Abstract
The APOE ε4 allele is a risk factor for late-life pathological changes that is also associated with anatomical and functional brain changes in middle-aged and elderly healthy subjects. We investigated structural and functional effects of the APOE polymorphism in 18 young healthy APOE ε4-carriers and 18 matched noncarriers (age range: 20–35 years). Brain activity was studied both at rest and during an encoding memory paradigm using blood oxygen level-dependent fMRI. Resting fMRI revealed increased “default mode network” (involving retrosplenial, medial temporal, and medial-prefrontal cortical areas) coactivation in ε4-carriers relative to noncarriers. The encoding task produced greater hippocampal activation in ε4-carriers relative to noncarriers. Neither result could be explained by differences in memory performance, brain morphology, or resting cerebral blood flow. The APOE ε4 allele modulates brain function decades before any clinical or neurophysiological expression of neurodegenerative processes.
Keywords: hippocampus, memory, neuroimaging, resting connectivity
Apolipoprotein E (apoE, protein; APOE, gene) is a very-low-density lipoprotein that removes cholesterol from the blood and carries it to the liver for processing (1). In the central nervous system, apoE has a key role in coordinating the mobilization and redistribution of cholesterol, phospholipids, and fatty acids, and it is implicated in mechanisms such as neuronal development, brain plasticity, and repair functions (2). The human APOE gene, which is encoded on chromosome 19, has 3 allelic variants (ε2, ε3, and ε4). The ε4 allele has been associated with a higher risk of cardiovascular disease (3), both early-onset (4) and late-onset (5) Alzheimer's disease (AD), poor outcome from traumatic brain injury (6), and age-related cognitive impairment (7).
Neuroimaging studies of the APOE polymorphism in healthy subjects have largely focused on gray matter (GM) alterations in middle or late life, particularly in brain regions associated with the greatest AD pathological findings. Even in asymptomatic subjects, hippocampal and frontotemporal GM reduction has been observed in APOE ε4-carriers relative to noncarriers (8). Moreover, a reduction of resting glucose metabolism was reported in young and middle-aged cognitively normal APOE ε4-carriers in brain regions known to be affected by AD, including the posterior cingulate, parietal, temporal, and prefrontal cortices (9–11). fMRI task-based studies (mainly investigating memory processes) have shown greater activation in middle-aged and elderly APOE ε4-carriers relative to noncarriers (12–16). Although these studies suggest an influence of the APOE ε4 allele on brain structure and metabolism, they do not make clear at what age these influences initially manifest. Furthermore, although differences in structure, resting metabolism, and function have each been reported in ε4-carriers relative to noncarriers, it remains to be established to what extent these characteristics interact.
Thus far, reports of structural and functional effects of the APOE ε4 allele in young adults are limited (17–20). Only 2 small fMRI studies have tested for early life associations of the APOE polymorphism with changes in brain function. Filbey et al. (18) reported greater activation in 8 APOE ε4-carriers compared with 8 noncarriers in medial frontal and anterior cingulate areas using a working memory paradigm. Mondadori et al. (17) reported reduced activation with an associative learning paradigm in 13 ε4-carriers relative to 11 ε2-carriers and 10 ε3-homozygotes. Both studies therefore suggest that the APOE genotype influences brain functions even early in adulthood.
Here, we used a multimodal MRI protocol to investigate structural and functional neurophysiological characteristics of 18 APOE ε4-carriers and 18 noncarriers, with ages ranging from 20 to 35 years old. Our first aim was to measure differences in spontaneous fluctuations in resting brain function in ε4-carriers relative to noncarriers using resting-state fMRI. Brain regions showing a strong temporal coherence (coactivation) in low-frequency fluctuations (less than 0.1 Hz) are defined as “resting state networks” (RSNs), and they reflect intrinsic properties of functional brain organization (21). We were specifically interested in studying the effect of the APOE ε4 allele on an RSN called the “default mode network” (DMN), which includes the prefrontal, anterior and posterior cingulate, lateral parietal, and inferior/middle temporal gyri; cerebellar areas; and thalamic nuclei and extending to mesial temporal lobe (MTL) regions (22). The DMN is affected by neurodegenerative processes (23); both AD patients (24) and people with amnestic mild cognitive impairment (aMCI) (25) are reported to have reduced coactivation of hippocampal and posterior cingulate regions.
Our second aim was to test for the effects of APOE genotype on task-related activations. Because the MTL and hippocampi, in particular, are the earliest brain regions to show pathological signs in AD (26), and because APOE receptors in the brain are mainly expressed in the hippocampal-entorhinal cortex complex (27), we selected a blood oxygen level-dependent (BOLD) fMRI task that preferentially activates MTL regions: the “novel vs. familiar” memory-encoding paradigm has been widely used to demonstrate robust hippocampal activation (28, 29).
Finally, we tested whether resting or task-related BOLD differences between ε4-carriers and noncarriers were associated with underlying subject-specific anatomical or resting brain perfusion (30) measures.
Results
Participants.
APOE ε4-carriers and noncarriers were matched for age, gender, and years of education, and 2 individuals in each group had a family history of dementia (either first or second degree). There were no differences in memory recognition performance or reaction times on the fMRI task (Table 1).
Table 1.
Sociodemographic and memory features of the 2 study groups
| APOE ε4-noncarriers | APOE ε4-carriers | P | |
|---|---|---|---|
| Sociodemographics | |||
| Age, y | 28.6 (±3.9) | 28.4 (±4.9) | 0.91 |
| Education, y | 19.5 (±1.5) | 19.6 (±2.0) | 0.93 |
| Gender, male/female | 10/8 | 11/7 | 1 |
| Family history of dementia | 2 | 2 | 1 |
| Memory test, % of corrected responses | |||
| Global performance | 83.8% | 84.4% | 0.86 |
| Familiar images | 97.8% | 97.8% | 1 |
| Novel images | 75.5% | 75.8% | 0.96 |
| Distractors | 94.6% | 95.4% | 0.58 |
| Reaction time, s | |||
| Familiar blocks | 0.75 (±0.14) | 0.80 (±0.22) | 0.44 |
| Novel blocks | 0.99 (±0.26) | 1.09 (±0.42) | 0.39 |
Values denote mean (± SD) or numbers of subjects.
