One of the most important recent discoveries in the field of genome biology has been the demonstration that genomes are nonrandomly organized in the cell nucleus (1, 2). Examples now abound of chromosomes or genes localizing to a particular location in the nucleus or undergoing positional changes as they became activated or repressed. Much of what we know about how genomes are organized in space and time comes from observation of single genes and is based on cytological, often descriptive, studies. The Holy Grail in this field is to elucidate what the mechanisms are that determine where a gene or a chromosome localizes within the cell nucleus. In a recent issue of PNAS, using a combined cytological and computational approach, Rajapakse et al. (3) make the first step toward this goal, and their findings support the provocative idea that genomes are self-organizing entities.
There are several clear indications for the nonrandomness of genome organization in the mammalian cell nucleus (1, 2). For one, specific chromosomes occupy distinct positions with respect to the periphery of the nucleus. In line with an abundance of transcriptionally silent heterochromatin at the nucleus' edge, gene-poor chromosomes, and those with low overall transcriptional activity, preferentially associate with the nuclear envelope, whereas gene-rich, highly transcribed, chromosomes are situated in the center (1, 2). The same correlation has been made for single genes in numerous reported cases where inactive genes move from the periphery to the center, although this simple rule does not apply to all, and probably not even the majority, of genes (4, 5). Increasing evidence also points to an important role of regulatory interactions among multiple chromosomes (5). For example, activation of the IFN-beta gene located on human chromosome 9 involves its physical association with regulatory enhancers on chromosomes 4 and 18 (6). The mechanisms that drive any of these nonrandom genome organization events are unknown.
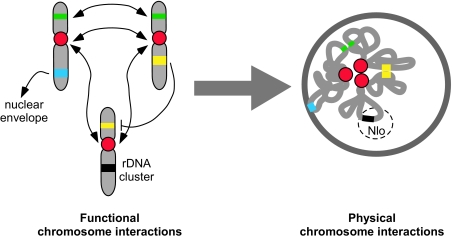
Self-organization is an intriguing and attractive mechanism to bring about the patterns of spatial genome organization observed in the mammalian nucleus (2). In a self-organization model the arrangement of chromosomes and genes is largely driven by the sum of all interactions acting on a given genomic region such as the association of a regulatory element on one chromosome with a target gene on another, tethering of genes to the nuclear periphery or clustering of active genes in the nuclear interior (Fig. 1). Each interaction imposes a constraint on a given chromosome, reducing its degrees of spatial freedom and thus contributing toward the formation of a global localization pattern. The difficulty with evaluating self-organization models has been that available experimental data on gene-positioning patterns have been limited to single genes and have thus not been sufficient to critically test a self-organization model that would apply to the full genome.
Fig. 1.
Self-organization in the genome. Functional interactions among chromosomes give rise to chromosome localization patterns. Interactions include association of centromeres (red), clustering of coregulated genes (green), association of a regulatory element and its target genes (yellow), interaction of a genome region with the nuclear envelope (blue) or clusters of rDNA (black) genes. Each interaction constrains the chromosome's motion, and the sum of all interactions ultimately determines the position of a chromosome relative to all others. The interchromosomal interactions may also indirectly determine the position of a chromosome relative to the center/periphery of the nucleus. Nlo, nucleolus.
Taking a global approach, Rajapakse et al. (3) now provide evidence for self-organizing in the genome. They explore this problem in a well established mouse hematopoietic differentiation system, in which multipotential precursor cells are differentiated either into erythrocytes or neutrophils depending on growth conditions. Rajapakse et al. first defined the genome-wide expression profile for both lineages at multiple time points during differentiation, determined on what chromosomes coregulated genes reside and asked whether there is a link between the presence of coregulated genes on a given chromosome and the chromosome's position relative to all other chromosomes. To simplify this analysis they used the arrangement of chromosomes in so-called rosettes as a surrogate. Chromosome rosettes form for a short period during cell division when condensed chromosomes align in the cell in a circle and they are useful since they allow relatively facile mapping of the spatial arrangement of all chromosomes relative to all others. Applying advanced matrix-based computational tools to this comparative analysis, Rajapakse et al. find a sound correlation between the spatial organization of chromosomes during differentiation and the set of coregulated genes, strongly suggesting that the gene expression activity of a given chromosome contributes to its position.
Self-organization models are notoriously difficult to test because it is not possible to experimentally manipulate a single component of a specific pathway without nonspecifically affecting the entire system. One approach to validate self-organization models is by computational simulation. When Rajapakse et al. (3) used an oscillator-based self-organization model to measure to what degree each chromosome associates with all other chromosomes in rosettes as a function of the expressed genes on a chromosome, they found that the chromosome distribution patterns and expression profiles are tightly linked and that the resulting chromosome association network is very similar to that predicted by a self-organization model.
