Abstract
Persistently activated STAT3 contributes to cell survival in many different human cancers. Cancer cell secretion of IL-6 is a frequent basis for persistent STAT3 activation; we show that antibodies against IL-6 or gp-130, the signaling unit of the IL-6 receptor, can abruptly remove persistently activated STAT3 causing prompt disappearance of cysteine proteases of serpin B3/B4 mRNAs, known as squamous cell carcinoma antigens 1 and 2. STAT3 occupies the promoter of serpin B3/B4 before removal and siRNA removal of B3/B4 mRNA caused cell death in HN13 head and neck cancer cells. Thus persistently activated STAT3 is a required part of the continuous activation of B3/B4 genes, which protects tumor cells from dying.
Keywords: STAT3, serpin B3/B4, squamous cancer
Introduction
Persistently tyrosine phosphorylated, therefore potentially transcriptionally active, STAT3 (pSTAT3) exists in a wide variety of human tumors [1, 2]. In cultured cells src transformation utilizes and requires STAT3 phosphorylation [3–5]. One result of persistently active STAT3 is protection of cells against induced apoptosis [1, 6]. The exact transcriptional targets of STAT3 that are responsible for its contribution to the oncogenic state in human cancer cells are not clear. The positive transcriptional activity of STAT3 on a variety of potentially antiapoptotic gene promoters has been well proven in transfection analyses [1, 6, 7]. These results imply that persistently active STAT3 could be important in contributing to the cancer phenotype. But these transfection experiments don’t address what genes the persistently active STAT3 actually drives in the cancer cells. The removal of STAT3 by antisense techniques [1, 8] or in some cases by drugs that affect STAT3 [4, 9], but possibly other intracellular proteins as well, leads to apoptosis or growth restraint but only after a lengthy lag period (~24–48 h). Such results leave unanswered the immediate, direct role of persistently active STAT3.
In an effort to establish the most proximate transcription targets of STAT3 in cancer cells that contribute to its oncogenic potential, we have sought an experimental system that would quickly remove phosphorylated STAT3 with reasonable specificity. Gutkind and colleagues [10] showed secretion by human tumor cells of IL-6 [11–13], a potent activator of STAT3, through the gp-130 receptor to be the cause of persistently active STAT3 in SCCHN (squamous cell carcinoma of head and neck). Antibodies against IL-6 or gp-130 decreased persistently active STAT3 in these cells.
We have extended the Gutkind result to other cancer cell lines, highlighting IL-6 as the potential activating agent of STAT3 in additional tumor cells. A microarray analysis of mRNA after removal of active STAT3 in tumor cells showed serpins B3/B4 mRNAs to decrease abruptly. These proteins appear to be important for survival of cells and are shown to be direct transcriptional targets of STAT3.
Materials and Methods
Cell Culture, antibodies and reagents
HN13 and HN30 cells were gifts from J. S. Gutkind; MCF7 and 231 cells were a gift of D. Foster. DU145 and 293T cells were from ATCC. Cells were grown (37°C, 5% CO2; DMEM with 10% bovine calf serum). Recombinant hIL-6 and hOSM, anti-hIL-6, anti-hOSM and anti-hgp130 antibodies were from R&D Systems. Activated STAT3 was detected with antiphosphotyrosine antibody (Cell Signaling Technology) and STAT3 specific antibody (Cell Signaling Technology). P6, a specific inhibitor of Jak1, was from J. Bromberg.
Whole Cell extracts, SDS/PAGE and western blotting on 10% SDS-polyacrylamide gels was as described [14]. Nuclear extracts were prepared and EMSA carried out as described [15].
