Abstract
The gene encoding the cytochrome P450 CYP121 is essential for Mycobacterium tuberculosis. However, the CYP121 catalytic activity remains unknown. Here, we show that the cyclodipeptide cyclo(l-Tyr-l-Tyr) (cYY) binds to CYP121, and is efficiently converted into a single major product in a CYP121 activity assay containing spinach ferredoxin and ferredoxin reductase. NMR spectroscopy analysis of the reaction product shows that CYP121 catalyzes the formation of an intramolecular C-C bond between 2 tyrosyl carbon atoms of cYY resulting in a novel chemical entity. The X-ray structure of cYY-bound CYP121, solved at high resolution (1.4 Å), reveals one cYY molecule with full occupancy in the large active site cavity. One cYY tyrosyl approaches the heme and establishes a specific H-bonding network with Ser-237, Gln-385, Arg-386, and 3 water molecules, including the sixth iron ligand. These observations are consistent with low temperature EPR spectra of cYY-bound CYP121 showing a change in the heme environment with the persistence of the sixth heme iron ligand. As the carbon atoms involved in the final C-C coupling are located 5.4 Å apart according to the CYP121-cYY complex crystal structure, we propose that C-C coupling is concomitant with substrate tyrosyl movements. This study provides insight into the catalytic activity, mechanism, and biological function of CYP121. Also, it provides clues for rational design of putative CYP121 substrate-based antimycobacterial agents.
Keywords: C-C coupling, cyclopeptide
Over the last twenty years, tuberculosis has made a dramatic come-back worldwide and is currently responsible for >2 million deaths annually (World Health Organization fact sheet on tuberculosis at http://www.who.int/mediacentre/factsheets/fs104/en/index.html). The major causes are the high susceptibility of immunocompromised individuals to tuberculosis and the emergence of numerous multidrug-resistant (MDR) strains of Mycobacterium tuberculosis, the pathogen responsible for human tuberculosis (1). This situation has created an urgent need for novel strategies and drugs to combat this human disease.
The complete sequence of the M. tuberculosis genome revealed various features that help elucidate the physiology of this pathogen (2). For example, twenty cytochrome P450 (P450) enzymes are encoded in the genome, presumably linked to the extensive lipid metabolism found in this organism. P450s are hemoproteins that catalyze numerous oxidation reactions generally involving hydrophobic substrates (3). Although these enzymes have essential functions in eukaryotes, they have long been suspected to be involved only in catabolic pathways and secondary metabolite synthesis in prokaryotes (4). However, the antimycobacterial and antitubercular activities of azoles—a group of non-selective P450 inhibitors— strongly suggest essential functions for M. tuberculosis P450s (5–8). Recently, the gene encoding the P450 CYP121, rv2276, was shown to be essential for M. tuberculosis viability: Munro and coworkers (9) were unable to obtain a chromosomal rv2276 knockout mutant of M. tuberculosis unless a complementing vector carrying rv2276 was present in trans. Furthermore, the correlation between the minimum inhibitory concentration values determined for azole drugs and the binding constant values of these azoles to CYP121 indicates that CYP121 may be the major target of these azoles in vivo (9). However, despite structural similarities with the macrolide oxygenase P450eryF (10), the substrate of, and the transformation catalyzed by, CYP121 remain unknown. The elucidation of the reaction catalyzed by CYP121 would allow a better understanding of M. tuberculosis physiology and the design of selective inhibitors.
The gene encoding CYP121 is associated in an operon-like structure with rv2275 (2, 11), suggesting that the two corresponding proteins might be involved in the same metabolic pathway. Interestingly, Rv2275 was recently shown to catalyze the formation of tyrosyl-containing cyclodipeptides (12). More than 90% of the cyclodipeptides synthesized by recombinant Escherichia coli expressing rv2275 are cyclo(l-Tyr-l-Tyr) (cYY), with each other cyclodipeptide synthesized representing <5%. These findings suggest that cYY is the main product of Rv2275 activity. We therefore investigated the relationships between CYP121 and cYY.
Results
Spectroscopic Characterization of cYY Binding to CYP121.