The computationally derived results by Rajapakse et al. (3) do not stand alone in support of genome self-organization, but are consistent with several experimental observations. In the same differentiation system used here, a number of homologous chromosome pairs have previously been found to be preferentially proximal to each other in rosettes. The frequency of homologue proximity varied with cell type and was related to the number of coregulated genes residing on the chromosome (7). Furthermore, in erythrocytes, coregulated genes have been reported to coalesce near each other, possibly being attracted by shared transcription and RNA-processing sites that contain high levels of specific factors required for expression of these genes (8, 9). In yeast, tRNA genes located on distinct chromosomes cluster in 3D space and in yeast and mammalian cells rDNA genes on multiple chromosomes coalesce to form transcription hot spots (10).
Self-organization is a good candidate mechanism to bring about spatial genome organization because it accounts for several prominent features of chromosome and gene-positioning patterns. One of the most important ones is the well established probabilistic nature of positioning patterns, whereby a given chromosome can occupy distinct positions in individual cells, even within the same cell population. We now know that gene expression profiles are not uniform among single cells (11) and it is likely that differences in the sets of expressed genes in a cell compared with its neighbor contributes to the differences in positioning among cells. A self-organizing mechanism can also explain the changes in genome-positioning patterns observed during differentiation, development, and disease that may be driven by changes in gene expression programs. Interestingly, self-organization may also play a larger role in nuclear organization because recent findings on the biogenesis of nuclear bodies have suggested that they are formed by self-organization (12), indicating that nuclear architecture is altogether largely driven by nuclear function in a self-organization process (13).
Chromosome distribution patterns and expression profiles are tightly linked.
Rajapakse et al. (3) have made an important first step toward understanding the molecular mechanisms of spatial genome organization. But first steps are always difficult and there are some limitations in this initial analysis. One is that transcriptionally silent mitotic chromosomes and not interphase chromosome patterns were analyzed. Although the authors previously validated that homologue proximity observed in rosettes was also present in interphase nuclei (7), there is debate about how closely rosettes reflect interphase genome organization. The analysis also relies heavily on novel computational tools and a highly simplified model that only accommodates a very small fraction of parameters that influence genome organization and is at best only a first approximation of biological reality. Finally, although the conclusions of the study are consistent with the model, they have not been experimentally tested. It is fair to argue that these first steps in understanding the mechanisms of genome organization are somewhat unsteady, but they are undoubtedly in the right direction. They blaze the trail in that the use of global analyses, rather than single-gene interrogations, and the application of advanced computational tools will soon become the standard in genome organization studies.
Footnotes
The author declares no conflict of interest.
See companion article on page 6679 in issue 16 of volume 106.
References
- 1.Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8:104–115. doi: 10.1038/nrg2041. [DOI] [PubMed] [Google Scholar]
- 2.Misteli T. Beyond the sequence: Cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 3.Rajapakse I, et al. The emergence of lineage-specific chromosomal topologies from coordinate gene expression. Proc Natl Acad Sci USA. 2009;106:6679–6684. doi: 10.1073/pnas.0900986106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kosak ST, Groudine M. Form follows function: The genomic organization of cellular differentiation. Genes Dev. 2004;18:1371–1384. doi: 10.1101/gad.1209304. [DOI] [PubMed] [Google Scholar]
- 5.Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Apostolou E, Thanos D. Virus infection induces NF-kappaB-dependent interchromosomal associations mediating monoallelic IFN-beta gene expression. Cell. 2008;134:85–96. doi: 10.1016/j.cell.2008.05.052. [DOI] [PubMed] [Google Scholar]
- 7.Kosak ST, et al. Coordinate gene regulation during hematopoiesis is related to genomic organization. PLoS Biol. 2007;5:e309. doi: 10.1371/journal.pbio.0050309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown JM, et al. Chromatin environment and splicing factor aggregations are stochastic modulators of association between active genes. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne CS, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 10.Thompson M, Haeusler RA, Good PD, Engelke DR. Nucleolar clustering of dispersed tRNA genes. Science. 2003;302:1399–1401. doi: 10.1126/science.1089814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 12.Kaiser TE, Intine RV, Dundr M. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 13.Misteli T. The concept of self-organization in cellular architecture. J Cell Biol. 2001;155:181–185. doi: 10.1083/jcb.200108110. [DOI] [PMC free article] [PubMed] [Google Scholar]