RT-PCR
For RT-PCR, RNA was extracted using the RNeasy kit (Qiagen) and reverse transcribed (Superscript, GIBCOBRL) to generate cDNA which was then subjected to 30 cycles of PCR amplification. The probes used for RT-PCR were as follows: serpinB3: F: ATGAATTCACTCAGTGAAGCC, R: GTGTAGGACTCCAGATAGCAC, serpinB4: F: CTGATGCATATGAGCTGAAGATCG, R: GTGTAGGACTTTAGATACTGA, GAPDH: F: GGGAGCCAAAAGGGTCATCATC, R: GTGTCGCTGTTGAAGTCAGAGG. Real time PCR in triplicate was by the SYBR Green PCR Mastermix (Applied Biosystems) and a UBI thermal cycler. Expression of each mRNA was normalized to relative levels of GAPDH. Completed PCR reactions were run on an agarose gel to confirm the generation of only the correct size amplification products. The primer sequences used were as follows: Serpin B3: F: TTACCTCGGTTCAAAGTGGAAGAG, R: AATCCTACTACAGCGGTGGCAG, Serpin B4: F: GCAAATGCTCCAGAAGAAAGTCG, R: GCCAATAGTCCCATCAGGAAATAGG, GAPDH: F: CTCCAGAACATCATCC, R: CGACGCCTGCTTCACCACCTTCTT.
Chromatin Immunoprecipitation (ChIP)
ChIP assays on HN13 cells used the ChIP assay kit (Upstate Biotechnology). Growing cells were incubated overnight in conditioned medium with or without P6 compound (50μM); STAT3-DNA complexes were precipitated with an anti-STAT3 antibody (Upstate Biotechnology). Polyclonal IgG antibody was used as a negative control. Precipitated DNA was amplified by PCR using primers flanking two putative gamma-activated sites (GAS) between −78 and −329 of the proximal serpin B3/B4 promoters. The primers were: F:GCTGGAGTAGAATACAAAGGGATGG and R:CTCAACTTCAGATTTCACAAGCGG.
siRNA experiments
Using a Silencer siRNA construction kit (Ambion) siRNAs {GGCAAAGAUCUAAGCAUGA and GGGCAGUGGGAGAAGAAAU; AAGCAAUUCGCAGAGCTAGA} [16] complementary to both serpin B3/B4 mRNAs were prepared and purified. HN13 or HN30 cells were transfected with 10–20 μM siRNA via a Lipofectamine RNAiMAX protocol (manufacturer’s instructions ) for 24 or 48 h. The floating cells with pyknotic nuclei were counted.
Results
Decrease of Persistent pSTAT3 in Several Tumor Cell Types by anti-IL-6 or gp-130 Antibodies
Initial experiments employed the human SCCHN cell line, HN13, a prostate cancer cell line, DU145, and two breast cancer cell lines, MCF7 and 231, all known to contain varying levels of persistently phosphorylated STAT3 {pSTAT3 [10, 17, 18] and data not shown}. Growth medium from each of these cell lines was used to treat for 10 min 293T cells that normally do not contain persistently active STAT3. The “conditioned medium” (CM) from 231, DU145 and HN13 cells resulted in strong pSTAT3 stimulation and that from the MCF7 cells a somewhat weaker stimulation. Addition of hIL-6 (human IL-6) specific antibody to the CM prior to adding it to the 293T cells resulted in significant reduction in STAT3 activation (Fig. 1A). In a complementary approach, washing the cell monolayers and adding fresh medium to DU145 and 231 cells led to a prompt decease in pSTAT3 that returned after 3 h (Fig. 1B, first and second panels respectively). Anti-IL-6 prevented the return of pSTAT3 levels up to 16 h (Fig. 1C). Anti-OSM (oncostatin M) antibody did not suppress pSTAT3 (Fig. 1D). Thus IL-6 is a major contributor to STAT3 activation in these four cell lines of squamous carcinoma cells [10], Moreover, STAT3 activity was suppressed up to 24 h in HN13, DU145 cells and in 231 cells (tested only for 1 h) by antibody to gp-130, the signaling component of the IL-6 receptor (Fig. 2A).
Fig. 1.
Effects of Conditioned Medium: STAT3 activation in human tumor cells occurs through the action of IL-6. A. 293T cells were treated for 10 min with 2 day conditioned medium (CM) from 231, DU145, HN13 and MCF7 cell lines respectively (left panel, lanes 1–5). pSTAT3 assayed via western blot. hIL-6 specific antibody added to the CM (right panel). B. DU145 cells (top panels, left) or 231 cells (top panel, right) were washed and re-supplied with fresh medium (10 min – 3 h). Western blot for pSTAT3 and total STAT3 (upper and lower panels). C. HN13 cells washed as in B but medium plus anti-IL-6 antibody added. D. pSTAT3 or total STAT3 in HN13; cells treated with IL-6 or OSM antibody for 1 h.
