- 13.Froehlich AC, Liu Y, Loros JJ, Dunlap JC. White Collar-1, a circadian blue light photoreceptor, binding to the frequency promoter. Science. 2002;297:815–819. doi: 10.1126/science.1073681. [DOI] [PubMed] [Google Scholar]
- 14.He Q, et al. White collar-1, a DNA binding transcription factor and a light sensor. Science. 2002;297:840–843. doi: 10.1126/science.1072795. [DOI] [PubMed] [Google Scholar]
- 15.Christie JM. Phototropin blue-light receptors. Annu Rev Plant Biol. 2007;58:21–45. doi: 10.1146/annurev.arplant.58.032806.103951. [DOI] [PubMed] [Google Scholar]
- 16.Zoltowski BD, et al. Conformational switching in the fungal light sensor Vivid. Science. 2007;316:1054–1057. doi: 10.1126/science.1137128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linden H, Macino G. White collar 2, a partner in blue-light signal transduction, controlling expression of light-regulated genes in Neurospora crassa. EMBO J. 1997;16:98–109. doi: 10.1093/emboj/16.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Q, Liu Y. Molecular mechanism of light responses in Neurospora: From light-induced transcription to photoadaptation. Genes Dev. 2005;19:2888–2899. doi: 10.1101/gad.1369605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belden WJ, Loros JJ, Dunlap JC. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol Cell. 2007;25:587–600. doi: 10.1016/j.molcel.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 20.Idnurm A, Heitman J. Light controls growth and development via a conserved pathway in the fungal kingdom. PLoS Biol. 2005;3:e95. doi: 10.1371/journal.pbio.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu YK, Sun KH, Shen WC. Blue light negatively regulates the sexual filamentation via the Cwc1 and Cwc2 proteins in Cryptococcus neoformans. Mol Microbiol. 2005;56:480–491. doi: 10.1111/j.1365-2958.2005.04549.x. [DOI] [PubMed] [Google Scholar]
- 22.Terashima K, Yuki K, Muraguchi H, Akiyama M, Kamada T. The dst1 gene involved in mushroom photomorphogenesis of Coprinus cinereus encodes a putative photoreceptor for blue light. Genetics. 2005;171:101–108. doi: 10.1534/genetics.104.040048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Idnurm A, et al. The Phycomyces madA gene encodes a blue-light photoreceptor for phototropism and other light responses. Proc Natl Acad Sci USA. 2006;103:4546–4551. doi: 10.1073/pnas.0600633103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Silva F, Torres-Martínez S, Garre V. Distinct white collar-1 genes control specific light responses in Mucor circinelloides. Mol Microbiol. 2006;61:1023–1037. doi: 10.1111/j.1365-2958.2006.05291.x. [DOI] [PubMed] [Google Scholar]
- 25.Silva F, et al. A RING-finger protein regulates carotenogenesis via proteolysis-independent ubiquitylation of a White Collar-1-like activator. Mol Microbiol. 2008;70:1026–1036. doi: 10.1111/j.1365-2958.2008.06470.x. [DOI] [PubMed] [Google Scholar]
- 26.Blumenstein A, et al. The Aspergillus nidulans phytochrome FphA represses sexual development in red light. Curr Biol. 2005;15:1833–1838. doi: 10.1016/j.cub.2005.08.061. [DOI] [PubMed] [Google Scholar]
- 27.Purschwitz J, et al. Functional and physical interaction of blue- and red-light sensors in Aspergillus nidulans. Curr Biol. 2008;18:255–259. doi: 10.1016/j.cub.2008.01.061. [DOI] [PubMed] [Google Scholar]
- 28.Bayram O, Biesemann C, Krappmann S, Galland P, Braus GH. More than a repair enzyme: Aspergillus nidulans photolyase-like CryA Is a regulator of sexual development. Mol Biol Cell. 2008;19:3254–3262. doi: 10.1091/mbc.E08-01-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bayram O, et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320:1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- 30.Corrochano LM. Fungal photoreceptors: Sensory molecules for fungal development and behaviour. Photochem Photobiol Sci. 2007;6:725–736. doi: 10.1039/b702155k. [DOI] [PubMed] [Google Scholar]
- 31.Herrera-Estrella A, Horwitz BA. Looking through the eyes of fungi: Molecular genetics of photoreception. Mol Microbiol. 2007;64:5–15. doi: 10.1111/j.1365-2958.2007.05632.x. [DOI] [PubMed] [Google Scholar]
- 32.Rodríguez-Romero J, Corrochano LM. The gene for the heat-shock protein HSP100 is induced by blue light and heat-shock in the fungus Phycomyces blakesleeanus. Curr Genet. 2004;46:295–303. doi: 10.1007/s00294-004-0534-4. [DOI] [PubMed] [Google Scholar]
- 33.Galland P. Phototropism of the Phycomyces sporangiophore: A comparison with higher plants. Photochem Photobiol. 1990;52:233–248. [Google Scholar]
- 34.Sambrook J, Russell DW. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 35.Navarro-Sampedro L, Yanofsky C, Corrochano LM. A genetic selection for Neurospora crassa mutants altered in their light regulation of transcription. Genetics. 2008;178:171–183. doi: 10.1534/genetics.107.079582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.