Abstract
Homologous flavoproteins from the photolyase (PHR)/cryptochrome (CRY) family use the FAD cofactor in PHRs to catalyze DNA repair and in CRYs to tune the circadian clock and control development. To help address how PHR/CRY members achieve these diverse functions, we determined the crystallographic structure of Arabidopsis thaliana (6-4) PHR (UVR3), which is strikingly (>65%) similar in sequence to human circadian clock CRYs. The structure reveals a substrate-binding cavity specific for the UV-induced DNA lesion, (6-4) photoproduct, and cofactor binding sites different from those of bacterial PHRs and consistent with distinct mechanisms for activities and regulation. Mutational analyses were combined with this prototypic structure for the (6-4) PHR/clock CRY cluster to identify structural and functional motifs: phosphate-binding and Pro-Lys-Leu protrusion motifs constricting access to the substrate-binding cavity above FAD, sulfur loop near the external end of the Trp electron-transfer pathway, and previously undefined C-terminal helix. Our results provide a detailed, unified framework for investigations of (6-4) PHRs and the mammalian CRYs. Conservation of key residues and motifs controlling FAD access and activities suggests that regulation of FAD redox properties and radical stability is essential not only for (6-4) photoproduct DNA repair, but also for circadian clock-regulating CRY functions. The structural and functional results reported here elucidate archetypal relationships within this flavoprotein family and suggest how PHRs and CRYs use local residue and cofactor tuning, rather than larger structural modifications, to achieve their diverse functions encompassing DNA repair, plant growth and development, and circadian clock regulation.
Keywords: blue-light photoreceptor, circadian clock, electron transfer, flavoprotein, FAD
In response to sunlight, homologous flavoproteins from the photolyase (PHR)/cryptochrome (CRY) family, found in bacteria to humans, use the same FAD cofactor to carry out their dissimilar functions (1, 2). CRYs are regulatory proteins that control growth and development in plants and tune biological clocks in animals (3). In contrast, PHRs are DNA repair proteins that revert UV-induced photoproducts into normal bases to maintain genetic integrity (4). Most prokaryotes have a single PHR, the class I or DNA PHR that repairs cyclobutane pyrimidine dimers (CPD), but some eukaryotes including plants possess two (Fig. S1): class II PHR for CPD repair and (6-4) PHR to repair pyrimidine–pyrimidone (6-4) photoproducts. Sunlight exposure makes DNA repair systems including PHRs essential for plants (5, 6): mutant plants with defective class II CPD PHR (UVR2) or (6-4) PHR (UVR3) exhibit impaired UV resistance (UVR) (7, 8).
CPD and (6-4) PHRs share many functional similarities, but have evolved distinct substrate specificities and mechanisms (9–12). CPD and (6-4) photoproducts (Fig. S2) both arise from UV-induced [2+2] cyclo-addition reactions between adjacent pyrimidines; yet the CPD reaction product between 2 C5 C6 double bonds is stable, whereas the product of the C5
C6 double bonds is stable, whereas the product of the C5 C6 bond with the C4 carboxyl or amino group rapidly rearranges into the (6-4) photoproduct (13). To maintain genetic integrity, (6-4) PHR must therefore catalyze not only covalent bond cleavage like the CPD PHR reaction, but also amino or hydroxy group transfer (14). Thus, the (6-4) PHR mechanism is complex and hypothesized to require light energy and simple electron donation from FAD (9, 14).
C6 bond with the C4 carboxyl or amino group rapidly rearranges into the (6-4) photoproduct (13). To maintain genetic integrity, (6-4) PHR must therefore catalyze not only covalent bond cleavage like the CPD PHR reaction, but also amino or hydroxy group transfer (14). Thus, the (6-4) PHR mechanism is complex and hypothesized to require light energy and simple electron donation from FAD (9, 14).
Mammalian CRYs most closely resemble (6-4) PHRs (Fig. 1 and Fig. S1) (15); yet human cells do not exhibit light-activated photoproduct repair (photoreactivation) (16). Instead, mammalian CRYs are critical components of circadian clock circuitry (17–19) that act in conjunction with PERIOD proteins to repress activity of the heterodimeric CLOCK/BMAL1 transcription factor (20, 21). Mammalian CRYs are crucial for maintaining robust circadian rhythms: deletion produces complete arrhythmicity (18, 19). In vertebrates having both (6-4) PHR and clock CRY proteins, (6-4) PHR does not disturb the circadian clock, and conversely clockwork CRYs do not appear to directly contribute to DNA repair (22, 23). Clockwork CRYs from different species have also been implicated in diverse processes, including nonvisual photoreception, sun compass orientation, and time–place learning (24–31). Yet, delays in mechanistic characterization of clock CRYs, caused in part by technical difficulties in protein expression and thus structure determination, have hampered biological understanding. In contrast, structures representative (Fig. S1) of plant-specific (32) and DASH CRYs (33), class I CPD PHRs (34–37), and other DNA base repair enzymes (38) have aided functional investigations. In the (6-4) PHR/clock CRY (6-4/clock) cluster, sequence analyses alone have failed to distinguish protein functions, so 3D structural analyses are needed to advance understanding.
Fig. 1.
Sequence alignment among (6-4) PHRs and clock CRYs in the α-helical domain, highlighting sequence conservation, functional motifs, and site-directed mutants. Sequence-conserved (white on red) and similar (red on white) residues are boxed. At64PHR secondary structure and amino acid numbering are shown at the top. Yellow circles show residues binding FAD with main and side chains (red and blue rims, respectively). Gray circles show the Trp triad. Squares show mutational sites for CLOCK/BMAL1 repression assays (see Fig. 5).