Fig. 2.

Anti-gp-130 antibody decreases active STAT3. A. HN13, DU145 cells treated for 1 or 24 h; 231 cells for 1 h with anti-gp-130 antibody; pSTAT3 and total STAT3 determined. B. Nuclear extracts from HN13 cells untreated (lane 1) or treated for 1 h with anti-gp-130 (lane 2) or anti-IL-6 (lane 3) were subjected to EMSA analysis with a labeled STAT3 binding site; lane 4 with anti-STAT3 antibody.
Nuclear extracts of HN13 cells treated with either anti-gp-130 (Fig. 2B, lane 2) or anti-IL-6 (Fig. 2B, lane 3) had greatly reduced STAT3 DNA binding activity as tested by EMSA with a STAT DNA binding site (Fig. 2B). These experiments confirm and extend the conclusion that human tumor cells from various origins contain persistently phosphorylated STAT3 due to autocrine secretion of IL-6 [10, 18–21] and the pSTAT3 can be decreased abruptly by appropriate antibody treatment.
Detection of mRNA Decreases in Cells Treated with gp-130 Antiserum
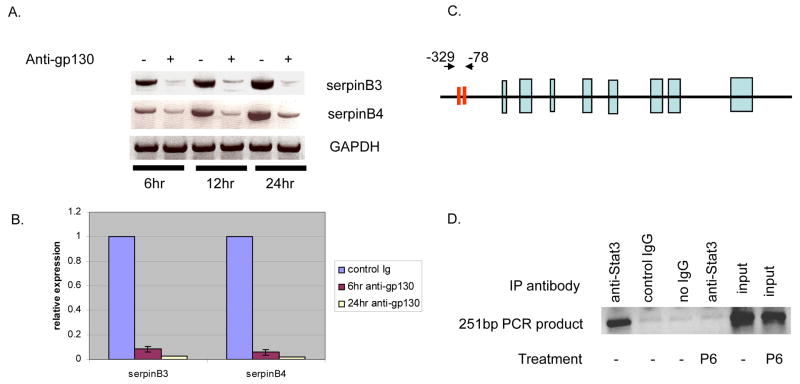
In order to determine if any mRNAs disappeared after STAT3 removal, a microarray experiment on mRNA concentrations in HN13 cells treated for 6, 12 or 24 h with gp-130 antisera or control antibody was carried out. By this assay, only after the 24 h time point were there a few mRNAs with substantial, >3–4 fold decreases, and about 40 mRNAs with a >2-fold decrease in the antibody treated sample (supplementary Table). Two mRNAs that both decreased substantially and potentially had some relevance to head and neck cancer encoded the protease inhibitors serpin B3/B4. These proteins are part of a large family of inhibitors of various proteases {both serine and cysteine proteases [22]}, including some intracellular proteases [22], that could contribute to protection against tumor cell death. Further studies were therefore carried out first to validate the loss of serpin B3/B4 mRNAs. Both conventional RT-PCR and real time PCR showed a more prompt decrease in serpin B3/B4 mRNA content after anti-gp130 treatment than did the microarray analysis (Fig. 3A and 3B, respectively). Thus the level of serpin B3/B4 mRNAs appeared dependent on pSTAT3. (The decrease in serpin B3/B4 mRNA by PCR was compared to a number of other mRNAs; supplementary Fig. 1.) We note that the microarray analysis at 6 and 12 h failed to uncover the changes seen in PCR at the 6 h time points for both the serpins and for the methyl transferase (NMMT).
Fig. 3.
Decrease of serpin B3/B4 mRNA after anti-gp-130 treatment of HN13 cells; a Chromatin Immunoprecipitation (ChIP) shows STAT3 binding to the serpin B3 promoter. A. A fraction of the RNA used (Table 1, supplementary data) was subjected to PCR for serpin B3/B4 mRNA. B. QPCR on cDNA from A. C. Diagram of serpin B3 gene showing proposed STAT3 sites (red bars). D. HN13 cells were treated or not with the Jak1 inhibitor P6 (50μM); ChIP carried out with STAT3 antibody or isotope matched control antibody. Input: 10% of starting extract.
