Abstract
Mutations in breast cancer susceptibility gene 1 and 2 (BRCA1 and BRCA2) predispose individuals to breast and ovarian cancer development. We previously reported an in vivo interaction between BRCA1 and BRCA2. However, the biological significance of their association is thus far undefined. Here, we report that PALB2, the partner and localizer of BRCA2, binds directly to BRCA1, and serves as the molecular scaffold in the formation of the BRCA1-PALB2-BRCA2 complex. The association between BRCA1 and PALB2 is primarily mediated via apolar bonding between their respective coiled-coil domains. More importantly, BRCA1 mutations identified in cancer patients disrupted the specific interaction between BRCA1 and PALB2. Consistent with the converging functions of the BRCA proteins in DNA repair, cells harboring mutations with abrogated BRCA1-PALB2 interaction resulted in defective homologous recombination (HR) repair. We propose that, via its direct interaction with PALB2, BRCA1 fine-tunes recombinational repair partly through its modulatory role in the PALB2-dependent loading of BRCA2-RAD51 repair machinery at DNA breaks. Our findings uncover PALB2 as the molecular adaptor between the BRCA proteins, and suggest that impaired HR repair is one of the fundamental causes for genomic instability and tumorigenesis observed in patients carrying BRCA1, BRCA2, or PALB2 mutations.
Keywords: BRCA1, BRCA2, FANCN
Breast cancer and ovarian cancer is estimated to be responsible for more than one-fifth of cancer mortality (1). Because germ-line mutations in breast cancer susceptibility gene 1 and 2 (BRCA1 and BRCA2) account for the development of a significant portion of hereditary breast and ovarian cancer, the understanding of their roles in tumor suppression is crucial to the improvement of therapeutic interventions. Because accumulation of genetic aberrations are often observed in cells derived from familial breast cancer patients with BRCA1 or BRCA2 mutations, the BRCA proteins have been considered as the caretakers of genomic integrity. Accordingly, tumor cells derived from these patients exhibit hypersensitivity to DNA damaging agents and display genomic instability (2–4).
Given the similar phenotypes in BRCA1 and BRCA2 patients, and the spectrum of deficits observed in cells deficient in these proteins, one would envision that the BRCA proteins might work in synchrony in certain cellular process(es) essential for tumor suppression. Consistent with this notion, interaction between the 2 BRCA proteins has been reported (5). However, exactly how their interaction is regulated and the biological significance for such interaction remains largely unexplored.
BRCA1 participates in numerous cellular processes (3, 6–8). In particular, BRCA1 has been proposed to have diverse roles to promote cell survival in response to genotoxic stress. Recent elucidation of multiple BRCA1 complexes in vivo suggests a multifactorial model by which BRCA1 mediates distinct processes that include checkpoint activation, damage signaling, and DNA repair (9, 10). However, BRCA2 has a pivotal role in the initiation of DNA repair, namely by loading of repair protein RAD51 onto single-stranded DNA for homologous recombination (HR) (11–13). More recently, Xia et al. (14) identified PALB2, the partner and localizer of BRCA2, as an essential component that is required for the loading of the BRCA2-RAD51 repair complex onto DNA. Similar to BRCA1 and BRCA2, PALB2 mutations have also been implicated in the predisposition of individuals to breast cancer development (15–20). That patients harbor PALB2 mutation carries normal BRCA1 and BRCA2 suggests that these 3 proteins might be functionally linked.
In the current study, we provide direct evidence to support that PALB2 serves as the bridging molecule that connects BRCA1 and BRCA2. Our data suggest that PALB2 is an integral component of the BRCA1-BRCA2-RAD51 axis, which is critical for the maintenance of genomic stability via recombinational repair.
Results
BRCA1 Is a PALB2 Interacting Protein.
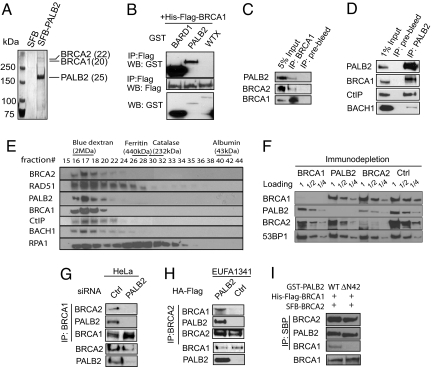
To identify proteins that interact with PALB2, we adopted a tandem affinity purification (TAP) scheme using lysate derived from 293T cells stably expressing streptavidin binding peptide-Flag-S protein (SFB)-tagged PALB2. Mass spectrometry analyses of proteins that copurified with PALB2 revealed peptides that corresponded not only to BRCA2, but also BRCA1 (Fig. 1A). By employing insect SF9 cells and baculovirus expression system, we confirmed that PALB2 does indeed interact directly with BRCA1 (Fig. 1B; Fig. S1b). To examine whether endogenous PALB2 and BRCA1 interact in vivo, we performed coimmunoprecipitation (coIP) experiment using 293T cell lysate. Consistent with TAP results, both PALB2 and BRCA2 were found in BRCA1 precipitates (Fig. 1C). Conversely, BRCA1 and BRCA1-interacting proteins CtIP and BACH1 were also observed in PALB2 immunoprecipitates (Fig. 1D), suggesting that PALB2 is a component of BRCA1 complexes in vivo.
Fig. 1.