Here, we present the crystallographic structure of Arabidopsis thaliana (6-4) PHR with bound FAD cofactor and phosphate. The overall structure is similar to that of CPD PHRs. However, combined structural and mutational analyses reveal distinct functional regions and motifs: the 3D adjacent phosphate-binding and Pro-Lys-Leu (PKL) protrusion motifs; the sulfur loop near the external end of the Trp electron-transfer pathway; and the C terminus, absent in previous structures. Because of the high sequence similarities within the 6-4/clock cluster, our structure provides a detailed, unified framework for understanding (6-4) PHR substrate recognition and repair mechanisms and for functional mapping of mammalian CRYs.
Results and Discussion
Conserved and Distinguishing Features of the (6-4) Photolyase Structure.
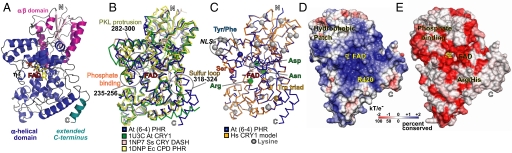
To address questions regarding (6-4) PHRs and their relationships to other PHR/CRY family members, we determined the crystallographic structure of A. thaliana (6-4) PHR (At64PHR), which confers UV protection to plants and shares >65% sequence similarities to human and mouse CRY1 and CRY2 (Fig. 1). The At64PHR structure, refined to 2.7-Å resolution with good residual errors (Rwork = 20.2% and Rfree = 23.8%) and geometry (Table S1), revealed the conserved fold and detailed structure, including the C-terminal extension, active-site channel, cofactor environment, Trp electron-transfer pathway, phosphate-binding site, and novel functional motifs (Fig. 2). Overall, the N-terminal α/β domain exhibits a variation of the Rossman nucleotide-binding fold and is tethered via a long connecting loop to the C-terminal helical domain (Fig. 2A). The helical domain has a conserved binding site for an unusual U-shaped conformation of the FAD cofactor beneath a long positively charged groove. Superposition of the At64PHR structure with other PHR/CRY structures (Fig. 2B) highlights the following novel features: the 3D adjacent phosphate-binding (Asp-235–Ala-256) and PKL protrusion motifs (Tyr-282–Leu-300); the sulfur loop (Met-318–Cys-324) near the external end of the Trp electron-transfer pathway; and a C-terminal helix (Asp-508–Asp-522, turquoise in Fig. 2A), not characterized in previous structures. The At64PHR structure, combined with sequence and phylogenetic analyses (Fig. 1 and Fig. S1) provided an informed basis for a detailed 3D model of human CRY1 (Fig. 2C) and for functional testing of the identified motifs (Fig. 2 B, D, and E) in the 6-4/clock cluster.
Fig. 2.
At64PHR structure revealed conserved fold with motifs unique to 6-4/clock CRYs. (A) N-terminal α/β (magenta) and C-terminal α-helical (blue) domains connected by a long, belt-like, Pro-rich loop, and followed by an extra C-terminal helix (cyan). (B) Superimposed structures of At64PHR (blue), plant-specific CRY1 (green), CRY DASH (tan), and CPD PHR (yellow) highlighting novel functional motifs: phosphate binding, PKL protrusion, and sulfur loop. (C) 3D homology model for human CRY1 (dark blue) based on At64PHR structure (gold), with balls marking Ser phosphorylation site (red), Trp electron-transfer triad (gold), hydrophobic switch (blue), and residues tuning FAD (green). Lys residues (gray balls) show core of NLS and potential targets for ubiqutination. (D) Electrostatic potential surface for At64PHR shows positively charged DNA-binding groove and nearby hydrophobic patch. (E) Conservation map for 6-4/clock cluster highlights features universal to 6-4/clock cluster: groove, FAD environment, and phosphate-binding motif. In D and E At64PHR is tilted backward from the shared orientation of A–C to see into the DNA-binding groove.
The phosphate-binding motif diverges from other known PHR/CRY structures via the short α9 helix to partially constrict entrance to the substrate-binding cavity with 310 helix η4 (Fig. 2A). This 310 helix is centered on Pro-245; the Lys-246 to Glu-243 salt bridge orients the intervening Lys-244 side chain inwards (Fig. 3) to hydrogen bond with FAD adenine N7. This motif encircles a phosphate ion (Fig. 4), which hydrogen bonds to the Glu-243 main chain and Trp-238 side chain, conserved in the 6-4/clock cluster. Conformational changes in the phosphate-binding motif upon phosphate binding or release thus have the potential to indirectly tune both FAD redox properties and substrate binding.
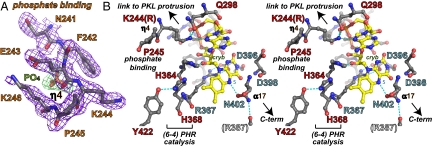
Fig. 3.
Phosphate and FAD binding sites. (A) Omit electron density map for At64PHR residues (purple contours) binding phosphate anion (green contours). (B) Stereoview of FAD-binding site residues in At64PHR specific to the 6-4/clock cluster (red labels) and universal to the PHR/CRY family (blue labels) shown with hydrogen bonds (dashed lines) to FAD (yellow with red oxygen and blue nitrogen atoms).
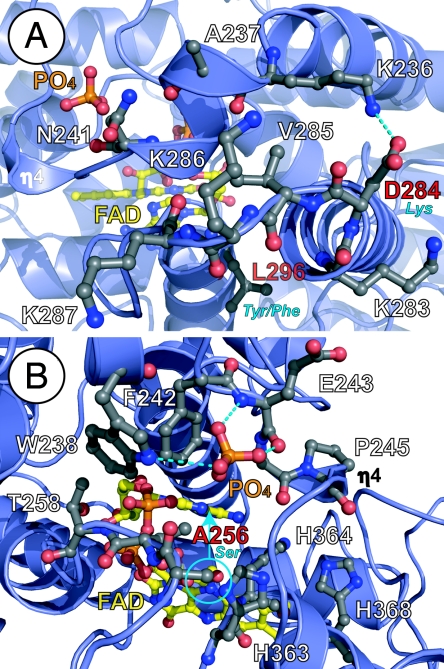
Fig. 4.