ChIP Analysis Shows pSTAT3 Occupation on Serpin B3 Promoter
To more directly implicate pSTAT3 in transcriptional control of serpin mRNAs, a ChIP assay of STAT3 bound to putative STAT sites in the suggested promoter region of the serpin B3/B4 genes [23] was carried out. Two putative sites with canonical STAT binding sequence lie between −78 and−329 upstream of the mRNA start site (Fig. 3C) of both genes [23, 24]. The promoters of serpin B3/B4 have 95% sequence identity [23]; and the sequences chosen as primers of the chromatin precipitated DNA fragments are the same for both promoters. ChIP assay (see Methods) with anti-STAT3 antibody or an irrelevant antibody from cells treated or not with a broad JAK inhibitor (P6) [25] that blocks all STAT action. (No sample from cells treated with anti-gp130 or IL-6 antibody was included because of the prohibitive cost of anti-gp130 or anti-IL6 antibodies to treat sufficient cells required for ChIP assays). Clear evidence (Fig. 3D, lane 1) was found that pSTAT3 does occupy the putative binding region in the serpin B3/B4 promoter of growing HN13 cells. This binding disappeared in cells treated with P6 (Fig. 3D, lane 4) strongly supporting the conclusion that serpin B3/B4 are transcriptional targets of persistently active STAT3.
siRNA Removal of Serpin B3/B4 Leads to Killing of HN13 Cells
To determine whether serpin B3/B4 were important for the survival of SCCHN, two specific siRNA oligonucleotides, complementary to both serpin B3/B4 mRNA [16], were introduced into HN13 and HN30 cells to decrease specifically the serpin mRNAs. Within 24 to 48 h of the introduction of serpin specific siRNAs, a high proportion of both the HN30 and HN13 cells were affected (i.e. released from culture plates with pyknotic nuclei) (Fig. 4).
Fig. 4.
Serpin B3/B4 specific siRNA causes HNSCC cells to die. A. HN13 and HN30 cells were transfected with serpin B3/B4 specific siRNA (20μM) or nonspecific siRNA cell attachment and exfoliated cells were counted after 24 or 48 h (in B). B. The data shown is representative of three experiments.
Discussion
The concept of “target genes” activated by specific induced transcription factors such as the STATs or NF-κB, the SMADs or steroid hormone receptors (all acutely induced, mammalian, positive-acting transcription factors) is rooted in experiments where cultured cells respond to cytokine or hormone inducers by promptly increasing the amount of particular mRNAs. Subsequently, binding sites are shown to be occupied by the transcription factor in chromatin in vivo.
Such “acute induction” experiments are universally accepted to prove a putative role for a particular induced transcription factor in “driving” the transcription of a target gene [26]. In tumor cells, persistent activation of a given transcription factor is very often taken to imply ongoing function of the transcription factor in elevating the level of target gene mRNAs whose activation was first proved by the simpler cell culture assays described above. However, the role of such a persistently activated transcription factor in controlling the mRNA profile of the tumor cell requires further experimentation [27] before assigning the cause of activation of specific genes in tumor cells. Demonstrating a continuing direct role of a transcription factor depends on the abrupt and specific removal of that factor and an evaluation of the immediate effects on transcription or at least on the short term mRNA composition of the cells.
Our purpose in the present experiments was to ascertain whether the ongoing pSTAT3 activity was responsible for driving the constant transcription of any genes that contributed to its oncogenic properties. Although direct readout of the micro array results at early times (6 or 12 h) was not successful, further analysis with RT-PCR of genes that appeared to be down regulated >3 fold at 24 h, allowed us to identify a handful of mRNAs that were fairly promptly (~6 h) decreased after blockade of STAT3 activation (supplementary Fig. 1). Two of these mRNAs, serpin B3/B4 (and presumably their encoded protein products) appear likely to play a continuous role in the survival of HN13 cells.