UV-Vis spectroscopy was used to probe the interaction between cYY and CYP121. Increasing concentrations of cYY were added to a purified CYP121 solution and the mixture followed spectroscopically. Binding of cYY to CYP121 was substrate-like with a shift of the major Soret band from 415 nm to ≈395 nm and the concomitant appearance of the charge-transfer band at ≈645 nm (Fig. 1). These spectral variations as a function of cYY concentration led us to determine a binding constant value for the interaction between cYY and CYP121: it was 21.3 ± 3.5 μM (Fig. S1). Such spectral variations are associated with the change of the heme iron from the low spin (LS) to the high spin (HS) form (13). However, this transition was not complete as a significant LS signal persisted even at saturating cYY concentrations. 9-GHz EPR spectroscopy was used to study the changes in the electronic environment induced by cYY binding to the CYP121 heme. The spectrum recorded at 20 K for ligand-free enzyme (Fig. 2, black traces) was very similar to that previously described, showing the enzyme predominantly in the LS state with effective g-values of 2.49 (gz), 2.25 (gy) and 1.89 (gx) (10, 14). Upon addition of cYY, the major signal remained typical of LS ferric heme iron (Fig. 2, blue traces). However, the effective g-values of the LS species shifted (2.46, 2.25 and 1.90) and the resonances at gx and gz sharpened, indicative of a modification of the heme iron environment.
Fig. 1.
Spectral titration of CYP121 with cYY. The spectral changes induced on titration of CYP121 (2 μM) with cYY (0–463 μM) are shown. Spectra corresponding to 0 and 463 μM cYY are in black. Arrows indicate directions of change in spectra upon addition of cyclodipeptide. The chemical structure of cYY is shown with the diketopiperazine (DKP) ring in bold.
Fig. 2.
EPR features of the LS ferric heme of cYY-bound CYP121. 9-GHz EPR spectra of substrate-free CYP121 (black) and CYP121 after addition of a 2-fold excess of cYY (blue) were recorded at 20 K under non-saturating conditions using a microwave power of 0.05 mW and a modulation amplitude of 16 G.
CYP121 Catalyzes the C-C Coupling of cYY Aryl Groups.
P450s activity is dependent on the supply of electrons to the iron from an electron transport chain. Prokaryotic P450s preferentially use a two-component system consisting of ferredoxin and ferredoxin reductase (15). We investigated CYP121 activity in a model system involving spinach ferredoxin and ferredoxin reductase. Incubation of CYP121 and cYY in the presence of the electron transport chain and NADPH (see Materials and Methods for details on concentrations of the reactants) resulted in complete consumption of cYY in <10 min (Fig. 3; retention time ≈34 min, [MH]+ ion at m/z 327) with the concomitant appearance of a less hydrophobic product (P1) with a retention time ≈26 min and [MH]+ ion at m/z 325 (2-unit different to cYY). The appearance of P1 was dependent on the presence of both cYY and CYP121 (Fig. S2). The absence of one of the electron transport chain components or the addition of a P450 inhibitor abolished both cYY consumption and P1 formation (Fig. S2). The consumption of cYY was estimated to be 3.8 nmoles min−1 nmole−1, under our experimental conditions.
Fig. 3.
cYY conversion by CYP121. CYP121, an electron transport chain, NADPH and cYY were incubated at 30 °C for various times (0, 1, 3, 10, and 30 min) then acidified and analyzed by LC-MS. Chromatograms were monitored at 214 nm and are shown for elution times from 10 to 45 min. NADPH and cYY were identified by comparison with standards. The chemical structure of P1 is shown.
We then investigated the chemical structure of P1 by NMR. Purified P1 was recovered from large-scale incubation. The NMR spectra of P1 exhibited a single set of 1H and 13C resonances for the two residues indicative of a C2-symmetric structure (Table S1). 1H and 13C resonances were readily assigned using 1D 1H, 1D 13C, 2D 1H-1H COSY, 2D 1H-13C HSQC, and 2D 1H-13C HMBC experiments (Table S1). The aromatic resonances on the NMR spectra consisted of three CH groups and three quaternary carbons, in agreement with a covalent modification of the phenyl rings. Indeed, the analysis of homonuclear coupling constants and of characteristic long-range scalar correlations on the 13C-1H HMBC spectrum indicated the presence of a C-C bond between the two aromatic cycles in the ortho position with respect to the OH group (Cε1 position; Fig. 3). These results are consistent with the mass spectrometry data showing a 2-unit difference between cYY and P1.
General Structural Features of cYY-Bound CYP121.