Resting State fMRI.
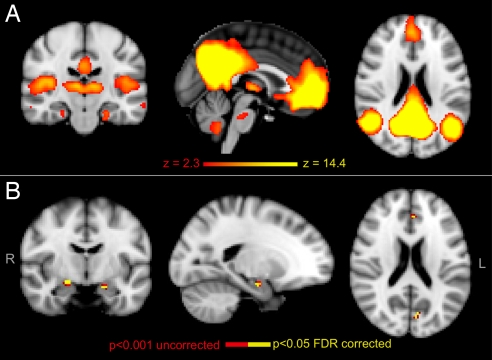
Probabilistic independent component analysis (ICA) (31) defined 25 components representing group-averaged networks of brain regions with BOLD fMRI signals that were temporally correlated. The DMN was identified as including BOLD fMRI signal in the prefrontal, anterior and posterior cingulate, lateral parietal, and inferior/middle temporal gyri; cerebellar areas; and thalamic nuclei and extending to MTL regions (Fig. 1A) (22). To investigate group differences in the DMN, individual DMN maps were estimated. A significantly increased degree of coactivation (measured by the regression coefficient as part of the “dual-regression” approach) within the DMN in ε4-carriers relative to noncarriers was found bilaterally in MTL regions [head of the hippocampus and amygdala; degree of coactivation increased by a factor of 5.3 (left) and 3.7 (right)] and in the medial-prefrontal (increase in coactivation by a factor of 2.5) and retrosplenial (increase in coactivation by a factor of 2.1) cortices (Fig. 1B). There were no brain regions where ε4-carriers had decreased coactivation relative to noncarriers. For comparison, we also investigated group differences in “visual cortex” (VC) and “sensorimotor” (SM) RSNs (31). No significant differences between ε4-carriers and noncarriers were found in the VC RSN, but ε4-carriers had an area of increased coactivation in the SM RSN localized to the right medial postcentral gyrus (peak coordinates in standard space: x = 10, y = −34, z = 54) but with a degree of increased coactivation (increase by a factor of 1.4) notably lower than the observed differences in the significant clusters within the DMN.
Fig. 1.
Results of analysis of resting fMRI. (A) Spatial map representing the DMN for all 36 subjects. Regions belonging to this network include prefrontal, anterior and posterior cingulate, lateral parietal, and inferior/middle temporal gyri; cerebellar areas; and thalamic nuclei and extending to MTL regions. (B) DMN comparison between the 2 groups revealed increased coactivation in retrosplenial, medial-prefrontal, and MTL (head of the hippocampus and amygdala) regions of the DMN in APOE ε4-carriers relative to noncarriers. Red-to-yellow colors define increases in coactivation. R, right hemisphere; L, left hemisphere.
Memory Task fMRI.
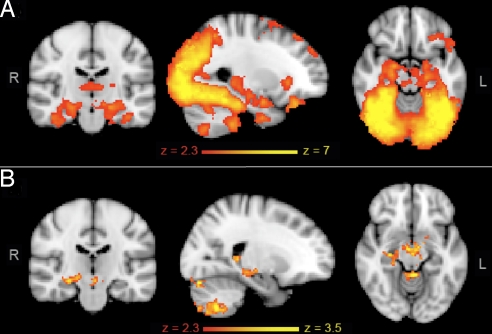
For all subjects, the novel vs. familiar contrast revealed increases in BOLD fMRI signal intensity in the hippocampus, temporal fusiform cortex, and parahippocampal gyrus bilaterally and in association areas in the frontal, parietal, and occipital lobes (Fig. 2A), as previously described (16, 32). A voxel-wise analysis revealed greater right hippocampal activation in APOE ε4-carriers relative to noncarriers for this contrast [reporting here and below, maximum z score, cluster size in voxels, and peak coordinates in standard space: 3.2, 241, and (22, −20, −10), respectively] (Fig. 2B) or when compared with the BOLD signal at rest [3.1, 333, and (26, −10, −20), respectively]. To investigate further this apparent laterality of effects in the hippocampus, possibly attributable to the nonverbal nature of the stimuli, subject-specific hippocampal masks were used to extract the BOLD signal change for each contrast in each hemisphere. Hippocampal signal change was, on average, 32% (range: 20–40%) greater in APOE ε4-carriers than in noncarriers in the significant contrasts (Table 2). Region-of-interest (ROI) analyses detected a significant bilateral effect, such that the group difference in the left hippocampus was just subthreshold in the whole-brain analysis; thus, hippocampal group differences are regarded as bilateral. No difference in the mean size of the mask used to extract BOLD signal change was observed between the 2 groups [left hippocampus: ε4-carriers (3.97 ± 0.46 mL), noncarriers (4.11 ± 0.40 mL), P = 0.36; right hippocampus: ε4-carriers (4.43 ± 0.64 mL), noncarriers (4.54 ± 0.60 mL), P = 0.59].
Fig. 2.
fMRI results for the novel vs. familiar contrast in the encoding task. (A) Mean activation for the novel vs. familiar contrast for all 36 subjects. Activation was found bilaterally in primary and secondary visual cortices as well as in regions involved in memory processes: hippocampus, temporal fusiform cortex, parahippocampal gyrus. (B) Regions of significantly increased BOLD for ε4-carriers relative to noncarriers. Activation in the right hippocampus and cerebellum was greater in the APOE ε4-carriers than in the noncarriers (P < 0.05, corrected for multiple comparisons). Red-to-yellow colors define increases in brain activation. R, right hemisphere; L, left hemisphere.
Table 2.