The serpins are a family of protease inhibitors originally grouped together as serine protease inhibitors, most of which are secreted [22]. The clade B serpins include a number of proteins including serpin B3/B4 that have no signal sequence and are retained inside the cell. Both serpin B3/B4 were recognized in the early 1990’s [28] as circulating “squamous cell carcinoma antigens” (SCCA1 and 2) [29] that were present in a substantial fraction of sera from patients bearing squamous cell cancers of the cervix, head and neck, esophagus and lung as well as in patients with hepatocellular carcinoma [30]. Presumably the presence of these serpins in serum comes from dying cancer cells. The nature of these circulating tumor cell antigens only became clear when the cDNAs encoding the proteins were cloned [22, 31]. Serpin B3/B4 are unusual because they inhibit papain-like cysteine proteases such as the cathepsins in addition to serine proteases. The cathepsins, particularly B, D and L, have been implicated in cancer (see Lah et al. [32]).
Serpins B3/B4 have a potential role in cancer cells as exhibited by overexpression, leading to protection from chemical or X-ray induced apoptosis [33, 34]. Recently a single C. elegans type B serpin has been shown to block lysosomal hydrolytic enzyme induction of necrotic cell death incident to several different toxic shocks (heat, anoxia, hypotonic shock and cation channel hyperactivity) [35].
While it is by no means clear exactly how and against which mode of cell death – apoptosis or lysosomal injury – elevated serpin B3/B4 might act in protecting cancer cells, it seems certain that these proteins do protect cancer cells of squamous origin from ultimate death. Continuing transcription of serpin B3/B4 mRNA by STAT3 thus is one logical role of STAT3 in protection against cell death of certain tumors, suggesting once again that compounds that could inhibit the persistently active STAT3 in human cancers might be effective antitumor agents.
Supplementary Material
Acknowledgments
This work was supported by NIH grants AI32489 and AI32440 to J. E. Darnell. S. T. Ahmed was the recipient of a Cancer Research Institute postdoctoral fellowship.
Footnotes
Supplementary information is available at BBRC’s website.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Yu H, Jove R. The STATs of cancer -- new molecular targets come of age. Nature Reviews Cancer. 2004;4:97–105. doi: 10.1038/nrc1275. [DOI] [PubMed] [Google Scholar]
- 2.Levy D, Darnell JE., Jr STATs: Transcriptional control and biologic impact. Nature Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 3.Yu CL, Meyer D, Campbell GS, Larner AC, Carter-Su C, Schwartz J, Jove R. Enhanced DNA-binding of a Stat3-related protein in cells transformed by the Src oncoprotein. Science. 1995;269:81–83. doi: 10.1126/science.7541555. [DOI] [PubMed] [Google Scholar]
- 4.Turkson J, Bowman T, Garcia R, Caldenhoven E, Groot RPD, Jove R. Stat3 activation by src induces specific gene regulation and is required for cell transformation. Mol Cell Biol. 1998;18:2545–2552. doi: 10.1128/mcb.18.5.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bromberg JF, Horvath CM, Besser D, Lathem WW, Darnell JE., Jr Stat3 activation is required for cellular transformation by v-src. Mol Cell Biol. 1998;18:2553–2558. doi: 10.1128/mcb.18.5.2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen Y, Devgan G, Darnell JE, Jr, Bromberg JF. Constitutively activated Stat3 protects fibroblasts from serum withdrawal and UV-induced apoptosis and antagonizes the proapoptotic effects of activated Stat1. Proc Natl Acad Sci USA. 2001;98:1543–1548. doi: 10.1073/pnas.041588198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res. 2005;65:939–947. [PubMed] [Google Scholar]