Environmental tuning of FAD for diverse functions of the 6-4/clock cluster. (A) The phosphate-binding motif (top) with bound phosphate (orange) and PKL protrusion motif (bottom) partially block substrate access (from left) to FAD (yellow). The PKL protrusion motif varies in sequence at CRY NLS (KVRK/R) equivalent to At64PHR 284-DVKK-287 (labeled in white, with sequence difference in red/blue) and the hydrophobic switch (bottom center), where At64PHR Leu-296 (red label) becomes aromatic (blue label), to distinguish CRY1(Tyr) from CRY2(Phe). (B) Conserved phosphate-binding motif (Asp-235–Ala-256) in 6-4/clock cluster encircles and hydrogen bonds (dashed lines) to phosphate anion (orange) in At64PHR, but can likely bind phosphorylated serine (blue circle) of clock CRYs (equivalent to At64PHR Ala-256, red label). Conformational changes in this motif may impact substrate access to FAD (from right) and binding near the signature His–His–Leu–Ala–Arg-His motif (lower right, with His residues labeled) required for catalysis. Helices and loops are shown with blue ribbons and loops, respectively, and pertinent side chain with ball-and-stick models, are colored by atom type.
The adjacent PKL protrusion motif (Figs. 1 and 2B) is centered on the projecting cis-peptide-linked double Pro-290 Pro-291 that also constricts access to the cavity. The PKL protrusion motif replaces an α-helix in other PHR/CRY structures. This motif begins with a Lys-rich, solvent-exposed, flexible loop (B values 50–75 Å2) closed by ring stacking of Tyr-282 with His-288. The motif ends in a hydrophobic surface patch centered on Leu-296 (Figs. 2D and 4). The PKL protrusion motif thus has the potential to link intermolecular interactions with substrate access.
The sulfur loop (Met-318–Cys-324) protrudes outward between FAD and the C terminus (Fig. 2B), near the Trp triad electron-transfer pathway (see below and Fig. 2C). The sulfur-containing Met and Cys residues at the beginning and end of this motif are invariant in the 6-4/clock cluster (Fig. 1), but are not conserved in other branches of the PHR/CRY family. Finally, unlike previous PHR/CRY structures, the At64PHR structure has an additional ordered helix (α22) at the C terminus (Fig. 2 A and B).
(6-4) Photoproduct Binding and Repair.
The At64PHR structure exhibits new active-site structural features relevant to photoproduct binding and repair. The At64PHR cavity for binding the damaged photoproduct is narrower and deeper than that of the CPD PHRs (34–36): the phosphate-binding and PKL protrusion motifs constrict the approach to FAD (Fig. 2) and several bulky residues line the channel. Within the cavity, the invariant His-His-Leu-Ala-Arg-His motif specific to the 6-4/clock cluster (Fig. 1) is important for catalysis. The second (At64PHR His-364) and final (His-368) His residues of this motif (Fig. 4B) are critical for DNA repair (9, 12), as shown by the loss of activity upon mutation (Fig. S3). Catalytic His-364 is oriented for electron transfer by a hydrogen bond to the FAD hydroxyl group, whereas His-368 (replacing the conserved Met characteristic of CPD PHRs) hydrogen-bonds with Tyr-422 (Fig. 3B). Intervening Leu-365 (Arg in most other PHR/CRY family members) and Leu-409 flank the His-368 ring to form the wall of the cavity leading to FAD. Substitution of the intervening Leu-365 by Ala, to remove van der Waals' contacts between protein and substrate, diminishes binding affinity (9). Directly across the positively charged DNA-binding channel (Fig. 2D) from the phosphate-binding motif, Arg-420 (Fig. 2E) is conserved in most PHRs and CRYs, but is replaced with His in clockwork CRYs of the 6-4/clock cluster (Fig. 1). In the structure of the CPD PHR complex with DNA (34), the equivalent Arg substitutes for the flipped photoproduct in stabilizing the complementary DNA strand. Nevertheless, this conserved Arg is not essential for DNA repair as mutant At64PHR proteins bearing either His (like clock CRY) or Ala substitutions for Arg-420 retain the ability to recognize and repair the 6-4 photoproduct (Fig. S4). Two aromatic residues (Trp-301 and Trp-408) proposed to play a key role in substrate recognition by CPD PHR (34, 35, 37) are conserved in At64PHR to form perpendicular walls of the substrate-binding cavity. Interestingly, to accommodate the His-368 and Leu-409 side chains, the At64PHR Trp-408 indole ring is flipped 180° relative to the equivalent ring in CPD PHR structures, but matching the similarly-flipped Tyr in the crystal structure of the cyanobacterial CRY DASH (33). Thus, as also found in other proteins (39), both the conformation and the type of a residue can be key to diversity within a conserved protein framework.
Cofactor Binding and Tuning.
In At64PHR flavin isoalloxazine and adenine moieties of FAD are in close proximity (Fig. 3B) forming a U-shaped conformation, which resembles that in CPD PHR structures. This highlights their shared use of light-activated reduced FAD in photoproduct repair. The isoalloxazine ring is anchored by main-chain hydrogen bonds from Asp-396 and Asp-398, and tuned by the Asn-402 side chain, which hydrogen bonds to redox-active FAD N5. The Arg-367 to Asp-396 salt bridge across the isoalloxane ring system orients the Arg side-chain guanidinium moiety to stabilize the FAD semiquinone radical at the C4a position (Fig. 3B). This salt bridge is invariant in all PHR/CRY family members. Residues Thr-258 through Ser-261 form main-chain hydrogen bonds with the FAD phosphate oxygens. Additionally, the Thr-257, Ser-261, and Trp-361 side chains hydrogen-bond with the FAD phosphate, ester, and sugar moieties, respectively. Unique to the At64PHR structure, and presumably all members of the 6-4/clock cluster (by sequence identity), is the charged hydrogen bond from Lys-244 (Arg in mouse/human CRYs) of the phosphate-binding motif to solvent-exposed FAD adenine N7. Lys-244 is well-ordered in all 3 At64PHR molecules (Fig. 3A), and the Lys side-chain NZ atom is further anchored by hydrogen bonds with the Gln-298 side-chain amide and FAD phosphate O2. Thus, proteins of the 6-4/clock cluster exhibit novel cofactor-tuning interactions with the FAD adenine N7.