Changes in hippocampal signal intensities during the encoding task in the 2 study groups
| BOLD percentage signal change | APOE ε4-noncarriers | APOE ε4-carriers | P |
|---|---|---|---|
| Left hippocampus, familiar vs. rest | 0.19 (± 0.07) | 0.22 (± 0.10) | 0.26 |
| Left hippocampus, novel vs. rest | 0.25 (± 0.10) | 0.33 (± 0.12) | 0.04 |
| Left hippocampus, familiar vs. novel | 0.14 (± 0.08) | 0.13 (± 0.09) | 0.97 |
| Left hippocampus, novel vs. familiar | 0.18 (± 0.07) | 0.28 (± 0.16) | 0.03 |
| Right hippocampus, familiar vs. rest | 0.19 (± 0.07) | 0.23 (± 0.12) | 0.27 |
| Right hippocampus, novel vs. rest | 0.25 (± 0.11) | 0.36 (± 0.11) | 0.006 |
| Right hippocampus, familiar vs. novel | 0.14 (± 0.09) | 0.14 (± 0.12) | 0.95 |
| Right hippocampus, novel vs. familiar | 0.19 (± 0.08) | 0.31 (± 0.13) | 0.003 |
Values are thresholded to 0 and denote mean (± SD).
Greater activation in the dentate nucleus, anterior midbrain, and lateral and cortical portions of the cerebellum was also found in APOE ε4-carriers when novel images were compared with familiar images (Fig. 2B) [3.8, 1,737, and (36, −58, −46); 3.54, 1,588, and (40, −65, −56)]. There were no brain regions in which noncarriers showed greater activation than ε4-carriers. There were no significant group differences for the familiar vs. novel or rest vs. novel contrasts. Absolute and relative motion parameters were similar between the 2 groups, and inclusion in the model did not alter the results.
Brain Morphology and Physiology.
No group differences were observed in whole-brain volume or in segmented GM, white matter, cerebrospinal fluid, or hippocampal volumes (Table 3). Moreover, no differences were found between the 2 study groups in GM concentration, using FMRIB Software Library (FSL) voxel-based morphometry (VBM) (33). Similarly, no significant differences were observed in resting brain perfusion using an ROI-based analysis of the hippocampi and 4 main lobes (frontal, temporal, parietal, and occipital).
Table 3.
Brain volumetric and perfusion measurements of the 2 study groups
| APOE ε4-noncarriers | APOE ε4-carriers | P | |
|---|---|---|---|
| Brain volumetry | |||
| Whole brain volume, mL | 1487.5 (± 159.3) | 1515.8 (± 122.2) | 0.55 |
| Gray matter* | 47.1 (± 1.7) | 47.2 (± 1.4) | 0.86 |
| White matter* | 33.4 (± 0.9) | 33.3 (± 1.5) | 0.74 |
| Cerebrospinal fluid* | 19.4 (± 1.9) | 19.5 (± 1.4) | 0.93 |
| Left hippocampus* | 0.27 (± 0.02) | 0.26 (± 0.03) | 0.15 |
| Right hippocampus* | 0.31 (± 0.02) | 0.29 (± 0.04) | 0.11 |
| Brain perfusion values, units of mL·100 g·min | |||
| Whole brain | 21.4 (± 3.9) | 22.5 (± 5.7) | 0.50 |
| Frontal lobe | 18.6 (± 3.8) | 19.6 (± 4.4) | 0.49 |
| Parietal lobe | 28.0 (± 5.8) | 29.3 (± 7.9) | 0.57 |
| Temporal lobe | 24.1 (± 4.6) | 25.3 (± 6.6) | 0.54 |
| Occipital lobe | 23.9 (± 5.8) | 25.5 (± 7.4) | 0.48 |
| Left hippocampus | 31.1 (± 7.6) | 30.9 (± 8.9) | 0.95 |
| Right hippocampus | 31.1 (± 7.2) | 32.9 (± 9.5) | 0.51 |
Values denote mean (± SD). *, Values are expressed as percentage of whole-brain volume.
GM and resting brain perfusion maps were separately added as covariates in the memory-encoding fMRI analysis model. BOLD group contrast differences observed between APOE ε4-carriers and noncarriers were not affected by adding these covariates, and hippocampal, anterior midbrain, and cerebellar activation survived as clear differences between the 2 groups. Group differences in DMN coactivation also survived when GM and resting cerebral blood flow values were added as covariates.
Discussion
In young adults, the APOE genotype is associated with differences in resting and task-related BOLD fMRI. APOE ε4-carriers have increased hippocampal coactivation within the DMN during rest and greater hippocampal activation during a memory-encoding task in the absence of any differences in brain volume, resting brain perfusion, or memory performance. These effects of the ε4 allele are shown together early in adulthood, before any degenerative processes are postulated to have taken place or cognitive impairments can be detected.
Increased Coactivation in the DMN in APOE ε4-Carriers.
Using a data-driven approach, we have shown that APOE influences hippocampal function even in the absence of a specific task. Interestingly, previous investigations of the DMN in AD and aMCI patients reported a reduction in coactivation of the hippocampus (24, 25). The difference between our results and the previous observations in patients with AD or aMCI may relate to hippocampal atrophy (30), normal aging, or factors related to disease progression. Further studies are required to establish the link between RSNs and structural changes associated with age or neuropathological findings. However, the increases in coactivation we observe suggest that the function of those areas subject to the disease process in AD are modulated by the presence of the APOE ε4 allele even in young adults.
Increased Task-Based Activation in APOE ε4-Carriers.
Because the APOE ε4 allelic variant is the best-established genetic risk factor for AD and also is associated with hippocampal structural and metabolic changes in midlife, we specifically targeted the MTL using an encoding memory task. The novel vs. familiar memory-encoding paradigm robustly activated hippocampal regions in our young sample, with the magnitude of signal change being similar to that reported in previous studies (34).
The greater task-related activation observed in APOE ε4-carriers relative to noncarriers is consistent with previous studies in middle-aged or elderly subjects investigating memory processes (12–16). In particular, Bookheimer et al. (12), Bondi et al. (14), and Fleisher et al. (16) observed increased hippocampal activation in APOE ε4-carriers using a task similar to that used in our study. Here, we suggest that the genotypic contribution to the hippocampal activation manifests decades before potential neurodegenerative or age-related processes.