- 8.Xi S, Gooding WE, Grandis JR. In vivo antitumor efficacy of STAT3 blockade using a transcription factor decoy approach: implications for cancer therapy. Oncogene. 2005;24:970–979. doi: 10.1038/sj.onc.1208316. [DOI] [PubMed] [Google Scholar]
- 9.Siddiquee K, Zhang S, Guida WC, Blaskovich MA, Greedy B, Lawrence HR, Yip MLR, Jove R, McLaughlin MM, Lawrence NJ, Sebti SM, Turkson J. Selective chemical probe inhibitor of STAT3, identified through structure-based virtual screening, induces antitumor activity. Proc Natl Acad Sci USA. 2007;104:7391–7396. doi: 10.1073/pnas.0609757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sriuranpong V, Park JI, Amornphimoltham P, Patel V, Nelkin BD, Gutkind JS. Epidermal growth factor receptor-independent constitutive activation of STAT3 in head and neck squamous cell carcinoma is mediated by the autocrine/paracrine stimulation of the Interleukin 6/gp130 cytokine system. Cancer Res. 2003;63:2948–2956. [PubMed] [Google Scholar]
- 11.Chen Z, Malhotra PS, Thomas GR, Ondrey FG, Duffey DC, Smith CW, Enamorado I, Yeh NT, Kroog GS, Rudy S, McCullagh L, Mousa S, Quezado M, Herscher LL, Waes CV. Expression of proinflammatory and proangiogenic cytokines in pateints with head and neck cancer. Clin Cancer Res. 1999;5:1369–1379. [PubMed] [Google Scholar]
- 12.Mann EA, Spiro JD, Chen LL, Kreutzer DL. Cytokine expression by head and neck squamous cell carcinomas. Am J Surg. 1992;164:567–573. doi: 10.1016/s0002-9610(05)80708-1. [DOI] [PubMed] [Google Scholar]
- 13.Woods KV, El-Naggar A, Clayman GL, Grimm EA. Variable expression of cytokines in human head and neck squamous cell carcinoma cell lines and consistent expression in surgical specimens. Cancer Res. 1998;58:3132–3141. [PubMed] [Google Scholar]
- 14.Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, Hoeve Jt, Ren Z, Mao X, Chen X, Shuai K, Darnell JE., Jr Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle. Proc Natl Acad Sci USA. 2005;102:3966–3971. doi: 10.1073/pnas.0501063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang X, Darnell JE., Jr Functional importance of Stat3 tetramerization in activation of the alpha 2-macroglobulin gene. J Biol Chem. 2001;276:33576–33581. doi: 10.1074/jbc.M104978200. [DOI] [PubMed] [Google Scholar]
- 16.Collins CS, Hong J, Sapinoso L, Zhou Y, Liu Z, Micklash K, Schultz PG, Hampton GM. A small interfering RNA screen for modulators of tumor cell motility identifies MAP4K4 as a promigratory kinase. Proc Natl Acad Sci USA. 2006;103:3775–3780. doi: 10.1073/pnas.0600040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mora LB, Buettner R, Seigne J, Diaz J, Ahmad N, Garcia R, Bowman T, Falcone R, Fairclough R, Cantor A, Muro-Cacho C, Livingston S, Karras J, Pow-Sang J, Jove R. Constitutive activation of Stat3 in human prostate tumors and cell lines: direct inhibition of Stat3 signaling induces apoptosis of prostate cancer cells. Cancer Res. 2002;62:6659–6666. [PubMed] [Google Scholar]
- 18.Berishaj M, Gao SP, Ahmed S, Leslie K, Al-Ahmadie H, Gerald WL, Bornmann W, Bromberg JF. Stat3 is tyrosine-phosphorylated through the interleukin-6/glycoprotein 130/Janus kinase pathway in breast cancer. Breast Cancer Res. 2007;9:R32. doi: 10.1186/bcr1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siegall CB, Schwab G, Nordan RP, Fitzgerald DJ, Pastan I. Expression of the Interleukin 6 receptor and interleukin 6 in prostrate carcinoma cells. Cancer Res. 1990;50:7786–7788. [PubMed] [Google Scholar]
- 20.Palmer J, Hertzog PJ, Hammacher A. Differential expression and effects of gp130 cytokines and receptors in prostrate cancer cells. Int J Biochem and cell B. 2004;36:2258–2269. doi: 10.1016/j.biocel.2004.04.017. [DOI] [PubMed] [Google Scholar]