Identification of PALB2 as an integral component of the BRCA protein complex. (A) Cell lysate prepared from 293T cells expressing SFB-PALB2 was subjected to TAP. PALB2-associated proteins were subsequently separated by SDS/PAGE and visualized by silver staining. Mass spectrometry analyses revealed the identities of the PALB2-associated proteins and their respective numbers of peptides obtained were shown in brackets. (B) SF9 cells were coinfected with baculoviruses expressing His-Flag-BRCA1 in addition to those expressing GST-PALB2, GST-BARD1, or GST-WTX. Cell lysates were subjected to IP by using anti-Flag (M2) beads. Immunoblotting was conducted as indicated. (C) CoIP of BRCA1 with PALB2 and BRCA2 was carried out using anti-BRCA1 antibody and immunoblotted with indicated antibodies. (D) Detection of PALB2-associated proteins; 293T cell lysates were subjected to IP using anti-PALB2 serum, and immunoblotting was carried out using antibodies as indicated. (E) Gel filtration chromatography analysis was performed using 293T cell lysate. Proteins eluted from the indicated fractions were separated by SDS/PAGE and analyzed by Western blotting using antibodies as indicated. (F) The 293T cell lysates were subjected to 3 rounds of immunodepletion using rabbit polyconal antibodies specifically raised against BRCA1, PALB2, or BRCA2. The resulting supernatants were examined for the presence of remaining BRCA1, PALB2, BRCA2, and 53BP1 content by immunoblotting using corresponding antibodies. (G) HeLa cells transfected with control or PALB2 siRNAs were subjected to IP using anti-BRCA1 antibody. Immunoblotting of whole-cell extracts or IPed samples was performed using antibodies as indicated. (H) Anti-BRCA2 IP was performed using lysates prepared from PALB2-deficient EUFA1341 cells or derivative cells reconstituted with HA-Flag-tagged PALB2. Immunoblotting of whole-cell extracts or IPed samples was performed using antibodies as indicated. (I) PALB2 bridges the BRCA1 and BRCA2 interaction in vitro. SF9 cells were infected with baculoviruses expressing His-Flag-BRCA1 and SFB-BRCA2, together with viruses expressing either GST-PALB2 or GST-PALB2 ΔN42. IP was carried out using streptavidin beads and immunoblotted with antibodies as indicated.
BRCA1 Exists in a Protein Complex with PALB2 and BRCA2.
Because it has been reported that a significant portion of PALB2 associates with BRCA2 (14), and BRCA1 also interacts with BRCA2 (5), we asked whether BRCA1, PALB2, and BRCA2 coexist in a protein complex. We performed gel filtration chromatography using nuclease-treated 293T cell lysate to identify possible native protein complexes that contain BRCA1, PALB2, and BRCA2. All of the 3 proteins were found in overlapping fractions that corresponded to ≈2 MDa (Fig. 1E). These results indicated that BRCA1, BRCA2, and PALB2 possibly exist as a protein complex in vivo. Also, CtIP and BACH1, as well as RAD51 and RPA, were also concentrated in these fractions.
To further substantiate that PALB2 and the BRCA proteins form a single complex, serial immunodepletion experiment was also performed. Consistent with previous observations (14), PALB2 depletion resulted in a significant reduction (≈50%) in the amount of BRCA2 (Fig. 1F). Also, PALB2 and BRCA2 protein levels were also reduced on BRCA1 depletion. Together, these results suggest that at least a fraction of BRCA1 associates with PALB2 and BRCA2 as a single protein complex.
One common feature of many DNA damage/repair proteins is their ability to localize and concentrate at sites of DNA breaks, forming discrete foci that colocalize with the double-stranded DNA break marker pH2AX. Because we found that BRCA1 associates with PALB2 and BRCA2 in vivo, we examined the extent of colocalization among these 3 proteins at DNA breaks in U2OS cells after ionizing radiation (IR). As shown in Fig. S1c, foci structures that contained BRCA1 and BRCA2, BRCA1 and PALB2, or BRCA1 and RAD51 were easily observed. Quantification analyses indicate that most of the PALB2, BRCA2, and RAD51 foci colocalize with that of BRCA1 (≈80%; Fig. S1d), which is similar to the extent of colocalization between PALB2 and RAD51 in PALB2-deficient EUFA1341 cell that has been reconstituted with flag-PALB2. These results suggest that the protein complex containing BRCA1, PALB2, and BRCA2 is concentrated at sites of DNA breaks, and may have a role in DNA damage signaling and/or DNA repair.
PALB2 Serves As the Molecular Scaffold for the Formation of BRCA Protein Complex.
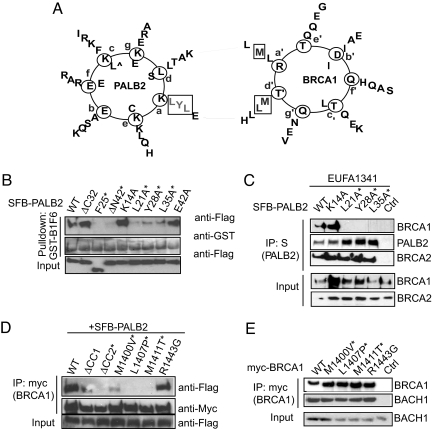
To study the functional significance of this complex formation, we first mapped the regions required for the BRCA1/PALB2 interaction. SFB-tagged wild-type PALB2 and a series of deletion mutants that span the entire PALB2 ORF (Fig. S2a) were subjected to coIP with full-length myc-BRCA1. Results showed that the PALB2 N terminus (P2F1; residues 1–319) is responsible for BRCA1 binding (Fig. S2b). Because a coiled-coil domain (residues 9–42) resides within this region, we tested whether it might be responsible for the association of PALB2 with BRCA1. Indeed, deletion mutant lacking the coiled-coil structure (P2ΔN42) was defective in BRCA1 binding (Fig. S2c), suggesting that BRCA1 binds directly to the very N terminus of PALB2.