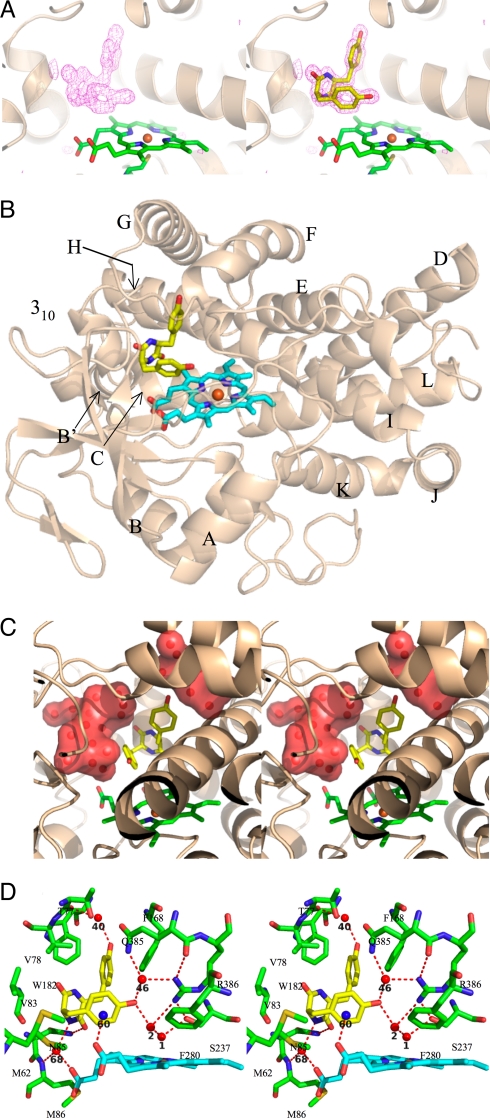
To investigate further the molecular basis of the unusual reaction catalyzed by CYP121, the crystal structure of cYY-bound CYP121 was solved and refined at high resolution (1.4 Å). For structural comparison, we used the crystal structure of native CYP121 solved and refined at the same resolution in our laboratory (Table S2) that is very similar to that previously reported (PDB entry 1N40) (10). The omit map calculated from the data collected for the CYP121-cYY complex shows unambiguous electron density for the cYY in the heme distal pocket (Fig. 4A). The cYY molecule occupies the binding pocket with a full occupancy according to the B-factor of the ligand (10 Å2): it was of the same order as the neighboring residues after refinement. No significant structural changes were observed between the ligand-free and substrate-bound CYP121 crystal structures either for the Cα trace (Fig. S3A; the root mean square deviation after superimposition of ligand-free and substrate-bound complex is 0.16 Å for 388 Cα atoms) or for amino acid side chains of the substrate-binding pocket (Fig. S3B).
Fig. 4.
Crystal structure of cYY-bound CYP121. (A) Fo-Fc electron density map contoured at 4σ level (pink) was calculated after 20 cycles of refinement upon removal of the ligand. The overall structure of CYP121 is in cartoon mode colored in wheat, the heme and Cys-345 are represented as sticks and colored in green, with oxygen, nitrogen and sulfur in red, blue and pale yellow, respectively. (Left) Representation of the active site after removal of cYY. (Right) The refined cYY is represented as sticks and colored in yellow (carbon atoms), red (oxygen atoms), and blue (nitrogen atoms). (B) The complex is shown in cartoon representation, with the heme and cYY represented as sticks. The CYP121 backbone is in wheat. The heme is in cyan with the iron in orange, and the cYY is in yellow, with oxygen atoms in red and nitrogen atoms in blue. The helices are labeled according to the previously published CYP121 crystal structure (10). (C) A stereoview of the active site highlighting the water-filled pockets surrounding the cYY. CYP121 is shown in cartoon representation in wheat. The heme and cYY are represented as sticks. The heme is in cyan with the iron in orange, and the cYY is in yellow, with oxygen atoms in red and nitrogen atoms in blue. Water molecules are represented as red spheres and water-filled pockets are shown in surface mode in red. (D) A stereoview in stick representation of the active site of CYP121 in complex with cYY. Cyclodipeptide is shown in yellow, the heme is in cyan with the iron represented as an orange sphere, CYP121 residues are in green and oxygen and nitrogen atoms are in red and blue, respectively. Water molecules are represented as red spheres and the cation presumed to interact with cYY is shown as a blue sphere. Probable H-bonds between CYP121, cyclodipeptide and non-bonded molecules are indicated as dotted lines. The numbering of non-bonded molecules is according to the pdb file.
The cYY Anchoring to CYP121.