Greater activation for the novel vs. familiar contrast was also observed in cerebellar and anterior midbrain regions. However, whereas the genotype difference in hippocampal activation was also present for the novel vs. rest condition, the midbrain and cerebellar activation difference was not. Hippocampal-midbrain interactions defined by fMRI have been reported recently during encoding (35). Moreover, the cerebellum has a well-established role in higher cognitive functions, including memory processes (36, 37), possibly subserving long-term memory encoding and information storage (36). However, because the midbrain and cerebellum are not directly affected by AD pathological changes, these contributions to BOLD signal increase in ε4-carriers may be secondary to effects in the hippocampus.
Brain Morphology and Resting Perfusion.
The magnitude of BOLD signal changes is determined by the interaction between increases in cerebral blood flow, cerebral blood volume, and rate of oxygen consumption (38, 39) and is affected by differences in resting cerebral blood flow and local GM structure. We found no differences in GM volume or resting perfusion between APOE ε4-carriers and noncarriers using either whole-brain or ROI-based approaches, consistent with interpretation of these results as suggestive of activation-related neurophysiological changes. Relative to the structural or perfusion measures used here, BOLD fMRI appears to be the most sensitive imaging marker of characteristics associated with APOE ε4 allele variation. However, previous studies in older subjects have reported reduced hippocampal volume in ε4-carriers relative to noncarriers (8). Moreover, reduced left entorhinal cortex thickness was reported in a large study of child and adolescent ε4-carriers relative to noncarriers (19), although this difference was influenced by larger volumes in ε2-carriers. Older ε4-carriers have also been reported to have increased resting cerebral blood flow relative to noncarriers (40). Sample size or age may have contributed to the lack of significant difference in morphological features or resting perfusion between ε4-carriers and noncarriers. Any subthreshold differences were tested for in our fMRI analysis by adding GM and resting perfusion maps as covariates; these did not change the results.
Physiological Interpretation.
Greater task-related activation in APOE ε4-carriers relative to noncarriers in hippocampal regions has previously been interpreted as reflecting greater cognitive “effort” by ε4-carriers to obtain the same level of performance as their noncarrier counterparts (14).
This interpretation seems more difficult to apply to young healthy adults who are many decades away from potentially manifesting cognitive decline. It is possible that results reflect neuronal mechanisms to compensate for processes, such as reduced synaptic plasticity (41–43), neuronal growth (44, 45), or altered long-term potentiation (LTP) (46, 47) in people carrying the ε4 allele. LTP in the hippocampus is a widely accepted model for the cellular basis of learning and memory (48), and it has been reported to be selectively affected by the apoE4 isoform (46, 47). Moreover, increased levels of fatty acids, known to regulate LTP processes (49), have been observed in hippocampal regions in mice genetically engineered to have an AD-like condition (50). Sanchez-Mejia et al. (50) suggested that an excess of arachidonic acid, a subtype of fatty acid, may lead to overstimulation of the brain cells, potentially determining a chronic increase in excitatory neuronal activity. Our observation of increased hippocampal BOLD signal both at rest and during a memory task could be explained by such a mechanism. Our finding of increased coactivation of a unilateral medial parietal area with the SM RSN in ε4-carriers relative to noncarriers suggests that the influence of the APOE genotype may not be exclusive to the hippocampus and/or memory system. A greater insight into the effects of this gene and the different allelic variants on neurophysiological processes certainly warrants further investigation.
Conclusion.
We have shown that the influence of the APOE ε4 allele on neurophysiological characteristics in young adults can be detected by measuring both resting and task-related function. BOLD fMRI thus emerges as the most sensitive measure of genotypic differences in this age range rather than either brain morphology or resting perfusion measures. It is important for future studies of larger samples to replicate these results and establish how these measures change with age. Longitudinal studies are required to determine whether increased coactivation within the DMN at rest, increased task-related BOLD signal, or both together are associated with a higher risk of developing later pathological changes. Finally, these data indicate that the APOE ε4 allelic variant plays a fundamental role in brain function, notably (but not exclusively) within the memory system, decades before any clinical expression of neurodegenerative and/or aging processes occurs. A brain-specific effect exerted by the APOE ε4 allele on the hippocampus may confer systematic vulnerability in the memory system in people carrying this allelic variant.
Materials and Methods
Participant Recruitment.
A total of 98 right-handed subjects aged 20 to 35 years old were recruited in Oxfordshire, United Kingdom. Exclusion criteria were current or past history of neurological or psychiatrical disorders, memory complaints, head injury, substance abuse (including alcohol), corticosteroid therapy, and youth diabetes therapy. Subjects were screened for APOE genotype (using a cheek swab sample) and then selected for the study on the basis of either having an ε4 allele (carrier) or being matched for gender, age, and educational level with an ε4-carrier (noncarrier). The study was approved by the local ethics committee, and informed consent was obtained from all subjects.
APOE Genotyping.
DNA was extracted from cheek swab samples of subjects according to standard procedures to allow PCR for the characterization of APOE genotype. The genotyping process was conducted at the Wellcome Trust Centre for Human Genetics in Oxford. The genotyping process was successful for 84 of 98 subjects. Fourteen samples could not be genotyped because the procedure failed to detect the APOE allelic variants. The APOE genotype distribution of our sample reflected the distribution expected in the normal population (χ2 = 2.424, df = 5, P = 0.96) (51). Observed genotypic frequencies were as follows: ε2ε2 [1 of 84 (1.19%)], ε2ε3 [6 of 84 (7.14%)], ε2ε4 [4 of 84 (4.76%)], ε3ε3 [54 of 84 (64.28%)], ε3ε4 [18 of 84 (21.42%], and ε4ε4 [1 of 84 (1.19%)].
Because the APOE ε2 allelic variant is relatively rare and has been reported to have a protective effect against AD (52) and cardiovascular diseases (53), and because it is also associated with increased longevity (54), we included only people carrying the APOE allelic combinations ε3ε4 and ε4ε4 (“ε4-carriers” group) and ε3ε3 (“noncarriers” group). Eighteen ε4-carriers and 18 matched noncarriers consented to undergo the neuroimaging protocol.
Neuroimaging Protocol.
Scanning was performed at the University of Oxford Centre for Clinical Magnetic Resonance Research using a 3-T Siemens Trio scanner with a 12-channel head coil. The neuroimaging protocol comprised functional, perfusion, and structural sequences as follows.
fMRI (Task).