A second cofactor, serving as a light-harvesting antenna, has been identified in some prokaryotic CPD PHRs. Two distinct antenna binding sites, for 5,10-methenyltetrahydrofolic acid (MTHF) and 8-hydroxy-5-deazaflavin (8-HDF), were identified from crystal structures of Esherichia coli and Anacystis nidulans CPD PHRs, respectively (35, 36). Other flavins including FMN and FAD can also bind in the 8-HDF site (40, 41). In the At64PHR structure, both potential antenna-binding sites are present, but display some sequence and structural changes. Tyr-117 (6-4/clock Tyr or Phe) replaces the carboxylate side chain required for MTHF binding (Glu-109 in E. coli CPD PHR) (42), and the binding groove is ≈2 Å wider. In contrast, the binding site for the 8-HDF isoalloxazine ring in A. nidulans CPD PHR is well conserved in the At64PHR structure (and 6-4/clock sequences), although the Tyr-117 side chain partially occupies the position for the flexible 8-HDF ribityl side chain. Thus, our structure supports a flavin, rather than MTHF antenna for the 6-4/clock cluster.
The Trp triad responsible for electron transfer for photoactivation, or light-induced reduction of FAD, in CPD PHR (4) is conserved in At64PHR (inside Trp-406 adjacent to FAD, middle Trp-383, and outside Trp-329), but with modifications (Fig. S5). In particular, the outside Trp (Trp-329) is buried, not solvent-exposed as in other structurally-characterized PHR/CRY family members. Strikingly both the outside (Trp-329) and middle (Trp-383) tryptophans of this electron-transfer pathway form side-chain hydrogen bonds from their ring nitrogen atoms to the sulfur atoms of Met-318 and Cys-324, respectively. Met-318 and Cys-324 lie at the ends of the sulfur loop motif and are unique to and conserved in the 6-4/clock cluster. These electron-rich sulfur atoms have the potential to influence electron transfer and the stability of radical intermediates along the Trp electron-transfer pathway to FAD.
Structural Implications for Circadian Clock Cryptochromes.
The high sequence similarity (Fig. 1) and phylogenetic clustering (Fig. S1) of the 6-4/clock cluster allow us to use the At64PHR structure to better model (Fig. 2C) and evaluate structure-based functions for vertebrate CRYs, including intermolecular interactions governing assembly, disassembly, cellular localization, and degradation (via ubiquitination; ref. 43) of clock components. Residues conserved in the 6-4/clock cluster (Fig. 2E) form the phosphate-binding motif, line the substrate-binding cavity above FAD, contribute positive charge to the DNA binding groove, and generate a hydrophobic patch (Fig. 2D), suggesting that these regions are important for clockwork CRY functions. However, structural variations within these conserved regions may point to their functional specialization. Specific similarities and differences in the phosphate-binding and PKL protrusion motifs constricting access to FAD, and in the FAD environment itself, suggest functional specialization of mammalian CRYs: (i) a surface-exposed signal for nuclear localization, (ii) regulation by phosphorylation, (iii) FAD tuning by the protein environment, and (iv) a hydrophobic switch differentiating CRY1 and CRY2.
CRYs have a positively charged monopartite nuclear localization signal (NLS sequence KVRK/R; Fig. 1) directing nuclear translocation of the CRY/PERIOD complex for repression of CLOCK/BMAL1 function (44, 45). In (6-4) PHRs, the equivalent sequence (At64PHR 284-DVKK-287) is surface-exposed on the PKL protrusion motif (Fig. 4A). Asp-284 replaces a positively charged Lys to disrupt the NLS, highlighting potential differences in the mechanism of nuclear entry between At64PHR and mammalian CRYs.
Phosphate Binding and Phosphorylation.
The bound phosphate anion (Fig. 3A) hydrogen-bonded with the Glu-243 backbone and Trp-238 side chain (Fig. 4) helps to create the “constriction” of access to FAD. This phosphate-binding motif is conserved in the 6-4/clock cluster (Figs. 1 and 2E) and, in vivo, may recognize a phosphorylated amino acid. Both PHRs and CRYs can become phosphorylated (46–48). Intriguingly, the phosphate-binding motif is 3D adjacent to the conserved serine phosphorylation site of mammalian CRYs identified through MAPK treatment (47). This serine, equivalent to At64PHR Ala-256, is also found in some (6-4) PHRs (Figs. 1 and 4B). A phospho-Ser at this position may mimic the interactions of the phosphate anion with the phosphate-binding motif, enforcing its phosphate-binding conformation and subsequent functional consequences. Ser phosphorylation and phosphate ion binding near conserved Ala-256 may offer a control mechanism to tune FAD (Figs. 3A and 4), ultimately leading to change in protein function. Phosphate binding by the Glu-243 main chain helps position Lys-244 to salt bridge with the adenine moiety of FAD (Fig. 3). In the phosphate-free form of clock CRYs, the serine could hydrogen-bond with the neighboring His-363, the first residue of the signature His–His–Leu–Ala–Arg–His motif where the second His (His-364) is hydrogen-bonded to 2′ hydroxyl group of the flavin linker (Figs. 3B and 4B).