Next, we sought to define the PALB2-binding region on BRCA1. We made use of a panel of myc-tagged deletion mutants that span the BRCA1 coding sequence (Fig. S3a). Pull-down assays indicated that full-length BRCA1 specifically associated with PALB2 in vitro (Fig. S3b). Further GST pull-down mapped the PALB2-binding region to residues 1314–1557 (Fig. S3 a and b). A more refined series of deletion mutants (Fig. S3c) narrowed down the binding region to residues 1364–1437 (Fig. S3d). Interestingly, this region (residues 1364–1437) of BRCA1 also harbors a putative coiled-coil domain. Internal deletion mutants of the predicted BRCA1 coiled-coil domains (B1ΔCC1 and B1ΔCC2) were generated (Fig. S3c). CoIP experiments demonstrated that although the wild-type and the B1ΔCC1 coIPed with PALB2, the B1ΔCC2 did not (Fig. S3e), indicating that the second coiled-coil domain (CC2) on BRCA1 is essential for its interaction with PALB2.
Intriguingly, the interaction domains between BRCA1 and PALB2 identified in this study (Fig. S2 and Fig. S3) overlap with the BRCA1-BRCA2 interaction domain identified previously (5). This observation prompted us to speculate that PALB2 may be the protein that links BRCA1 and BRCA2. To test this possibility, lysates prepared from HeLa cells transfected with control or PALB2 siRNA were subjected to IP by using anti-BRCA1 antibodies. Results showed that BRCA2 coIPed with BRCA1 only in the presence of PALB2 (Fig. 1G), suggesting that the interaction between BRCA1 and BRCA2 requires PALB2. Likewise, the BRCA1 and BRCA2 interaction was not observed in the PALB2 deficient EUFA1341 cells (Fig. 1H). However, when these cells were reconstituted with wild-type PALB2, the BRCA1/BRCA2 interaction was readily detected (Fig. 1H).
To further substantiate the notion that PALB2 bridges the interaction between BRCA proteins, coIP experiments were performed using recombinant His-Flag-BRCA1 and SFB-BRCA2 together with GST-PALB2 or GST-PALB2 ΔN42 expressed in insect cells. As shown in Fig. 1I, in the presence of wild-type PALB2, BRCA2 associated with both PALB2 and BRCA1. In contrast, BRCA1 failed to interact with BRCA2 in cells expressing the PALB2 ΔN42 mutant, which has compromised association with BRCA1 (Fig. S2c), although it exhibits intact interaction with BRCA2 (Fig. 1I). These data firmly established PALB2 as the bridging protein that mediates BRCA1/BRCA2 interaction.
Structural Requirements for the BRCA1-PALB2 Interaction.
Coiled-coil domains are known to mediate protein–protein interactions. Based on the results shown in Fig. S2 and Fig. S3, we suspected that the BRCA1-PALB2 interaction might be mediated by intermolecular interaction between their respective coiled-coil motifs. Therefore, we further studied this interaction, and generated alanine substitution mutations for residues at position “a” in the heptad-helical coiled-coil arrangement on PALB2 (K14A, L21A, Y28A, L35A, and E42A) (Fig. 2A). Pull-down experiments using GST-BRCA1 showed that the interactions between BRCA1 and the PALB2 L21A, Y28A, or L35A mutants were largely abolished (Fig. 2B), indicating that these residues are likely to be the contacting sites at the PALB2-BRCA1 interface. Reconstitution of these PALB2 mutants into PALB2-deficient EUFA1341 cells confirmed their compromised binding to BRCA1, whereas these mutants interacted normally with BRCA2 (Fig. 2C).
Fig. 2.
Determination of regions required for the BRCA1-PALB2 interaction. (A) Graphical projection of association between PALB2 (residues 9–42) and BRCA1 (residues 1393–1424) coiled-coil domains. Positions of the heptad repeat (positions a to g) were predicted by the Coil program (window = 28) (36). Boxed residues were experimentally demonstrated to be responsible for the hetero-oligomeric interaction between PALB2 and BRCA1. (B and C) PALB2 position a mutants disrupted the interactions of PALB2 with BRCA1, but not that with BRCA2. Lysates prepared from 293T cells expressing different SFB-PALB2 mutants were subjected to pull-down assay using beads coated with GST-B1F6 (B). Alternatively, PALB2-deficient EUFA1341 cells were reconstituted with SFB-tagged wild-type or mutants of PALB2, and IP was carried out using anti-Flag M2 beads (C). Immunoblotting was performed using indicated antibodies. Asterisks indicate mutants with disrupted binding with BRCA1. (D) Patient-derived BRCA1 mutations abolished the BRCA1-PALB2 association. CoIP experiments were performed using lysates derived from 293T cells expressing SFB-PALB2, together with myc-tagged wild-type or point mutants of BRCA1. Asterisks indicate mutants with disrupted binding to PALB2. (E) Interaction of wild-type or mutant BRCA1 with BACH1; 293T cell lysates expressing myc-tagged wild-type or mutant BRCA1 were subjected to IP using anti-myc antibodies, and immunoblotted using indicated antibodies.