The diketopiperazine (DKP) ring faces the 310 helix and B' helix. One tyrosine side chain points between helices F and G and the other tyrosine side chain approaches the iron from the side parallel to the heme and points toward the I helix (Fig. 4B). The DKP ring adopts a flattened-boat conformation with a dihedral angle of 15° between the two peptide bonds, similar to what has been observed in other disubstituted DKPs in solution (16). Furthermore, one cYY tyrosyl faces the DKP ring, a conformation observed for tyrosyl-containing DKPs in solution (17). In this conformation, cYY only partially fills the large active site cavity (1350 Å3; 10): numerous water molecules remain in this cavity after cYY binding, and define two water-filled pockets around cYY (Fig. 4C). The location and conformation of the substrate in the pocket place the hydroxyl group and the Cε1 carbon atom of one substrate tyrosyl close to the heme (6.25 Å and 6.07 Å from the iron, respectively). Binding of cYY to CYP121 involves numerous van der Waals contacts with hydrophobic side chains of Met-62, Val-78, Val-83, Phe-168, Trp-182, Ala-233, Phe-280 and the heme macrocycle. A large network of stacking interactions is observed between Trp-182, Phe-168, the two tyrosyls of cYY and Phe-280. Only one direct hydrogen bond between cYY and CYP121 is observed: it bridges one carbonyl of the DKP cycle and the Nδ2 of Asn-85 (Fig. 4D). There are water-mediated polar contacts between the hydroxyl of the substrate tyrosyl pointing between helices F and G and the main chain oxygen of Thr-77 and between one nitrogen of the DKP cycle and both the main chain nitrogen of Met-86 and one propionate group of the heme (Fig. 4D). Finally, a density peak is located 2.7 Å from one propionate group of the heme and at an equal distance (3.6–3.8 Å) from each of the six aromatic carbons of the tyrosyl (Fig. 4D). This geometry is consistent with a cation-π interaction and because our crystallization solution contained ammonium sulfate we modeled this density with a NH4+.
An Unusual H-Bonding Pattern at the Oxygen-Binding Site.
An intriguing feature of this structure is the water-mediated H-bonded network just above the heme upon cYY binding (Fig. 4D). The substrate hydroxyl (cYY-OH) interacts with the heme iron through two tightly H-bonded water molecules: WAT2 and WAT1 (distances between the heteroatoms of cYY-OH/WAT2/WAT1/Fe: 2.6 Å, 2.4 Å and 2.4 Å, respectively; B-factor values: 15 Å2, 23 Å2, 19 Å2 and 9 Å2, respectively). The sixth iron ligand WAT1 is not displaced, as indicated by its B-factor value and by the position of the iron in the plane of the heme both being similar to those in the X-ray structure of ligand-free CYP121. Furthermore, cYY-OH also interacts with the Nε2 of Gln-385 and one nitrogen of the Arg-386 guanidinium through WAT46, and this is stabilized on substrate binding (B-factor values of 15 Å2 and 9 Å2 in ligand-free and cYY-bound CYP121 structures, respectively). As in the ligand-free structure, the Ser-237 hydroxyl interacts with the sixth iron ligand and one nitrogen of the Arg-386 guanidinium.
We compared the active site pocket between the free and cYY-bound enzymes: the presence of cYY is associated with only subtle modifications of the hydrogen bond network above the heme (Fig. S4). The substrate hydroxyl substitutes a water molecule (WAT661) with distances of <0.7 Å and does not interact with nitrogen of Arg-386 guanidinium, whereas WAT661 is involved in such an interaction in the ligand-free CYP121 structure. WAT2 is shifted by ≈0.9 Å and becomes H-bonded only to one nitrogen of Arg-386, concomitant with its stabilization (B-factor values of 33 Å2 and 23 Å2 in ligand-free and cYY-bound CYP121 structures, respectively). All other interactions are conserved in the two structures.
Discussion
The substrate of, and the transformation catalyzed by, any particular P450 cannot generally be deduced from amino acid sequence unless sequence identity with a P450 of known function is ≥40%. The greatest similarities between the CYP121 amino acid sequence and other P450 sequences in databases were ≈34% identity, and thus were too low to predict a function for CYP121 confidently. Nevertheless, the structural similarities between CYP121 and the macrolide oxygenase P450eryF suggested that polyketides or macrolides may be substrates for CYP121 (10). Consequently, the identification in our study of the cyclodipeptide cYY as a substrate is unexpected. However, binding constant (21.3 μM) and substrate turnover (3.8 nmoles min−1 nmole−1) values are consistent with those reported for another P450 catalyzing C-C bond formation (18). Furthermore, very few byproducts were formed during the reaction indicating high specificity of cYY transformation by CYP121 (Fig. 3). Cyclodipeptides belong to the large family of the DKP molecules characterized by a piperazine 2–5 dione ring (Fig. 1). DKPs are not usual biological substrates for P450s: as far as we are aware, there are only a few descriptions of metabolic pathways involving the conversion of DKPs by P450s (19–21). In these cases, the P450s were suspected to catalyze the insertion of an oxygen atom. Thus, CYP121 exhibits a novel DKP-modifying activity that catalyzes the formation of a C-C bond between the two tyrosyl side chains of cYY. The resulting molecule is not found in Chemical Abstracts or in the Cambridge Structural Database (last interrogation in December 2008) and therefore is a chemical entity that has not previously been described.