Encoding memory was assessed from a single run using a single gradient echoplanar imaging (EPI) sequence covering the whole brain [repetition time (TR) = 3,000 ms, echo time (TE) = 28 ms, flip angle = 89°, field of view = 192 mm, voxel dimension = 3 mm isotropic, acquisition time = 9 min 6 s].
Perfusion MRI.
Whole-brain perfusion imaging was performed using pulsed arterial spin labeling (ASL) with a 3D gradient spin echo readout. ASL data were collected at multiple inflow periods, starting at 400 ms and ending at 2,400 ms in increments of 200 ms. This multiple inflow method allows for estimation of the time for the magnetically tagged blood to reach the imaging volume [i.e., arterial transit time (in units of seconds)], resulting in improved confidence in the resting perfusion quantitation (TR = 3,136 ms, TE = 40 ms, field of view = 200 × 200 × 130 mm, 26 slices, voxel dimensions = 3.125 × 3.125 × 5 mm, acquisition time = 11 min).
fMRI (Rest).
Whole-brain functional imaging was performed using a gradient echo EPI sequence (TR = 2,000 ms, TE = 28 ms, flip angle = 89°, field of view = 224 mm, voxel dimension = 3 × 3 × 3.5 mm, acquisition time = 6 min 4 s).
Structural MRI.
High-resolution 3D T1-weighted MRI scans were acquired using a magnetization-prepared rapid gradient echo sequence (TR = 2,040 ms, TE = 4.7 ms, flip angle = 8°, field of view = 192 mm, voxel dimension = 1 mm isotropic, acquisition time = 12 min).
Experimental Task.
Programming of the paradigms was carried out using Presentation (Neurobehavioral Systems). A set of 91 colored pictures was used representing animals and landscapes similar in complexity, brightness, and contrast; emotionally neutral; and with no persons represented. Eight pictures (4 animals and 4 landscapes) were randomly chosen to comprise the group that participants had to familiarizes themselves with. The 8 familiar images were displayed to each subject 8 times outside the scanner in a pseudorandom order on a personal computer (PC) screen [image presentation = 3,250 ms, interstimulus interval = 500 ms, between-block interval (fixation cross) = 5,000 ms]. Subjects were instructed to inspect each picture and select from a 2-button response according to whether the images contained animals. To ensure that participants had satisfactorily encoded the familiar images, a memory task (8 familiar plus 8 unfamiliar images with the same number of animals and landscapes presented in pseudorandom order) was performed. Subjects were asked to select between 2 buttons whether the images were familiar or different, and they all scored 100%. Inside the scanner but before data collection, the 8 familiar images were presented another 2 times to allow subjects to familiarize themselves with viewing the stimuli in the scanner. Foam padding and a head restraint were used to control head movement.
During the scanning session, images were displayed in pseudorandom order in a blocked design fashion with 6 blocks each of 8 familiar and novel images. Familiar images were presented in a different order each time, and novel images comprised a total of 48 images (24 animals and 24 landscapes) (image presentation = 3,250 ms, interstimulus interval = 500 ms, block duration = 30,000 ms). Between each block of images was 15,000 ms of rest, during which subjects passively viewed a fixation cross (a total of 12 rest blocks). Subjects were instructed to select from a 2-button response according to whether the images contained animals. Responses were monitored by the scanner operators to ensure compliance and accuracy and were registered by the software in a text file. Participants were also instructed to try to remember the images for a subsequent memory task.
Outside the scanner, ≈50 min after imaging, 83 images were presented on a PC screen for 4,000 ms each (interstimulus interval = 1,000 ms). The 48 novel images, the 8 familiar images, and 27 “distractors” (images never seen before: 13 animals and 14 landscapes) were displayed in pseudorandom order. Subjects had to select between 2 buttons according to whether the images had been seen inside the scanner or not.
Image Analysis.
Data analysis was carried out using FSL tools (www.fmrib.ox.ac.uk/fsl).
fMRI (Rest).
fMRI analysis at rest was carried out using Multivariate Exploratory Linear Optimized Decomposition into Independent Components (MELODIC) (31). For the resting-state scan, subjects were instructed to lie in dimmed light with their eyes open, to think of nothing in particular, and not to fall asleep. Individual prestatistical processing consisted of motion correction, brain extraction, spatial smoothing using a Gaussian kernel of full-width at half-maximum (FWHM) of 6 mm, and high-pass temporal filtering equivalent to 150 s (0.007 Hz). fMRI volumes were registered to the individual's structural scan and standard space images using FMRIB's Nonlinear Image Registration Tool (FNIRT). Preprocessed functional data containing 180 time points for each subject were temporally concatenated across subjects to create a single 4D data set.
The between-subject analysis of the resting data was carried out using a regression technique (dual regression) that allows for voxel-wise comparisons of resting functional connectivity. This approach proceeds in 3 stages. First, the concatenated multiple fMRI data sets are decomposed using ICA to identify large-scale patterns of functional connectivity in the population of subjects. In this analysis, the data set was decomposed into 25 components, in which the model order was estimated using the Laplace approximation to the Bayesian evidence for a probabilistic principal component model. RSNs of interest were selected using spatial correlation against a set of previously defined maps (31). The DMN was identified as the spatial map comprising prefrontal, anterior and posterior cingulate, lateral parietal, and inferior/middle temporal gyri; cerebellar areas; and thalamic nuclei and extending to MTL regions (21, 22). The VC and SM RSNs were identified as the spatial maps comprising occipital regions (VC) and pre- and postcentral gyri (SM), respectively (31). Second, the dual-regression approach is used to identify, within each subject's fMRI data set, subject-specific temporal dynamics and associated spatial maps. This involves (i) using the full set of group-ICA spatial maps in a linear model fit (spatial regression) against the separate fMRI data sets, resulting in matrices describing temporal dynamics for each component and subject, and (ii) using these time-course matrices in a linear model fit (temporal regression) against the associated fMRI data set to estimate subject-specific spatial maps. Finally, the different component maps are collected across subjects into single 4D files (1 per original ICA map, with the fourth dimension being subject identification) and tested voxel-wise for statistically significant differences between groups using nonparametric permutation testing (5,000 permutations) (55). This results in spatial maps characterizing the between-subject/group differences.