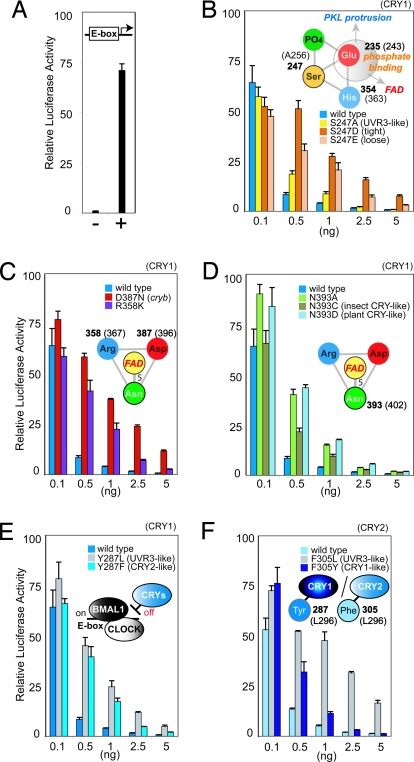
Phospho-Ser-mimicking mutations at this site in mouse CRY1 attenuate CRY repression of the CLOCK/BMAL1 complex (Fig. 5B). The shorter “tight” Asp mutant (Cry1S247D) is more effective than the longer “loose” Glu mutant (Cry1S247E) for inactivation of this CRY function, whereas the Ala mutant (Cry1S247A) functions more similarly to wild type. These results, plus the prior observation that the MAPK pathway may inhibit CRY repression function without altering the protein stability (47), implicate conformational flexibility of the phosphate-binding motif, without bound phosphate, in the repression function of circadian clock-regulating CRYs.
Fig. 5.
Activity of CRY mutants in repressing CLOCK/BMAL1 transcription. (A) Negative and positive controls for CLOCK/BMAL transcription assayed by luciferase activity after transient transfection. (B) Phospho-Ser mimicking mutations in the phosphate-binding motif interfere with repression by CRY1. (C) Salt bridge stabilizing FAD radical is essential. (D) Asn side-chain hydrogen bond to FAD N5 tunes repression by CRY1. (E and F) Tyr specific for CRY1 (E) and Phe specific for CRY2 (F) replace At64PHR Leu-296 hydrophobic switch in the PKL protrusion motif to discriminate function. (E) CRY1 mutations mimicking CRY2 lose repression, like those mimicking (6-4)PHR. (F) CRY2 mutations mimicking CRY1 maintain repression.
FAD Tuning in Cryptochrome Function.
We used mutational analyses to investigate functional roles of conserved and novel structural features for FAD tuning. Interestingly, even subtle modification of the invariant Asp–Arg salt bridge positioned to stabilize the FAD radical (Fig. 3B) has significant functional consequences. A Drosophila mutant designated cryb bearing a single substitution from the salt-bridging Asp to Asn fails to detect light (26). The equivalent mouse CRY1 mutant (Cry1D387N) produces a severe functional defect in the CLOCK/BMAL1 repression (Figs. 1 and 5C) (49). When we substituted the Arg salt-bridge partner to Lys in CRY1 (Cry1R358K), repression was also impaired (Figs. 1 and 5C), further reinforcing the crucial role of the salt bridge in CRY function. Because this twisted Arg to Asp salt bridge orients a terminal Arg nitrogen to stabilize FAD semiquinone radical formation at C4a (Fig. 3B), these mutational results suggest that FAD redox reactions and radical stability are universally important for function in both photosensing and transcriptional repression for all PHR/CRY family members.
Two specific residues in direct contact with FAD are not conserved between At64PHR and clockwork CRYs: Lys-244 (Arg in mammalian CRYs) makes a salt bridge with FAD adenine N7, and Thr-257 (Pro in mammalian CRYs) hydrogen-bonds to FAD phosphate. However, mutational swapping of Lys and Arg (or Thr and Pro) between At64PHR and mouse CRY1 at these positions did not significantly impact either the DNA repair or CLOCK/BMAL1 repression functions, respectively. This finding implies either other conserved residues interacting with FAD may contribute to clockwork CRY function or the Lys/Arg-244 interaction with FAD may only be relevant for integrating the phosphorylation state of the protein (see previous section).
The key residue interacting with the redox-active FAD N5 position of FAD is characteristic of each branch of the PHR/CRY family: Asn in class I CPD PHRs, DASH CRYs, all members of the 6-4/clocks cluster (At64PHR Asn-402), whereas the residue is substituted to Cys in insect-specific CRYs and to Asp in plant-specific CRYs (50–52). Our mouse CRY1 mutants, in which this Asn was substituted with Cys or Asp (Cry1N393C,Cry1N393D), mimicking insect- or plant-specific CRY, showed attenuated repression of CLOCK/BMAL1 (Fig. 5D), supporting the key role of this residue in modulating FAD function in circadian clock CRYs.
The Hydrophobic Switch for Clock Cryptochromes.
Modeling of human CRY sequences onto the backbone of the At64PHR crystal structure (Fig. 2C) produced significant clashes in hydrophobic packing near the At64PHR Leu-296 position in the PKL protrusion. Unlike (6-4) PHRs, the circadian clock-regulating CRYs conserve an aromatic residue here that distinguishes between the 2 types of mammalian CRYs: Tyr in CRY1 and Phe in CRY2 (Figs. 1 and 4A). This aromatic residue participates in a hydrophobic patch (Fig. 2D) buttressing the rim of the substrate-binding cavity, behind the constriction. Our substitution of this Tyr or Phe with Leu (Cry1Y287L or Cry2F305L) in mouse CRYs, to mimic the local structure of At64PHR, reduced repression of CLOCK/BMAL1 activity (Fig. 5 E and F). Furthermore, mutational swapping of the Tyr and Phe between CRY1 and CRY2 highlighted important differences (53) in their repression function. The Cry1Y287F mutant mimicking CRY2 lost as much repression activity as the Cry1Y287L mutant mimicking the (6-4) PHRs. Conversely, Cry2F305Y mimicking CRY1 was a better repressor than Cry2F305L mimicking the (6-4) PHRs (Fig. 5 E and F). Overall, these results suggest that subtle structural changes caused by a single (or few) amino acids tuning local regions, rather than drastic surface modifications, produce the specific functions that distinguish the DNA repair (6-4) PHRs from the circadian CRYs, and CRY1 from CRY2.