The catalytic activity of CYP121 together with the resolution of the X-ray structure of cYY-bound CYP121 raise intriguing questions about the catalytic mechanism and the final events leading to the formation of the C-C bond. One feature specific to the CYP121-cYY complex revealed by our study is the H-bonded network involving cYY, residues of the active site and water molecules. In particular, the X-ray structure of cYY-bound CYP121 and low temperature EPR experiments indicate that the sixth iron ligand is not displaced and participates in this H-bonded network. The difficulty of displacing the sixth iron ligand has previously been observed upon fluconazole binding to CYP121 (14). In P450s, substrate binding generally provokes the displacement of the sixth iron ligand, such that the iron becomes penta-coordinated and HS (22). In CYP121, a substrate-induced HS signal was clearly observed only during titration experiments performed at room temperature. The native CYP121 spin state has been reported to be slightly sensitive to temperature (23). A spin state thermal equilibrium for the CYP121 heme iron is consistent with the absence of any significant HS signal in the low-temperature EPR spectrum of cYY-bound CYP121.
C-C coupling reactions involve the two carbon atoms being activated and positioned in sufficient proximity to allow covalent linkage (18, 24–28). The crystal structure of cYY-bound CYP121 clearly shows a minimal distance of 5.4 Å between two carbon atoms involved in the final bond. In addition, the NMR structures of tyrosine-containing DKPs show a preferential conformation with the tyrosyl side chain facing the DKP ring, similar to that observed in the cYY-bound CYP121 structure (17). This preferential conformation does not allow a close approach of the other tyrosyl for C-C coupling because of steric hindrance. A possible explanation, in view of the steric constraints arising from the chemical nature of the substrate, is that the two tyrosyl side chains of the substrate rotate around their Cα-Cβ bond during catalysis. The X-ray structure of cYY-bound CYP121 clearly shows a large water-filled pocket just above the substrate tyrosyl side chain facing the DKP ring and approaching the heme (Fig. 4C). A closer examination of possible movement indicates that this tyrosyl side chain may adopt another tyrosine rotamer (change of χ1 value from 64° to −177°) without steric constraints. Any such movement of this substrate tyrosyl side chain would trigger the movement of the other substrate tyrosyl side chain, so that it comes to face the DKP ring. These movements would presumably be coordinated so that the two tyrosyl side chains are, at some point, sufficiently close together and in the appropriate orientation to form the C-C bond. A biradical mechanism has been suggested for other P450-catalyzed C-C coupling reactions: it involves the highly electrophilic compound I (CpdI; FeIV  O+·) as depicted in Fig. 5(28, 29). CpdI can promote the formation of two tyrosyl radicals, as demonstrated with the horseradish peroxidase, another heme-containing protein forming CpdI as the reactive species (30, 31). Moreover, one of the major reactions observed between two tyrosyl radicals is the formation of a tyrosyl cross link between the carbon atoms positioned ortho with respect to the hydroxyl (32). Consistent with the movements of these tyrosyl side chains, we propose that the activation of the carbon atoms may proceed through a radical mechanism (Fig. 5): reaction of CpdI with the tyrosyl approaching the heme and located ≈6 Å from the iron leads to the formation of the first tyrosyl radical and the FeIV(-OH) reactive species or compound II (CpdII; step 1) (22). This first radical then modifies the electronic properties of the aromatic ring and triggers movements of the tyrosyl side chains; during these movements, the radical could undergo intramolecular transfer to the other tyrosyl (step 2 and 3). The second radical is then generated from CpdII (step 4). Alternatively, the second radical may be generated on the second tyrosyl directly from the heme reactive species when this tyrosyl points toward the heme (step 2 and 3′). The final C-C bond is then formed during the movements of the tyrosyl side chains (step 5 and 6).
O+·) as depicted in Fig. 5(28, 29). CpdI can promote the formation of two tyrosyl radicals, as demonstrated with the horseradish peroxidase, another heme-containing protein forming CpdI as the reactive species (30, 31). Moreover, one of the major reactions observed between two tyrosyl radicals is the formation of a tyrosyl cross link between the carbon atoms positioned ortho with respect to the hydroxyl (32). Consistent with the movements of these tyrosyl side chains, we propose that the activation of the carbon atoms may proceed through a radical mechanism (Fig. 5): reaction of CpdI with the tyrosyl approaching the heme and located ≈6 Å from the iron leads to the formation of the first tyrosyl radical and the FeIV(-OH) reactive species or compound II (CpdII; step 1) (22). This first radical then modifies the electronic properties of the aromatic ring and triggers movements of the tyrosyl side chains; during these movements, the radical could undergo intramolecular transfer to the other tyrosyl (step 2 and 3). The second radical is then generated from CpdII (step 4). Alternatively, the second radical may be generated on the second tyrosyl directly from the heme reactive species when this tyrosyl points toward the heme (step 2 and 3′). The final C-C bond is then formed during the movements of the tyrosyl side chains (step 5 and 6).
Fig. 5.