These maps were thresholded using an alternative hypothesis test based on fitting a Gaussian/gamma mixture model to the distribution of voxel intensities within spatial maps (see ref. 31 for further details) and controlling the local false-discovery rate at P < 0.05.
fMRI (Task).
fMRI analysis was carried out using fMRI Expert Analysis Tool (FEAT) v. 5.98. Preprocessing consisted of head motion correction, brain extraction, spatial smoothing using a Gaussian kernel of FWHM of 5 mm, and high-pass temporal filtering equivalent to 130 s. Time-series statistical analysis was carried out with local autocorrelation correction. A boxcar convolved with a gamma hemodynamic response function, and its temporal derivative was used to model the data. The main contrast of interest was novel vs. familiar, but estimates of familiar vs. novel, novel vs. rest, and familiar vs. rest were also obtained for reference. fMRI volumes were registered to the individual's structural scan and standard space images using a nonlinear registration tool (FNIRT). These transformations into standard space were applied to images of contrasts of interest and their variances. Higher level (group level) analysis was carried out using FMRIB's Local Analysis of Mixed Effects (FLAME) (56). To reduce effects attributable to outliers, an automatic outlier deweighting tool was also applied (57).
The general linear model (GLM) included the 2 groups (APOE ε4-carriers and noncarriers). We tested for group averages and differences between groups for each of the contrasts of interest. The Z statistic images were thresholded using clusters determined by Z >2.3, and a family-wise error–corrected cluster significance threshold of P < 0.05 was applied to the superthreshold clusters.
Structural MRI.
The complementary approaches of ROI and VBM were used to study GM and to relate it to APOE polymorphism.
Whole-brain analysis was carried out with FSL-VBM, a voxel-based morphometry style analysis (33), using default settings. In brief, brain extraction and tissue-type segmentation were performed, and resulting GM partial volume images were aligned to standard space using FMRIB's Linear Image Registration Tool (FLIRT) and then nonlinear (FNIRT) registration tools. The resulting images were averaged, modulated, and smoothed with an isotropic Gaussian kernel of 4 mm to create a study-specific template. Finally, voxel-wise GLM was applied using permutation nonparametric testing (5,000 permutations), correcting for multiple comparisons across space.
Perfusion MRI.
Resting perfusion maps in units of mL·100g·min were calculated using tools developed in-house (58) and interrogated using ROIs as defined below.
ROIs.
Masks of the left and right hippocampi and frontal, temporal, parietal, and occipital lobes were created for each subject. Individual hippocampal ROIs were obtained using FMRIB's Integrated Registration and Segmentation Tool (FIRST), an automatic subcortical segmentation program. Boundary correction was used for the classification of the boundary voxels. ROIs were visually inspected in the coronal plane to ensure accuracy. ROIs were registered to functional coordinate space and used to extract BOLD percentage signal changes from fMRI data using Featquery (part of FSL). Only voxels with z > 0 were included. Hippocampal masks were registered to perfusion space to extract mean resting perfusion values for comparison between the 2 study groups. Lobar masks were created using the Harvard-Oxford brain atlas on a conventional standard image. Each mask was registered to each structural image and subsequently registered to each perfusion image.
Covariates.
To determine whether BOLD contrast differences between the 2 groups were influenced by structural differences, structural images were used as covariates to interrogate fMRI data. GM images of each subject were extracted using FMRIB's Automated Segmentation Tool (FAST), registered in standard space, smoothed to match the fMRI data, demeaned within each group, and added to the model used to analyze fMRI data. Resting perfusion maps were also used as covariates to interrogate fMRI data. Each perfusion map was registered in standard space and then demeaned within each group. Resting perfusion maps were used during group statistics in an analogous fashion to GM maps.
Statistics.
Statistical analyses of nonimaging variables were carried out using SPSS software (SPSS, Inc.). For sociodemographic variables, brain structure volumes, memory performance, and reaction times, t tests were used. Exact Fisher's and Yates continuity correction was used for categorical variables (gender and family history of dementia). It was confirmed by χ2 test that the genotype distribution of our sample reflected that expected in the normal white population.
Acknowledgments.
We thank Prof. Jonathan Flint and Amarjit Bhorma, Wellcome Trust Centre for Human Genetics, for genotyping APOE data; Dr. Natalie Voets for contributing to fMRI task development; Prof. Peter Jezzard and Dr. David Feinburg for the ASL pulse sequence; and Prof. Klaus Ebmeier and Dr. Kate Watkins for helpful discussions. The authors acknowledge academic collaborative contribution from GlaxoSmithKline to fund data acquisition. P.M.M. is a full-time employee of GlaxoSmithKline. N.F. is funded by the Gordon Small Charitable Trust. C.E.M. is partly funded by the TJ Crow Psychosis Trust.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Mahley RW, Rall SC., Jr Apolipoprotein E: Far more than a lipid transport protein. Annu Rev Genomics Hum Genet. 2000;1:507–537. doi: 10.1146/annurev.genom.1.1.507. [DOI] [PubMed] [Google Scholar]
- 2.Mahley RW. Apolipoprotein E: Cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 3.Lenzen HJ, Assmann G, Buchwalsky R, Schulte H. Association of apolipoprotein E polymorphism, low-density lipoprotein cholesterol, and coronary artery disease. Clin Chem. 1986;32:778–781. [PubMed] [Google Scholar]