Within the coiled-coil domain of BRCA1 required for PALB2-binding (residues 1364–1437), we identified 3 BRCA1 missense mutations found among cancer patients (the Breast cancer information core database and the Human mutation database). Strikingly, all of these patient mutations (M1400V, L1407P, and M1411T) coincided with position a or “d” in the heptad coiled-coil (CC2) on BRCA1 (Fig. 2A). CoIP experiments revealed that, unlike the control mutation R1443G, all of the 3 point mutations within the BRCA1 CC2 region resulted in attenuated PALB2 interactions (Fig. 2D), although they have negligible effect on their interactions with BACH1 (Fig. 2E). These results imply that the interaction between BRCA1 and PALB2 may have a role in the tumor suppression functions of these proteins.
Independent Requirements for PALB2 and BRCA1 Focal Accumulation at DNA Double-Strand Breaks (DSBs).
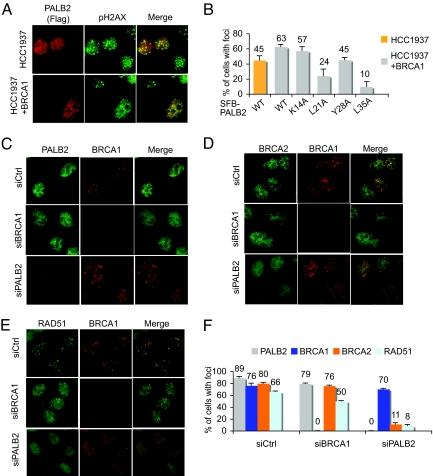
We next examined whether the physical interaction between BRCA1 and PALB2 might be important for their localization at sites of DNA breaks. IR-induced BRCA1 foci were observed for wild-type BRCA1, as well as all of the BRCA1 mutants within the CC2 region, regardless of their PALB2-binding activity (Fig. S4 a and c). Similarly, discrete foci were also detected for the BRCA1-binding defective mutants of PALB2 (Fig. S4b), although a reduction in the percentage of foci positive cells was observed for some of these mutants when compared with wild-type PALB2 (Fig. S4d). However, the PALB2 ΔF mutation (lacking residues 71–561), which retains BRCA1-binding motif, showed a drastic reduction in its foci formation capability (Fig. S4 b and d). Therefore, the foci forming abilities of these PALB2 mutants do not strictly correlate with their BRCA1-binding activities.
Exogenous PALB2 foci were also readily observed in HCC1937 cells (Fig. 3 A and B), which express only a truncated BRCA1 that lacks BRCT domain, and could not be recruited to site of DSB (Fig. S4e). This result indicates that sustained localization of PALB2 at DNA breaks does not require prior focal accumulation of BRCA1. It is noteworthy to mention that PALB2 foci forming ability, although not entirely BRCA1-dependent, as observed in and Fig. 3 A–C and Fig. S4, is modestly elevated in HCC1937 cells reconstituted with wild-type BRCA1 (HCC1937+BRCA1) (Fig. 3B), suggesting that BRCA1 may help stabilize PALB2 at DNA damage sites.
Fig. 3.
Distinct requirements for sustained localization of BRCA1 and PALB2 at DNA damage sites. (A) IR-induced PALB2 foci formation in HCC1937 and HCC937+BRCA1 cells was analyzed by coimmunostaining using anti-Flag and anti-pH2AX antibodies. (B) Quantification of IR-induced foci formation of wild-type or mutant PALB2 in HCC1937 or HCC1937+BRCA1 cells. (C–F) Representative pictures of coimmunostaining of PALB2 (C), BRCA2 (D), and RAD51 (E) with BRCA1 in U2OS cells treated with control, BRCA1, or PALB2 specific siRNAs. (F) Quantification of the percentage of foci positive cells as described in C–E.
Previous report implicated an essential role of PALB2 in the loading of BRCA2-RAD51 at DNA breaks (Fig. S5 a and b) (14, 21). In agreement with the proposed accessory role of BRCA1 in stabilizing PALB2 at DNA breaks, RAD51 foci were modestly reduced (≈2-fold) in HCC1937 cells, when compared with the same cells reconstituted with wild-type BRCA1 (Fig. S5 c and d) (22). To further explore whether BRCA1 might be essential for focal accumulation of PALB2, BRCA2, and RAD51 after DNA damage, we examined the localization of these proteins in U2OS cells with siRNA-mediated depletion of BRCA1. BRCA1 depletion led to a modest reduction in the foci forming ability of PALB2, BRCA2, and RAD51 (Fig. 3 C–F). However, in consistent with those observed in PALB2 deficient patient cells (Fig. S5 a and b), PALB2 depletion in U2OS cells resulted in a significant decrease in the number of cells containing damage-induced BRCA2 or RAD51 foci (Fig. 3 C–F), confirming that PALB2 is required for the stable accumulation of both BRCA2 and RAD51 at DNA damage sites. However, PALB2-depleted cells still displayed apparent normal BRCA1 foci formation after DNA damage (Fig. 3 C–F). Together, these results suggest that BRCA1 and PALB2 are recruited to DNA lesions via independent mechanisms.
The BRCA1-PALB2 Interaction Is Involved in HR Repair.