- 21.Campbell CL, Ziang Z, Savarese DM, Savarese TM. Increased expression of the interleukin 11 receptor and evidence of Stat3 in prostate carcinoma. Am J Pathol. 2001;158:25–32. doi: 10.1016/S0002-9440(10)63940-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Silverman GA, Whisstock JC, Askew DJ, Pak SC, Luke CJ, Cataltepe S, Irving JA, Bird PI. Human clade B serpins (ov-serpins) belong to a cohort of evolutionarily dispersed intracellular proteinase inhibitor clades that protect cells from promiscuous proteolysis. Cell Mol Life Sci. 2004;61:301–325. doi: 10.1007/s00018-003-3240-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamada K, Shinomiya H, Asano Y, Kihana T, Iwamoto M, Hanakawa Y, Hashimoto K, Hirose S, Kyo S, Ito M. Moleculer cloning of human squamous cell carcinoma antigen 1 gene and charcterization of its promoter. Bioch Biophy Acta. 2001;1518:124–131. doi: 10.1016/s0167-4781(01)00174-9. [DOI] [PubMed] [Google Scholar]
- 24.Sakaguchi Y. Structural analysis of human SCC antigen 2 promoter. Biochim Biophys Acta. 1999;1444:111–116. doi: 10.1016/s0167-4781(98)00259-0. [DOI] [PubMed] [Google Scholar]
- 25.Pedranzini L, Dechow T, Berishaj M, Comenzo R, Zhou P, Azare J, Bornmann W, Bromberg J. Pyridone 6, a pan-Janus-activated kinase inhibitor, induces growth inhibition of multiple myeloma cells. Cancer Res. 2006;66:9714–9721. doi: 10.1158/0008-5472.CAN-05-4280. [DOI] [PubMed] [Google Scholar]
- 26.Brivanlou AH, Darnell JE., Jr Signal transduction and the control of gene expression. Science. 2002;295:813–818. doi: 10.1126/science.1066355. [DOI] [PubMed] [Google Scholar]
- 27.Darnell JE., Jr Transcription factors as targets for cancer therapy. Nature Reviews Cancer. 2002;2:740–749. doi: 10.1038/nrc906. [DOI] [PubMed] [Google Scholar]
- 28.Kato H. In: Squamous cell carcinoma, Serological Cancer Markers. Sell S, editor. Humana Press; Totowa, NJ: 1992. pp. 437–451. [Google Scholar]
- 29.Schneider SS, Schick C, Fish KE, Miller E, Pena JC, Treter SD, Hui SM, Silverman GA. A serine proteinase inhibitor locus at 18q21.3 contains a tandem duplication of the human squamous cell carcinoma antigen gene. Proc Natl Acad Sci USA. 1995;92:3147–3151. doi: 10.1073/pnas.92.8.3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pontisso P, Calabarese F, Benvegnu L, Lise M, Belluco C, Ruvoletto MG, Falco SD, Marino M, Valente M, Nitti D, Gatta A, Fassina G. Overexpression of squamous cell carcinoma antigen variants in hepatocellular carcinoma. Brit J of Cancer. 2004;90:833–837. doi: 10.1038/sj.bjc.6601543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Suminami Y, Kishi F, Sekiguchi K, Kato H. Squamous cell carcinoma antigen is a new member of the serine protease inhibitors. Biochem Biophys Res Commun. 1991;181:51–58. doi: 10.1016/s0006-291x(05)81380-4. [DOI] [PubMed] [Google Scholar]
- 32.Lah TT, Cercek M, Blejec A, Kos J, Gorodetsky E, Somers R, Daskal I. Cathepsin B, a prognostic indicator in lymph node-negative breast carcinoma patients: comparison with cathepsin D, cathepsin L, and other clinical indicators. Clin Cancer Res. 2000;66:578–584. [PubMed] [Google Scholar]
- 33.Murakami A, Suminami Y, Hirakawa H, Nawata S, Numa F, Kato H. Squamous cell carcinoma antigen suppresses radiation-induced cell death. Brit J of Cancer. 2001;84:851–858. doi: 10.1054/bjoc.2000.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suminami Y, Nagashima S, Vujanovic NL, Hirabayashi K, Kato H, Whiteside TL. Inhibition of apoptosis in human tumour cells by the tumour-associated serpin, SCC antigen-1. Brit J Cancer. 2000;82:981–989. doi: 10.1054/bjoc.1999.1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Luke CJ, Pak SC, Askew YS, Naviglia TL, Askew DJ, Nobar SM, Vetica AC, Long OS, Watkins SC, Stolz DB, Barstead RJ, Moulder GL, Bromme D, Silverman GA. An intracellular serpin regulates necrosis by inhibiting the induction and sequelae of lysomoal injury. Cell. 2007;130:1108–1119. doi: 10.1016/j.cell.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.