- 4.Okuizumi K, et al. ApoE-epsilon 4 and early-onset Alzheimer's. Nat Genet. 1994;7:10–11. doi: 10.1038/ng0594-10b. [DOI] [PubMed] [Google Scholar]
- 5.Strittmatter WJ, et al. Binding of human apolipoprotein E to synthetic amyloid beta peptide: Isoform-specific effects and implications for late-onset Alzheimer disease. Proc Natl Acad Sci USA. 1993;90:8098–8102. doi: 10.1073/pnas.90.17.8098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Teasdale GM, Nicoll JA, Murray G, Fiddes M. Association of apolipoprotein E polymorphism with outcome after head injury. Lancet. 1997;350:1069–1071. doi: 10.1016/S0140-6736(97)04318-3. [DOI] [PubMed] [Google Scholar]
- 7.Deary IJ, et al. Cognitive change and the APOE epsilon 4 allele. Nature. 2002;418:932. doi: 10.1038/418932a. [DOI] [PubMed] [Google Scholar]
- 8.Wishart HA, et al. Regional brain atrophy in cognitively intact adults with a single APOE epsilon4 allele. Neurology. 2006;67:1221–1224. doi: 10.1212/01.wnl.0000238079.00472.3a. [DOI] [PubMed] [Google Scholar]
- 9.Reiman EM, et al. Preclinical evidence of Alzheimer's disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- 10.Small GW, et al. Cerebral metabolic and cognitive decline in persons at genetic risk for Alzheimer's disease. Proc Natl Acad Sci USA. 2000;97:6037–6042. doi: 10.1073/pnas.090106797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reiman EM, et al. Functional brain abnormalities in young adults at genetic risk for late-onset Alzheimer's dementia. Proc Natl Acad Sci USA. 2004;101:284–289. doi: 10.1073/pnas.2635903100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bookheimer SY, et al. Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med. 2000;343:450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burggren AC, Small GW, Sabb FW, Bookheimer SY. Specificity of brain activation patterns in people at genetic risk for Alzheimer disease. Am J Geriatr Psychiatry. 2002;10:44–51. [PubMed] [Google Scholar]
- 14.Bondi MW, Houston WS, Eyler LT, Brown GG. fMRI evidence of compensatory mechanisms in older adults at genetic risk for Alzheimer disease. Neurology. 2005;64:501–508. doi: 10.1212/01.WNL.0000150885.00929.7E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wishart HA, et al. Increased brain activation during working memory in cognitively intact adults with the APOE epsilon4 allele. Am J Psychiatry. 2006;163:1603–1610. doi: 10.1176/ajp.2006.163.9.1603. [DOI] [PubMed] [Google Scholar]
- 16.Fleisher AS, et al. Identification of Alzheimer disease risk by functional magnetic resonance imaging. Arch Neurol. 2005;62:1881–1888. doi: 10.1001/archneur.62.12.1881. [DOI] [PubMed] [Google Scholar]
- 17.Mondadori CR, et al. Better memory and neural efficiency in young apolipoprotein E epsilon4 carriers. Cereb Cortex. 2007;17:1934–1947. doi: 10.1093/cercor/bhl103. [DOI] [PubMed] [Google Scholar]
- 18.Filbey FM, Slack KJ, Sunderland TP, Cohen RM. Functional magnetic resonance imaging and magnetoencephalography differences associated with APOEepsilon4 in young healthy adults. NeuroReport. 2006;17:1585–1590. doi: 10.1097/01.wnr.0000234745.27571.d1. [DOI] [PubMed] [Google Scholar]
- 19.Shaw P, et al. Cortical morphology in children and adolescents with different apolipoprotein E gene polymorphisms: An observational study. Lancet Neurol. 2007;6:494–500. doi: 10.1016/S1474-4422(07)70106-0. [DOI] [PubMed] [Google Scholar]
- 20.Scarmeas N, et al. APOE related alterations in cerebral activation even at college age. J Neurol Neurosurg Psychiatry. 2005;76:1440–1444. doi: 10.1136/jnnp.2004.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raichle ME, et al. A default mode of brain function. Proc Natl Acad Sci USA. 2001;98:676–682. doi: 10.1073/pnas.98.2.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boly M, et al. Intrinsic brain activity in altered states of consciousness: How conscious is the default mode of brain function? Ann NY Acad Sci. 2008;1129:119–129. doi: 10.1196/annals.1417.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buckner RL, et al. Molecular, structural, and functional characterization of Alzheimer's disease: Evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 2005;25:7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greicius MD, Srivastava G, Reiss AL, Menon V. Default-mode network activity distinguishes Alzheimer's disease from healthy aging: Evidence from functional MRI. Proc Natl Acad Sci USA. 2004;101:4637–4642. doi: 10.1073/pnas.0308627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sorg C, et al. Selective changes of resting-state networks in individuals at risk for Alzheimer's disease. Proc Natl Acad Sci USA. 2007;104:18760–18765. doi: 10.1073/pnas.0708803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirano A, Zimmerman HM. Alzheimer's neurofibrillary changes. A topographic study. Arch Neurol. 1962;7:227–242. doi: 10.1001/archneur.1962.04210030065009. [DOI] [PubMed] [Google Scholar]
- 27.Andersen OM, Willnow TE. Lipoprotein receptors in Alzheimer's disease. Trends Neurosci. 2006;29:687–694. doi: 10.1016/j.tins.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 28.Squire LR, Zola-Morgan S. The medial temporal lobe memory system. Science. 1991;253:1380–1386. doi: 10.1126/science.1896849. [DOI] [PubMed] [Google Scholar]
- 29.Tulving E, Markowitsch HJ, Kapur S, Habib R, Houle S. Novelty encoding networks in the human brain: Positron emission tomography data. NeuroReport. 1994;5:2525–2528. doi: 10.1097/00001756-199412000-00030. [DOI] [PubMed] [Google Scholar]
- 30.Oakes TR, et al. Integrating VBM into the General Linear Model with voxelwise anatomical covariates. NeuroImage. 2007;34:500–508. doi: 10.1016/j.neuroimage.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Beckmann CF, DeLuca M, Devlin JT, Smith SM. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc London B. 2005;360:1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Golby A, et al. Memory encoding in Alzheimer's disease: An fMRI study of explicit and implicit memory. Brain. 2005;128(Pt 4):773–787. doi: 10.1093/brain/awh400. [DOI] [PubMed] [Google Scholar]