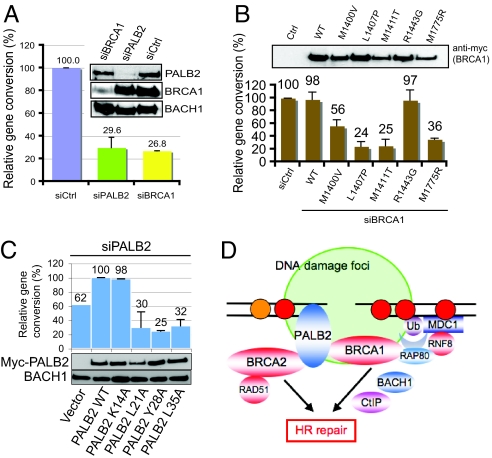
Because PALB2 has a critical role in HR repair through its ability to recruit BRCA2 and RAD51 to DNA breaks, we tested whether the interaction between BRCA1 and PALB2 may be involved in HR. Using U2OS cells with a single integration of the DR-GFP reporter, we found that gene conversion efficiency was reduced by ≈3- or 4-fold in PALB2 or BRCA1-depleted cells (Fig. 4A). To examine whether the direct interaction between PALB2 and BRCA1 has a role in the documented BRCA1-mediated HR repair, we introduced siRNA-resistant constructs of wild-type or mutant BRCA1 into cells after the depletion of endogenous BRCA1 by siRNA. FACS analyses indicated that reintroduction of wild-type BRCA1 successfully restored gene conversion to levels comparable with that of control cells (Fig. 4B; Fig. S6b). By contrast, BRCA1 mutants defective in PALB2-binding failed to fully rescue the gene conversion defect associated with BRCA1 depletion. Also, the ability of BRCA1 in promoting gene conversion closely correlates with its ability to interact with PALB2. Although the introduction of L1407P and M1411T failed to restore gene conversion in BRCA1-depleted cells, the M1400V mutant, which has some residual PALB2-binding activity (Fig. 2D), partially restored gene conversion activity (Fig. 4B; Fig. S6 b and c).
Fig. 4.
The BRCA1-PALB2 interaction is required for homologous recombination repair. (A) Gene conversion was assessed in U2OS DR-GFP cells treated with control, PALB2, or BRCA1-specific siRNAs. Percentage of GFP positive cells was determined by FACS analyses 48 h posttransfection. Relative gene conversion efficiency was normalized using control siRNA transfected cells, which was set as 100%. (B) Relative gene conversion was determined in BRCA1-depleted U2OS cells reconstituted with siRNA-resistant wild-type or mutant BRCA1. (C) Gene conversion was determined in PALB2-depleted cells transfected with empty vector, or reconstituted with siRNA-resistant wild-type or mutant PALB2. Relative gene conversion efficiency was normalized using cells reconstituted with wild-type PALB2, which was set as 100%. (D) A working model of the BRCA1/BRCA2/PALB2 complex in the DNA damage response.
To further corroborate the importance of the BRCA1-PALB2 complex formation in HR repair, we also assessed rates of gene conversion in PALB2-depleted U2OS cells reconstituted with either wild-type PALB2 or its BRCA1-binding defective mutants. Reintroduction of wild-type PALB2 restored efficient gene conversion, whereas the L21A, Y28A, and L35A mutants did not (Fig. 4C). As an extension to further validate the significance of the BRCA1-PALB2 interaction, mitomycin C (MMC) sensitivity assay was performed using PALB2-deficient EUFA1341 cells that have been reconstituted with wild-type or mutants PALB2. EUFA1341 cells reconstituted with PALB2 ΔN42, L21A, Y28A, or L35A mutants were hypersensitive toward MMC treatments, whereas the reintroduction of wild-type PALB2 or its control K14A mutant rescued the MMC hypersensitive phenotype (Fig. S6d). Together, these data suggest that the physical interaction between BRCA1 and PALB2 has an instrumental role essential for HR repair and cell survival.
Discussion
In this study, we have uncovered PALB2 as an integral component of the BRCA complex in vivo. Importantly, we found that PALB2 directly associates with BRCA1. We also provided biochemical evidences to demonstrate the existence of the trimeric BRCA complex containing BRCA1, PALB2, and BRCA2. We demonstrated that the specific interaction between BRCA1 and PALB2 is mediated by the coiled-coil motifs where mutations on the coiled-coil domains, including cancer patient derived BRCA1 mutations, would disrupt their complex formation. Functional analyses suggested that HR repair would be compromised when the PALB2-BRCA1 interaction is disrupted. These findings extend our earlier study (5), underscore the importance of the BRCA1-PALB2-BRCA2 complex in HR repair, and support the idea that distinct BRCA1 macromolecular complexes participate in various aspect in the DNA damage response.
BRCA1 has multiple roles in the cellular response to DNA damage (3, 6–10). Besides its functions in DNA damage checkpoint control, BRCA1 can also promotes cell survival via DNA repair (23, 24). Our findings that abrogation of the BRCA1-PALB2 interaction resulted in HR repair defects and compromised cell survival on DNA damage suggest that BRCA1 contributes to HR repair, at least in part, via the PALB2-BRCA2-RAD51 axis. That BRCA1, and its interaction with PALB2, is required for optimal formation of RAD51-ssDNA nucleoprotein filament (Fig. S7 a and b) is entirely consistent with this hypothesis. Given that BRCA1 also interacts with CtIP and BACH1, both of which have documented nonoverlapping roles at various steps of HR repair (25–29), it will be interesting to examine how each of these BRCA1 macrocomplexes work in concert in the maintenance of genome stability.