Proposed mechanism for the intramolecular C-C coupling reaction of cYY by CYP121. For simplification, the DKP ring is represented as a sphere.
The CYP121 gene is essential for M. tuberculosis viability (9), so the identification of the biological activity of CYP121 may lead to a better comprehension of the pathogen's physiology. This has implications for the fight against tuberculosis. Several observations argue for the CYP121 activity we describe being the biological function of CYP121. First, the gene encoding CYP121 is organized in an operon-like structure with rv2275 (11); rv2275 encodes a cyclodipeptide synthase (CDPS) which mainly synthesizes cYY (12). Second, the presence of the same chromosomal organization of rv2275 and the gene encoding CYP121 in all sequenced M. tuberculosis complex genomes suggests that it has biological significance. Third, several studies of the M. tuberculosis transcriptome indicate that rv2275 and rv2276 are not silent and, indeed, are strongly expressed in mycobacterial cells (33–35); furthermore, CYP121 has been detected in mycobacterial extracts (9). According to our results, the requirement for CYP121 might be linked either to a toxic effect of cYY in the cell or to an essential function of the product of cYY transformation by CYP121. Note that DKPs express a wide range of biological activities in prokaryotes, including enzyme inhibition and regulation of gene expression (36). Our study provides a valuable basis for investigating the role of cYY and its product of transformation by CYP121 in mycobacterial extracts; this should led to the identification of the essential function of CYP121 in M. tuberculosis.
Our work makes another potentially useful contribution to the fight against tuberculosis. The identification of a CYP121 substrate may be extremely valuable for the design of antimycobacterial drugs. In addition to being essential to M. tuberculosis viability, CYP121 may be the main target of azoles, a group of antimycobacterial agents (9). However, the use of azole-derived inhibitors in medicinal treatment often leads to serious side effects due to inhibition of host P450s. Designing P450 substrate analogues is a common means for obtaining selective inhibitors with fewer side effects (37). Our findings and the proposed mechanism, could be used to modify cYY in such a way that C-C coupling could be prevented leading to substrate analogues not transformed by CYP121. Analysis of the X-ray structure of cYY-bound CYP121 indicates that there are no steric constraints to the addition of small chemical functions to the carbon atoms involved in C-C coupling. Further analysis of the X-ray structures of ligand-bound CYP121 will allow the design and synthesis of more efficient CYP121 inhibitors with clinical potential for treating M. tuberculosis infections. Alternatively, analogs of the product of cYY transformation by CYP121 identified in this study might also be developed as original CYP121 inhibitors.
Materials and Methods
Production and Purification of CYP121.
CYP121 production, purification and characterization were generally as previously described (23) with some modifications (SI Materials and Methods).
MS.
Electrospray mass spectrometry (MS and LC-MS) was carried out using an Esquire HCT ion trap mass spectrometer (Bruker Daltonik GmbH, Germany) in the positive ion mode. Nebullization and desolvation conditions were optimized to obtain maximum sensitivity. For MS analyses, the CYP121 solution was desalted using Zip Tip C4 (Millipore, Waters) and was directly infused at a flow rate of 3 μL/min. For LC-MS analyses, acidified samples were injected onto a C18 Atlantis column (4.6 × 150 mm, 3 μm, 90 Å) equilibrated in 0.1% formic acid and elution was carried out at 600 μL/min with a linear gradient of 90% CH3CN in 0.1% formic acid. During elution, the flow was split with 8.3% directed to the electrospray mass spectrometer. HyStar/EsquireControl software was used for full scan MS acquisitions. Data analysis software was used for data processing.
The cYY Solutions.
cYY was obtained from NeoMPS and solutions (10–120 mM) were prepared in DMSO (Sigma-Aldrich). Concentrations of cYY solutions were determined by amino acid analyses on an AminoTac JLC-500/V amino acids analyzer (JEOL, Japan) using standard conditions and samples diluted 1 in 50 in water to reduce the DMSO concentration to <2%. A calibration table obtained with a standard amino acid mix (from Pierce) was used for quantification.
CYP121 Absorbance Spectroscopy.
A double-beam Uvikon 943 spectrophotometer and 1-cm pathlength quartz cells were used for absorbance experiments with CYP121. Spectral titration with cYY was done at 20 °C according to standard procedures (13). Difference spectra were used to calculate the overall absorbance variation for each cYY concentration. Thereafter, the calculated values were plotted against the relevant cYY concentrations. Data were fitted to a rectangular hyperbola that was used to determine the binding constant, Ks. Values reported are means of four independent experiments.
Electron Paramagnetic Resonance Spectroscopy.