General Implications.
These combined structural and mutational results reveal functional motifs characteristic of the phylogenetically defined 6-4/clock cluster, which encompasses (6-4) PHRs from all species, vertebrate clockwork CRYs and the related insect CRY2 proteins, and other closely-related homologs. Analyses of the At64PHR structure therefore help clarify enigmatic structure/function relationships within the PHR/CRY family, by identifying conserved vs. distinguishing features of (6-4) and CPD PHRs, and especially of (6-4) PHRs and circadian clock CRYs. Taken together, our analyses reveal that tuning of the FAD cofactor by its specific protein environment is pivotal to function for proteins of the 6-4/clock cluster. Circadian clock CRY function is also regulated by the hydrophobic switch between Phe and Tyr in the hydrophobic patch near the positively charged DNA-binding groove. This switch likely mediates CRY function through the structurally-identified constriction, formed by the phosphate-binding and PKL protrusion motifs. Besides these motifs, the CRY C terminus may assist in CLOCK/BMAL1 repression through FAD-regulated intermolecular interactions, which may explain its ability to impart repressive function to a noncircadian PHR (54). The combined results presented here reveal that the substrate recognition site, specific for (6-4) photoproducts, the cofactor binding sites, and the Trp electron-transfer pathway show differences from those of bacterial PHR/CRY, consistent with a distinct DNA repair mechanism. Our results furthermore show that the major functional differences within the PHR/CRY family are controlled, not by large structural modifications, but by local residue tuning that, in particular, impacts FAD chemistry and conformation at the binding cavity.
Materials and Methods
Full details of the methods used are presented in SI Text.
Cloning, Expression, and Purification.
A gene encoding At64PHR (UVR3) was isolated from cDNA and expressed in E. coli. At64PHR was purified with Blue Sepharose, DNA cellulose, hydroxyl apatite, and monoS column chromatography. Mutant clones were constructed with QuikChange.
Crystallization, Data Collection, and Structure Determination.
At64PHR crystals were grown in space group P212121 with unit cell dimensions a = 111.7 Å, b = 140.1 Å, c = 144.6 Å and 3 molecules per asymmetric unit. Crystals grew by hanging drop vapor diffusion at 4 °C from mixtures of protein [>25 mg/mL in 50 mM Tris·HCl (pH 8.0), 50 mM NaCl, 10% glycerol, 10 mM DTT] and well [100 mM Hepes (pH 6.6), 25 mM potassium acetate, 20% polyethylene glycol 6000] solutions. After X-ray diffraction data collection at Advanced Light Source beamline 8.3.1 (Berkeley, CA), phases were determined by molecular replacement using as a search probe a structure-based (6-4) PHR model overlaid with PHR and CRY crystal structures. Crystallographic statistics are summarized in Table S1.
Assays.
For DNA binding, base flipping, and DNA repair assays, deoxyoligonucleotides containing (6-4) photoproduct were synthesized and hybridized to complementary strand (9, 11, 55). To assay the activity of mouse CRY mutants in repressing BMAL/CLOCK transcription, 293T cells were reverse-transfected in 96-wellplates with 250 ng of plasmid DNA mixtures including 25 ng of Per1-Luciferase reporter construct, 50 ng of CMV-mBMAL1, 120 ng of CMV-hCLOCK, and 0–5 ng of CMV-CRY plus filler DNA. Twenty-four hours later, cell extracts were assayed for luciferase activity.
Supplementary Material
Acknowledgments.
We thank Dr. H. Nakamura for modeling advice; H. Le, E. Sato, C. Hitomi, and Drs. M. Ariyoshi and Y. Fujiwara for technical assistance; Drs. T. Ishikawa, S. Nakajima, and K. Yamamoto for UVR3 sequence information and help with repair assays; Dr. T. Oyama for the cDNA library; Drs. D. Shin, J. Huffman, and J. Tubbs for manuscript suggestions; and the Advanced Light Source, which is supported by U.S. Department of Energy Program Integrated Diffraction Analysis Technologies under Contract DE-AC02-05CH11231, for X-ray data collection facilities. This work was supported by National Institutes of Health Grants GM37684 (to E.D.G.), GM046312 (to J.A.T.), EY016807 (to S.P.), and 1F32GM082083-01 (to L.D.), Pew Scholars (S.P.), Asahi Glass Foundation (S.I.), Human Frontier Science Program (S.I. and J.A.T.), the Japan Society for the Promotion of Science fellowships (to K.H. and J.Y.), and The Skaggs Institute for Chemical Biology (K.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates have been deposited in in the Protein Data Bank, www.pdb.org (PDB ID code 3FY4).
This article contains supporting information online at www.pnas.org/cgi/content/full/0809180106/DCSupplemental.