Domain mapping studies revealed that the BRCA1-PALB2 interaction is mediated via their respective coiled-coil motifs, and is independent from the BRCA1 BRCT domain (Fig. 2; Fig. S3). This observation is striking, considering that distinct BRCA1 macrocomplexes, including those that contains CtIP, BACH1, and the CCDC98/RAP80 complex, respectively, are selectively formed via the BRCA1 BRCT domain. This result suggests that PALB2 might be present among the various BRCA1 macrocomplexes. Indeed, both CtIP and BACH1 were found in PALB2 precipitates (Fig. 1D). Thus, the existence of BRCA1-PALB2 complex that contains CtIP or BACH1 highlights the possibility that BRCA1 may coordinate and fine-tune DNA repair via its ability to monitor processes including DNA resection and RAD51 loading simultaneously. Indeed, besides its interaction with PALB2, a requirement of the BRCA1 BRCT domain for efficient RAD51 foci formation (Fig. S5 and Fig. S6a), and optimal HR repair (Fig. 4B; Fig. S6 b and c), was also observed, and parallels the repair deficits in CtIP and BACH1-depleted cells (Fig. S7). Also, codepletion of PALB2 and CtIP or BACH1 demonstrated further reductions in gene conversion rates, as compared with PALB2-depleted cells (Fig. S7e). All these results, together, imply that BRCA1 may have accessory functions at multiple steps during HR repair via its associations with several of these proteins directly involved in HR processes.
Notably, disruption of the BRCA1-PALB2 interaction, although resulted in profound effects in HR repair and MMC hypersensitivity, did not noticeably affect the intra-S-phase checkpoint (Fig. S8 a and b), revealing the function specificity for the complex formation. We now know that through its phosphorylation-dependent interaction with the CCDC98/RAP80 complex, the BRCA1 BRCT domain is required for its damage-induced focal accumulation at sites of DNA damage (30–34). Intriguingly, our data demonstrated that BRCA1 only has an accessory role in the focal concentration of PALB2 and RAD51 at DSBs (Fig. 3; Fig. S4, Fig. S5, Fig. S6a, and Fig. S8c). This observation suggests that other factors or activities might be more important for the sustained localization of PALB2 at DNA lesions. We speculate that, although BRCA1 is not essential for sustained localization of PALB2 at DNA breaks, BRCA1 may stabilize PALB2 at the sites of DNA damage by serving as an additional anchor point for PALB2 at DSBs (Fig. 4D).
Mutations in BRCA1 predispose individuals to early development of breast and ovarian cancers. Given the prominent role of BRCA1 in protecting genome integrity, one would speculate that clinical mutations found among BRCA1 patients might provide mechanistic insights into its tumor suppressing function in vivo. Accordingly, our study exploited the patient derived BRCA1 missense mutations residing within its PALB2-interacting coiled-coil motif. The altered BRCA1-PALB2 interaction (Fig. 2; Fig. S2 and Fig. S3) and, thus, the BRCA1-PALB2-BRCA2 complex formation, together with DNA repair deficits observed with these patient-derived BRCA1 mutations, strongly suggest that in vivo assembly of the BRCA complex through PALB2 likely constitutes an important event required for their tumor suppressor functions. These results may also explain the similar phenotype observed in patients harboring mutations in each of these 3 proteins. Together with the observation that the BRCA1 BRCT mutant M1775R also displays significant DNA repair deficits (Fig. 4B; Fig. S6 b and c), our data support the idea that various BRCA1 functions converge and contribute to its role in HR repair and its tumor suppressor activity.
Materials and Methods
Antibodies.
Monoclonal antibody against the FLAG epitope (M2) and Myc epitope (9E10) were purchased from Sigma and Covance, respectively. Mouse monoclonal anti-RPA1, RPA2 and BRCA1 (SD118) antibodies were purchased from Calbiochem. Anti-GST-HRP was purchased from Santa Cruz. Rabbit polyclonal anti-RAD51, anti-BRCA2, and anti-BRCA1 antibodies were generous gifts from David Livingston. Rabbit polyclonal anti-pH2AX, BACH1, CtIP, and 53BP1 antibodies used were described previously (9, 27, 35). Rabbit polyclonal anti-PALB2 antibodies were generated by immunizing rabbits with recombinant GST-PALB2 fragments (containing residues 463–814 and 611–764 of PALB2), which were expressed and purified from Escherichia coli.
Purification of PALB2-Associated Protein Complex.
293T cells expressing SFB-PALB2 were lysed with NETN (20 mM Tris·HCl, pH 8/100 mM NaCl/1 mM EDTA/0.5% Nonidet P-40) on ice for 20 min, followed by centrifugation at 13,000 × g for 20 min at 4 °C. Supernatant was incubated with streptavidin beads for 2 h at 4 °C. Bound complex was eluted with 2 mg/mL biotin diluted in NETN. Supernatant was further incubated with S protein conjugated agarose beads for 2 h at 4 °C. Beads were washed 3 times with NETN buffer, and proteins bound to the beads were eluted by boiling with SDS sample buffer. Proteins were resolved by SDS/PAGE, stained with silver. Visible bands were excised for mass spectrometry protein identification (Taplin biological mass spectrometry facility, Harvard University, Cambridge, MA).
Serial Immunodepletion Experiments.