9-GHz EPR spectra were recorded at 20K on a Bruker Elexsys E500 spectrometer fitted with a standard TE102 cavity equipped with a liquid helium cryostat (Oxford Instrument) and a microwave frequency counter (Bruker ER 049X). Frozen solutions of substrate-free and substrate-bound enzyme in 50 mM Tris-maleate pH 7.2 were analyzed in 4 mm-quartz tubes. For the substrate tests, 40 μL of native enzyme solution (at 0.30 mM) was mixed with 0.5 μL of concentrated cYY solution to obtain a 2-fold substrate excess in the final mix.
Enzyme Assay.
CYP121 activity against cYY was detected by incubating purified CYP121 (5 μM) and cYY (≈0.1 mM) at 30 °C with 1 μM spinach ferredoxin (Sigma-Aldrich), 0.1 unit spinach ferredoxin-NADP+ reductase (Sigma-Aldrich), and 1 mM NADPH (Sigma-Aldrich) in 60 mM K2HPO4/KH2PO4 at pH 7.2. The reaction was started by adding NADPH (100 mM stock solution) after 5 min incubation at 30 °C. At various times, 80 μL aliquots were withdrawn and the reaction was stopped by adding 5 μL 20% trifluoroacetic acid: these samples were then analyzed by LC-MS.
Large-Scale Incubation.
Large-scale incubation of cYY with CYP121 was carried out in a 20-mL reaction mixture consisting of 0.6 mM cYY (3.9 mg), 5 μM CYP121, 1 μM spinach ferredoxin (Sigma-Aldrich), 4 units of spinach ferredoxin-NADP+ reductase (Sigma-Aldrich), 4 mM D-glucose-6-phosphate (sodium salt, Sigma-Aldrich), 20 units of yeast glucose-6-phosphate dehydrogenase (Sigma-Aldrich), 0.4 mM NADP+, and 10 mM MgCl2 in 100 mM Tris·HCl pH 7.2. The reaction mix was incubated at 30 °C for 4 h with the addition of 4 mM D-glucose-6-phosphate every hour. The sample was then acidified and centrifuged and the constituents were separated by semipreparative reverse phase HPLC (C18 column 10 × 250 mm, 5 μm, Vydac) using a linear gradient from 0 to 25% CH3CN (1% CH3CN increase/min) in 0.1% trifluoroacetic acid at a flow rate of 3 mL/min, monitored with UV detection at 220 nm. The desired fractions were collected and lyophilized. Aliquots were analyzed by LC-MS to check purity and confirm the identity of the compounds collected.
NMR Spectroscopy.
The NMR experiments were recorded on a Bruker Avance III spectrometer equipped with a TCI cryoprobe and operating at a 1H frequency of 500 MHz. Lyophilized samples (≈1 mg) were dissolved in DMSO-d6 (Eurisotop) and spectra were recorded at 27 °C. 1H and 13C resonances were assigned through the analysis of 1D 1H, 1D 13C JMOD, 2D 1H-1H COSY, 2D 1H-13C HSQC and 2D 1H-13C HMBC (optimized for long-range heteronuclear couplings of 8 Hz). 1H and 13C chemical shifts were referenced to the DMSO solvent signal (2.50 and 39.5 ppm, respectively). NMR experiments were processed and analyzed with the Bruker TOPSPIN 2.0 program.
Crystallization and Collection of Diffraction Data.
Purified CYP121 (10 mg/mL; ≈230 μM) was crystallized by the sitting drop method using conditions similar to those previously described (38). Cocrystals of CYP121 in complex with cYY were obtained similarly but using a solution containing 2 mM cYY in addition to the protein. The crystals were cryo-cooled in liquid nitrogen, and all datasets were collected at the ESRF on beamline ID23–2 at 1.4 Å resolution. The data were processed with MOSFLM and scaled with SCALA from the CCP4 package (39). The structure of our native CYP121 was solved by molecular replacement with the program MOLREP from CCP4 (40) using the available structure of CYP121 as a search model (PDB entry 1N40) and our native dataset. The structure was refined with REFMAC from CCP4 (41) and inspected with the program TURBO-FRODO (42); it was found to be very similar to the published CYP121 structure (PDB entry 1N40; root mean square deviation on 388 Cα of 0.09 Å) with only slight differences at few amino acid side chains and at the dioxygen that is not observed in our structure (10). In the case of the complex, cYY was built at the end of the refinement process based on the Fo-Fc electron density map and the quality of the electron density was compatible with a complete building of the molecule. X-ray analysis statistics are summarized in Table S2.
Supplementary Material
Acknowledgments.