References
- 1.Cashmore AR. Cryptochromes: Enabling plants and animals to determine circadian time. Cell. 2003;114:537–543. [PubMed] [Google Scholar]
- 2.Sancar A. Regulation of mammalian circadian clock by cryptochrome. J Biol Chem. 2004;279:34079–34082. doi: 10.1074/jbc.R400016200. [DOI] [PubMed] [Google Scholar]
- 3.Lin C, Todo T. The cryptochromes. Genome Biol. 2005;6:220. doi: 10.1186/gb-2005-6-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weber S. Light-driven enzymatic catalysis of DNA repair: A review of recent biophysical studies on photolyase. Biochim Biophys Acta. 2005;1707:1–23. doi: 10.1016/j.bbabio.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 5.Jiang CZ, Yee J, Mitchell DL, Britt AB. Photorepair mutants of Arabidopsis. Proc Natl Acad Sci USA. 1997;94:7441–7445. doi: 10.1073/pnas.94.14.7441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ries G, et al. Elevated UV-B radiation reduced genome stability in plants. Nature. 2000;406:30–31. doi: 10.1038/35017595. [DOI] [PubMed] [Google Scholar]
- 7.Landry LG, et al. An Arabidopsis photolyase mutant is hypersensitive to ultraviolet-B radiation. Proc Natl Acad Sci USA. 1997;94:328–332. doi: 10.1073/pnas.94.1.328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakajima S, et al. Cloning and characterization of a gene (UVR3) requires for photorepair of 6-4 photoproducts in Arabidopsis thaliana. Nucleic Acids Res. 1998;26:638–644. doi: 10.1093/nar/26.2.638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitomi K, et al. Role of two histidines in the (6-4) photolyase reaction. J Biol Chem. 2001;276:10103–10109. doi: 10.1074/jbc.M008828200. [DOI] [PubMed] [Google Scholar]
- 10.Weber S, et al. Photoreactivation of the flavin cofactor in Xenopus laevis (6-4) photolyase: Observation of a transient tyrosyl radical by time-resolved electron paramagnetic resonance. Proc Natl Acad Sci USA. 2002;99:1319–1322. doi: 10.1073/pnas.032469399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hitomi K, et al. Binding and catalytic properties of Xenopus (6-4) photolyase. J Biol Chem. 1997;272:32591–32598. doi: 10.1074/jbc.272.51.32591. [DOI] [PubMed] [Google Scholar]
- 12.Schleicher E, et al. Electron nuclear double resonance differentiates complementary roles for active site histidines in (6-4) photolyase. J Biol Chem. 2007;282:4738–4747. doi: 10.1074/jbc.M604734200. [DOI] [PubMed] [Google Scholar]
- 13.Clivio P, Fourrey J-L, Gasche J, Favre A. DNA photodamage mechanistic studies: Characterization of a thietane intermediate in a model reaction relevant to “6-4 lesions”. J Am Chem Soc. 1991;113:5481–5483. [Google Scholar]
- 14.Kim S-T, Malhotra K, Smith CA, Taylor JS, Sancar A. Characterization of (6-4) photoproduct DNA photolyase. J Biol Chem. 1994;269:8535–8540. [PubMed] [Google Scholar]
- 15.Todo T, et al. Similarity among the Drosophila (6-4)photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science. 1996;272:109–112. doi: 10.1126/science.272.5258.109. [DOI] [PubMed] [Google Scholar]
- 16.Chao CC. Lack of DNA enzymatic photoreactivation in Hela cell-free extracts. FEBS Lett. 1993;336:411–416. doi: 10.1016/0014-5793(93)80848-o. [DOI] [PubMed] [Google Scholar]
- 17.Thresher RJ, et al. A role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 18.van der Horst GT, et al. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 19.Vitaterna MH, et al. Differential regulation of mammalian perios genes and circadian rhythmicity by cryptochrome 1 and 2. Proc Natl Acad Sci USA. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kume K, et al. mCRY and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 21.Lee C, Etchengaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- 22.Kobayashi Y, et al. Molecular analysis of zebrafish photolyase/cryptochrome family: Two types of cryptochromes present in zebrafish. Genes Cells. 2000;5:725–738. doi: 10.1046/j.1365-2443.2000.00364.x. [DOI] [PubMed] [Google Scholar]
- 23.Zhu H, Conte F, Green CB. Nuclear localization and transcriptional repression are confined to separable domains in the circadian protein CRYPTOCHROME. Curr Biol. 2003;13:1653–1658. doi: 10.1016/j.cub.2003.08.033. [DOI] [PubMed] [Google Scholar]
- 24.Van der Zee EA, et al. Circadian time-place learning in mice depends on Cry genes. Curr Biol. 2008;18:844–848. doi: 10.1016/j.cub.2008.04.077. [DOI] [PubMed] [Google Scholar]
- 25.Emery P, So WV, Kaneko M, Hall JC, Rosbash M. CRY, a Drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell. 1998;95:669–679. doi: 10.1016/s0092-8674(00)81637-2. [DOI] [PubMed] [Google Scholar]
- 26.Stanewsky R, et al. The cryb mutation identifies cryptochrome as a circadian photoreceptor in Drosophila. Cell. 1998;95:681–692. doi: 10.1016/s0092-8674(00)81638-4. [DOI] [PubMed] [Google Scholar]
- 27.Sauman I, et al. Connecting the navigational clock to sun compass input in monarch butterfly brain. Neuron. 2005;46:457–467. doi: 10.1016/j.neuron.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 28.Gegear RJ, Casselman A, Waddell S, Reppert SM. Cryptochrome mediates light-dependent magnetosensitivity in Drosophila. Nature. 2008;454:1014–1018. doi: 10.1038/nature07183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tu DC, Batten ML, Palczwski K, Van Gelder RN. Nonvisual photoreception in the chick iris. Science. 2004;306:129–131. doi: 10.1126/science.1101484. [DOI] [PubMed] [Google Scholar]