Cell lysate was subjected to immunodepletion using indicated antibodies coupled to protein A beads for 2 h. Supernatant was saved and immunodepleted for 2 additional rounds. Thereafter, cell lysates were boiled in SDS loading buffer and analyzed by immunoblotting.
Protein Production in Insect Cells.
Baculoviruses expressing His-Flag-BRCA1 or GST-BARD1 were gifts from Richard Baer. The coding sequences of full-length PALB2, BRCA2, WTX, and PALB2 N42 were transferred to pDEST20 vector for the expression of GST-fusion proteins in insect cells. Transposition occurred in DH10Bac competent cells and correct bacmids confirmed by PCR were transfected into SF9 cells for baculovirus production. Protein expression was confirmed by SDS/PAGE, Coomassie blue staining, and Western blotting. For coIP experiments, SF9 cells infected with corresponding baculoviruses were lysed in NETN for 20 min on ice, and the crude lysate was clarified by centrifugation (13,000 × g, 10 min). Supernatant was saved, and pellet was digested with Benzoase for 1 h at 4 °C and clarified again by centrifugation. Pooled supernatant was used for coIP.
Immunostaining.
Cells were treated with 10 Gy of gamma radiation. After recovery, cells were washed with PBS, fixed at room temperature with 3% paraformaldehyde for 12 min, permeabilized with 0.5% triton for 3 min, and then immunostained with appropriate antibodies for 30 min. Whenever transfection was needed, cells were transfected with indicated constructs using Lipofectamine 2000 (Invitrogen), and irradiated 24 h posttransfection. For detection of RPA foci, cells grown on coverslips were first permeabilized with 0.5% triton for 2.5 min, washed twice with PBS, and fixed with 3% paraformaldehyde for 15 min at room temperature before incubation with primary anti-RPA antibodies. After incubation with primary antibodies, cells were washed twice with PBS and immunostained with Rhodamine-conjugated goat anti-mouse and/or FITC-conjugated goat anti-rabbit antibodies for 30 min. Nuclei were counterstained with DAPI. Images were visualized and captured (Nikon Eclipse 800 microscope). For foci quantification, all of images were captured at identical exposure time, and 100 cells were counted for duplicated experiments.
Gene Conversion Assay.
U2OS cells (3 × 106) stably expressing DR-GFP substrate and pCBASce plasmid were electroporated with 8 μg pCBASce plasmid at 250V, 975 μF by using a Bio-Rad genepulsar II. For the reintroduction of BRCA1 or PALB2 after siRNA-mediated depletion, constructs containing either BRCA1 or PALB2 were electroporated together with the pCBASce construct into the cell. Cells were then plated onto 10-cm dishes and incubated in culture media for 48 h before FACS analyses. Cells were analyzed in a Becton-Dickinson FACScan on a green (FL1) versus orange (FL2) fluorescence plot. Results represent average of 2 to 3 independent experiments (mean ± SEM).
For more details, see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Prof. David Livingston (Dana-Farber Harvard Cancer Institute, Boston) for providing the pOZC-PALB2 construct and the various antibodies used in this study; Prof. Maria Jasin (Memorial Sloan-Kettering Cancer Center, New York) for the U2OS cells with DR-GFP integration, DR-GFP, and pCBASce plasmids; Prof. Johan P. de Winter (Vrije Universiteit Medical Center, Amsterdam) for EUFA1341 cells; Prof. Richard Baer (Columbia University, New York) for the BRCA1 and BARD1 baculoviruses; Drs. Gargi Ghosal, Xiaohua Xu, and Zihua Gong for their assistance in gel filtration chromatography; and Drs. Jingsong Yuan and Jun Huang for insightful discussion. S.M.H.S. is supported by a Postdoctoral fellowship from the Croucher Foundation; M.S.Y.H. by an Anna Fuller Fund fellowship; and J.C. by grants from the National Institutes of Health and an Era of Hope Scholar award from the Department of Defense. J.C. is a member of the Mayo Clinic Breast Specialized Programs of Research Excellence program.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0811159106/DCSupplemental.