- 33.Douaud G, et al. Anatomically related grey and white matter abnormalities in adolescent-onset schizophrenia. Brain. 2007;130(Pt 9):2375–2386. doi: 10.1093/brain/awm184. [DOI] [PubMed] [Google Scholar]
- 34.Gabrieli JD, Brewer JB, Desmond JE, Glover GH. Separate neural bases of two fundamental memory processes in the human medial temporal lobe. Science. 1997;276:264–266. doi: 10.1126/science.276.5310.264. [DOI] [PubMed] [Google Scholar]
- 35.Shohamy D, Wagner AD. Integrating memories in the human brain: Hippocampal-midbrain encoding of overlapping events. Neuron. 2008;60:378–389. doi: 10.1016/j.neuron.2008.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreasen NC, et al. Short-term and long-term verbal memory: A positron emission tomography study. Proc Natl Acad Sci USA. 1995;92:5111–5115. doi: 10.1073/pnas.92.11.5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fliessbach K, Trautner P, Quesada CM, Elger CE, Weber B. Cerebellar contributions to episodic memory encoding as revealed by fMRI. NeuroImage. 2007;35:1330–1337. doi: 10.1016/j.neuroimage.2007.02.004. [DOI] [PubMed] [Google Scholar]
- 38.Buxton RB, Uludag K, Dubowitz DJ, Liu TT. Modeling the hemodynamic response to brain activation. NeuroImage. 2004;23(Suppl 1):S220–S233. doi: 10.1016/j.neuroimage.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 39.Brown GG, et al. BOLD and perfusion response to finger-thumb apposition after acetazolamide administration: Differential relationship to global perfusion. J Cereb Blood Flow Metab. 2003;23:829–837. doi: 10.1097/01.WCB.0000071887.63724.B2. [DOI] [PubMed] [Google Scholar]
- 40.Fleisher AS, et al. Cerebral perfusion and oxygenation differences in Alzheimer's disease risk. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.01.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Arendt T, et al. Plastic neuronal remodeling is impaired in patients with Alzheimer's disease carrying apolipoprotein epsilon 4 allele. J Neurosci. 1997;17:516–529. doi: 10.1523/JNEUROSCI.17-02-00516.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Masliah E, et al. Neurodegeneration in the central nervous system of apoE-deficient mice. Exp Neurol. 1995;136:107–122. doi: 10.1006/exnr.1995.1088. [DOI] [PubMed] [Google Scholar]
- 43.Buttini M, et al. Expression of human apolipoprotein E3 or E4 in the brains of Apoe−/− mice: Isoform-specific effects on neurodegeneration. J Neurosci. 1999;19:4867–4880. doi: 10.1523/JNEUROSCI.19-12-04867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun Y, et al. Glial fibrillary acidic protein-apolipoprotein E (apoE) transgenic mice: Astrocyte-specific expression and differing biological effects of astrocyte-secreted apoE3 and apoE4 lipoproteins. J Neurosci. 1998;18:3261–3272. doi: 10.1523/JNEUROSCI.18-09-03261.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bellosta S, et al. Stable expression and secretion of apolipoproteins E3 and E4 in mouse neuroblastoma cells produces differential effects on neurite outgrowth. J Biol Chem. 1995;270:27063–27071. doi: 10.1074/jbc.270.45.27063. [DOI] [PubMed] [Google Scholar]
- 46.Trommer BL, et al. ApoE isoform-specific effects on LTP: Blockade by oligomeric amyloid-beta1–42. Neurobiol Dis. 2005;18:75–82. doi: 10.1016/j.nbd.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 47.Yun SH, et al. Blockade of nicotinic acetylcholine receptors suppresses hippocampal long-term potentiation in wild-type but not ApoE4 targeted replacement mice. J Neurosci Res. 2005;82:771–777. doi: 10.1002/jnr.20684. [DOI] [PubMed] [Google Scholar]
- 48.Bliss TV, Collingridge GL. A synaptic model of memory: Long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 49.Williams JH, Errington ML, Lynch MA, Bliss TV. Arachidonic acid induces a long-term activity-dependent enhancement of synaptic transmission in the hippocampus. Nature. 1989;341:739–742. doi: 10.1038/341739a0. [DOI] [PubMed] [Google Scholar]
- 50.Sanchez-Mejia RO, et al. Phospholipase A2 reduction ameliorates cognitive deficits in a mouse model of Alzheimer's disease. Nat Neurosci. 2008;11:1311–1318. doi: 10.1038/nn.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Menzel HJ, Kladetzky RG, Assmann G. Apolipoprotein E polymorphism and coronary artery disease. Arteriosclerosis. 1983;3:310–315. doi: 10.1161/01.atv.3.4.310. [DOI] [PubMed] [Google Scholar]
- 52.Benjamin R, et al. Apolipoprotein E genes in Lewy body and Parkinson's disease. Lancet. 1994;343:1565. doi: 10.1016/s0140-6736(94)92961-0. [DOI] [PubMed] [Google Scholar]
- 53.Wilson PW, et al. Apolipoprotein E alleles, dyslipidemia, and coronary heart disease. The Framingham Offspring Study. Jama. 1994;272:1666–1671. [PubMed] [Google Scholar]
- 54.Schachter F, et al. Genetic associations with human longevity at the APOE and ACE loci. Nat Genet. 1994;6:29–32. doi: 10.1038/ng0194-29. [DOI] [PubMed] [Google Scholar]
- 55.Nichols TE, Holmes AP. Nonparametric permutation tests for functional neuroimaging: A primer with examples. Hum Brain Mapp. 2002;15:1–25. doi: 10.1002/hbm.1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Woolrich MW, Behrens TE, Beckmann CF, Jenkinson M, Smith SM. Multilevel linear modelling for FMRI group analysis using Bayesian inference. NeuroImage. 2004;21:1732–1747. doi: 10.1016/j.neuroimage.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 57.Woolrich M. Robust group analysis using outlier inference. NeuroImage. 2008;41:286–301. doi: 10.1016/j.neuroimage.2008.02.042. [DOI] [PubMed] [Google Scholar]
- 58.MacIntosh BJ, et al. Measuring the effects of remifentanil on cerebral blood flow and arterial arrival time using 3D GRASE MRI with pulsed arterial spin labelling. J Cereb Blood Flow Metab. 2008;28:1514–1522. doi: 10.1038/jcbfm.2008.46. [DOI] [PubMed] [Google Scholar]