- 30.Mouritsen H, et al. Cryptochromes and neuronal-activity markers colocalize in the retina of migratory birds during magnetic orientation. Proc Natl Acad Sci USA. 2004;101:14299–14304. doi: 10.1073/pnas.0405968101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cermakian N, et al. Light induction of a vertebrate clock gene involves signaling through blue-light receptors and MAP kinases. Curr Biol. 2002;12:844–848. doi: 10.1016/s0960-9822(02)00835-7. [DOI] [PubMed] [Google Scholar]
- 32.Brautigam CA, et al. Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci USA. 2004;101:12142–12147. doi: 10.1073/pnas.0404851101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brudler R, et al. Identification of a new cryptochrome class: Structure, function, and evolution. Mol Cell. 2003;11:59–67. doi: 10.1016/s1097-2765(03)00008-x. [DOI] [PubMed] [Google Scholar]
- 34.Mees A, et al. Crystal structure of a photolyase bound to a CPD-like DNA lesion after in situ repair. Science. 2004;306:1789–1793. doi: 10.1126/science.1101598. [DOI] [PubMed] [Google Scholar]
- 35.Park HW, Kim S-T, Sancar A, Deisenhofer J. Crystal structure of DNA photolyase from Escherichia coli. Science. 1995;268:1866–1872. doi: 10.1126/science.7604260. [DOI] [PubMed] [Google Scholar]
- 36.Tamada T, et al. Crystal structure of DNA photolyase from Anacystis nidulans. Nat Struct Biol. 1997;4:887–891. doi: 10.1038/nsb1197-887. [DOI] [PubMed] [Google Scholar]
- 37.Torizawa T, et al. Investigation of the cyclobutane pyrimidine dimer (CPD) photolyase DNA recognition mechanism by NMR analyses. J Biol Chem. 2004;279:32950–32956. doi: 10.1074/jbc.M404536200. [DOI] [PubMed] [Google Scholar]
- 38.Hitomi K, Iwai S, Tainer JA. The intricate structural chemistry of base excision repair machinery: Implications for DNA damage recognition, removal, and repair. DNA Repair (Amst) 2007;6:410–428. doi: 10.1016/j.dnarep.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 39.Mylvaganam SE, Bonaventura C, Bonaventura J, Getzoff ED. Structural basis for the root effect in hemoglobin. Nat Struct Biol. 1996;3:275–283. doi: 10.1038/nsb0396-275. [DOI] [PubMed] [Google Scholar]
- 40.Klar T, et al. Natural and non-natural antenna chromophores in the DNA photolyase from Thermus thermophilus. Chembiochem. 2006;7:1798–1804. doi: 10.1002/cbic.200600206. [DOI] [PubMed] [Google Scholar]
- 41.Fujihashi M, et al. Crystal structure of archaeal photolyase from Sulfolobus tokodaii with two FAD molecules: Implication of a novel light-harvesting cofactor. J Mol Biol. 2007;365:903–910. doi: 10.1016/j.jmb.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Henry AA, Jimenez R, Hanway D, Romesberg FE. Preliminary characterization of light harvesting in E. coli DNA photolyase. Chembiochem. 2004;5:1088–1094. doi: 10.1002/cbic.200400013. [DOI] [PubMed] [Google Scholar]
- 43.Busino L, et al. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 44.Tamanini F, Yagita K, Okamura H, van der Horst GTJ. Nucleocytoplasmic shuttling of clock proteins. Methods Enzymol. 2005;393:418–435. doi: 10.1016/S0076-6879(05)93020-6. [DOI] [PubMed] [Google Scholar]
- 45.Hirayama J, Nakamura H, Ishikawa T, Kobayashi T, Todo T. Functional and structural analyses of cryptochrome. Vertebrate CRY regions responsible for interaction with the CLOCK:BMAL1 heterodimer and its nuclear localization. J Biol Chem. 2003;278:35620–35628. doi: 10.1074/jbc.M305028200. [DOI] [PubMed] [Google Scholar]
- 46.Shalitin D, et al. Regulation of Arabidopsis cryptochrome 2 by blue-light-dependent phosphorylation. Nature. 2002;417:763–767. doi: 10.1038/nature00815. [DOI] [PubMed] [Google Scholar]
- 47.Sanada K, Harada Y, Sakai M, Todo T, Fukada Y. Serine phosphorylation of mCRY1 and mCRY2 by mitogen-activated protein kinase. Genes Cells. 2004;9:697–708. doi: 10.1111/j.1356-9597.2004.00758.x. [DOI] [PubMed] [Google Scholar]
- 48.Teranishi M, Nakamura K, Morioka H, Yamamoto K, Hidema J. The native cyclobutane pyrimidine dimer photolyase of rice is phosphorylated. Plant Physiol. 2008;146:1941–1951. doi: 10.1104/pp.107.110189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan Y, Hida A, Anderson DA, Izumo M, Johnson CH. Cycling of CRYPTOCHROME proteins is not necessary for circadian-clock function in mammalian fibroblasts. Curr Biol. 2007;17:1091–1100. doi: 10.1016/j.cub.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xu L, et al. Active site of Escherichia coli DNA photolyase: Asn378 is crucial both for stabilizing the neutral flavin radical cofactor and for DNA repair. Biochemistry. 2008;47:8736–8743. doi: 10.1021/bi800391j. [DOI] [PubMed] [Google Scholar]
- 51.Berndt A, et al. A novel photoreaction mechanism for the circadian blue light photoreceptor Drosophila cryptochrome. J Biol Chem. 2007;282:13011–13021. doi: 10.1074/jbc.M608872200. [DOI] [PubMed] [Google Scholar]
- 52.Kottke T, Batschauer A, Ahmad M, Heberle J. Blue-light-induced changes in Arabidopsis cryptochrome 1 probed by FTIR difference spectroscopy. Biochemistry. 2006;45:2472–24729. doi: 10.1021/bi051964b. [DOI] [PubMed] [Google Scholar]
- 53.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chaves I, et al. Functional evolution of the photolyase/cryptochrome protein family: Importance of the C terminus of mammalian CRY1 for circadian core oscillator performance. Mol Cell Biol. 2006;26:1743–1753. doi: 10.1128/MCB.26.5.1743-1753.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yamamoto J, Hitomi K, Todo T, Iwai S. Chemical synthesis of oligodeoxyribonucleotides containing the Dewar valence isomer of the (6-4) photoproduct and their use in (6-4) photolyase studies. Nucleic Acids Res. 2006;34:4406–4415. doi: 10.1093/nar/gkl572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.