References
- 1.Jemal A, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Zhang H, Tombline G, Weber BL. BRCA1, BRCA2, and DNA damage response: Collision or collusion? Cell. 1998;92:433–436. doi: 10.1016/s0092-8674(00)80936-8. [DOI] [PubMed] [Google Scholar]
- 3.Wang W. Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat Rev Genet. 2007;8:735–748. doi: 10.1038/nrg2159. [DOI] [PubMed] [Google Scholar]
- 4.Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25:5864–5874. doi: 10.1038/sj.onc.1209874. [DOI] [PubMed] [Google Scholar]
- 5.Chen J, et al. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317–328. doi: 10.1016/s1097-2765(00)80276-2. [DOI] [PubMed] [Google Scholar]
- 6.Mullan PB, Quinn JE, Harkin DP. The role of BRCA1 in transcriptional regulation and cell cycle control. Oncogene. 2006;25:5854–5863. doi: 10.1038/sj.onc.1209872. [DOI] [PubMed] [Google Scholar]
- 7.Venkitaraman AR. Functions of BRCA1 and BRCA2 in the biological response to DNA damage. J Cell Sci. 2001;114:3591–3598. doi: 10.1242/jcs.114.20.3591. [DOI] [PubMed] [Google Scholar]
- 8.Kerr P, Ashworth A. New complexities for BRCA1 and BRCA2. Curr Biol. 2001;11:R668–R676. doi: 10.1016/s0960-9822(01)00389-x. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg RA, et al. Multifactorial contributions to an acute DNA damage response by BRCA1/BARD1-containing complexes. Genes Dev. 2006;20:34–46. doi: 10.1101/gad.1381306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lord CJ, Ashworth A. RAD51, BRCA2 and DNA repair: A partial resolution. Nat Struct Mol Biol. 2007;14:461–462. doi: 10.1038/nsmb0607-461. [DOI] [PubMed] [Google Scholar]
- 12.Pellegrini L, Venkitaraman A. Emerging functions of BRCA2 in DNA recombination. Trends Biochem Sci. 2004;29:310–316. doi: 10.1016/j.tibs.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Thorslund T, West SC. BRCA2: A universal recombinase regulator. Oncogene. 2007;26:7720–7730. doi: 10.1038/sj.onc.1210870. [DOI] [PubMed] [Google Scholar]
- 14.Xia B, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell. 2006;22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 15.Rahman N, et al. PALB2, which encodes a BRCA2-interacting protein, is a breast cancer susceptibility gene. Nat Genet. 2007;39:165–167. doi: 10.1038/ng1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Simpson S. PALB2-new breast-cancer susceptibility gene. Lancet Oncol. 2007;8:105. doi: 10.1016/s1470-2045(07)70025-6. [DOI] [PubMed] [Google Scholar]
- 17.Reid S, et al. Biallelic mutations in PALB2 cause Fanconi anemia subtype FA-N and predispose to childhood cancer. Nat Genet. 2007;39:162–164. doi: 10.1038/ng1947. [DOI] [PubMed] [Google Scholar]
- 18.Foulkes WD, et al. Identification of a novel truncating PALB2 mutation and analysis of its contribution to early-onset breast cancer in French-Canadian women. Breast Cancer Res. 2007;9:R83. doi: 10.1186/bcr1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Erkko H, et al. A recurrent mutation in PALB2 in Finnish cancer families. Nature. 2007;446:316–319. doi: 10.1038/nature05609. [DOI] [PubMed] [Google Scholar]
- 20.Tischkowitz M, et al. Analysis of PALB2/FANCN-associated breast cancer families. Proc Natl Acad Sci USA. 2007;104:6788–6793. doi: 10.1073/pnas.0701724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia B, et al. Fanconi anemia is associated with a defect in the BRCA2 partner PALB2. Nat Genet. 2007;39:159–161. doi: 10.1038/ng1942. [DOI] [PubMed] [Google Scholar]
- 22.Zhang J, et al. Chk2 phosphorylation of BRCA1 regulates DNA double-strand break repair. Mol Cell Biol. 2004;24:708–718. doi: 10.1128/MCB.24.2.708-718.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer Res. 2001;61:4842–4850. [PubMed] [Google Scholar]
- 24.Moynahan ME, Chiu JW, Koller BH, Jasin M. Brca1 controls homology-directed DNA repair. Mol Cell. 1999;4:511–518. doi: 10.1016/s1097-2765(00)80202-6. [DOI] [PubMed] [Google Scholar]
- 25.Takeda S, Nakamura K, Taniguchi Y, Paull TT. Ctp1/CtIP and the MRN complex collaborate in the initial steps of homologous recombination. Mol Cell. 2007;28:351–352. doi: 10.1016/j.molcel.2007.10.016. [DOI] [PubMed] [Google Scholar]
- 26.Yu X, Wu LC, Bowcock AM, Aronheim A, Baer R. The C-terminal (BRCT) domains of BRCA1 interact in vivo with CtIP, a protein implicated in the CtBP pathway of transcriptional repression. J Biol Chem. 1998;273:25388–25392. doi: 10.1074/jbc.273.39.25388. [DOI] [PubMed] [Google Scholar]
- 27.Yu X, Chini CC, He M, Mer G, Chen J. The BRCT domain is a phospho-protein binding domain. Science. 2003;302:639–642. doi: 10.1126/science.1088753. [DOI] [PubMed] [Google Scholar]
- 28.Gupta R, et al. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA-binding protein. Blood. 2007;110:2390–2398. doi: 10.1182/blood-2006-11-057273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cantor SB, et al. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell. 2001;105:149–160. doi: 10.1016/s0092-8674(01)00304-x. [DOI] [PubMed] [Google Scholar]
- 30.Kim H, Huang J, Chen J. CCDC98 is a BRCA1-BRCT domain-binding protein involved in the DNA damage response. Nat Struct Mol Biol. 2007;14:710–715. doi: 10.1038/nsmb1277. [DOI] [PubMed] [Google Scholar]
- 31.Liu Z, Wu J, Yu X. CCDC98 targets BRCA1 to DNA damage sites. Nat Struct Mol Biol. 2007;14:716–720. doi: 10.1038/nsmb1279. [DOI] [PubMed] [Google Scholar]
- 32.Sobhian B, et al. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science. 2007;316:1198–1202. doi: 10.1126/science.1139516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim H, Chen J, Yu X. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science. 2007;316:1202–1205. doi: 10.1126/science.1139621. [DOI] [PubMed] [Google Scholar]
- 34.Wang B, et al. Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science. 2007;316:1194–1198. doi: 10.1126/science.1139476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lou Z, et al. MDC1 regulates DNA-PK autophosphorylation in response to DNA damage. J Biol Chem. 2004;279:46359–46362. doi: 10.1074/jbc.C400375200. [DOI] [PubMed] [Google Scholar]
- 36.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.