We thank Florence Doucet-Populaire (Hôpital Antoine Béclerè, Clamart, France) for kindly providing us with M. tuberculosis H37Ra chromosomal DNA, Anabella Ivancich for helpful discussions on EPR spectroscopy, Marcel Delaforge for helpful discussions on P450s biochemistry and assistance with CO experiments, and Vincent Dive for thoughtful comments on the manuscript. This work was supported by the Commissariat à l'Energie Atomique (CEA) and the Centre National de la Recherche Scientifique. A.F. is a recipient of a CEA postdoctoral fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates of native CYP121 and CYP121 in complex with cyclo(l-Tyr-l-Tyr) have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 3G5F and 3G5H, respectively).
This article contains supporting information online at www.pnas.org/cgi/content/full/0812191106/DCSupplemental.
References
- 1.Corbett EL, et al. The growing burden of tuberculosis: Global trends and interactions with the HIV epidemic. Arch Intern Med. 2003;163:1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- 2.Cole ST, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 3.Isin EM, Guengerich FP. Complex reactions catalyzed by cytochrome P450 enzymes. Biochim Biophys Acta. 2007;1770:314–329. doi: 10.1016/j.bbagen.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 4.McLean KJ, et al. The preponderance of P450s in the Mycobacterium tuberculosis genome. Trends Microbiol. 2006;14:220–228. doi: 10.1016/j.tim.2006.03.002. [DOI] [PubMed] [Google Scholar]
- 5.Ahmad Z, Sharma S, Khuller GK. Azole antifungals as novel chemotherapeutic agents against murine tuberculosis. FEMS Microbiol Lett. 2006;261:181–186. doi: 10.1111/j.1574-6968.2006.00350.x. [DOI] [PubMed] [Google Scholar]
- 6.Ahmad Z, Sharma S, Khuller GK. The potential of azole antifungals against latent/persistent tuberculosis. FEMS Microbiol Lett. 2006;258:200–203. doi: 10.1111/j.1574-6968.2006.00224.x. [DOI] [PubMed] [Google Scholar]
- 7.Burguiere A, Hitchen PG, Dover LG, Dell A, Besra GS. Altered expression profile of mycobacterial surface glycopeptidolipids following treatment with the antifungal azole inhibitors econazole and clotrimazole. Microbiology. 2005;151:2087–2095. doi: 10.1099/mic.0.27938-0. [DOI] [PubMed] [Google Scholar]
- 8.McLean KJ, et al. Azole antifungals are potent inhibitors of cytochrome P450 mono-oxygenases and bacterial growth in mycobacteria and streptomycetes. Microbiology. 2002;148:2937–2949. doi: 10.1099/00221287-148-10-2937. [DOI] [PubMed] [Google Scholar]
- 9.McLean KJ, et al. Characterization of active site structure in CYP121: A cytochrome P450 essential for viability of Mycobacterium tuberculosis H37Rv. J Biol Chem. 2008;283:33406–33416. doi: 10.1074/jbc.M802115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leys D, et al. Atomic structure of Mycobacterium tuberculosis CYP121 to 1.06 Å reveals novel features of cytochrome P450. J Biol Chem. 2003;278:5141–5147. doi: 10.1074/jbc.M209928200. [DOI] [PubMed] [Google Scholar]
- 11.Roback P, et al. A predicted operon map for Mycobacterium tuberculosis. Nucleic Acids Res. 2007;35:5085–5095. doi: 10.1093/nar/gkm518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gondry M, et al. Cyclodipeptide synthases are a family of tRNA-dependent peptide bond-forming enzymes. Nat Chem Biol. 2009 doi: 10.1038/nchembio.175. in press. [DOI] [PubMed] [Google Scholar]
- 13.Schenkman JB, Jansson I. Spectral analyses of cytochromes P450. Methods Mol Biol. 2006;320:11–18. doi: 10.1385/1-59259-998-2:11. [DOI] [PubMed] [Google Scholar]
- 14.Seward HE, Roujeinikova A, McLean KJ, Munro AW, Leys D. Crystal structure of the Mycobacterium tuberculosis P450 CYP121-fluconazole complex reveals new azole drug-P450 binding mode. J Biol Chem. 2006;281:39437–39443. doi: 10.1074/jbc.M607665200. [DOI] [PubMed] [Google Scholar]
- 15.Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems–biological variations of electron transport chains. Biochim Biophys Acta. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- 16.MacDonald JC, Whitesides JM. Solid-state structures of hydrogen-bonded tapes based on cyclic secondary diamides. Chem Rev. 1994;94:2383–2420. [Google Scholar]
- 17.Kopple KD, Marr DH. Conformations of cyclic peptides. The folding of cyclic dipeptides containing an aromatic side chain. J Am Chem Soc. 1967;89:6193–6200. doi: 10.1021/ja01000a035. [DOI] [PubMed] [Google Scholar]
- 18.Zhao B, et al. Binding of two flaviolin substrate molecules, oxidative coupling, and crystal structure of Streptomyces coelicolor A3(2) cytochrome P450 158A2. J Biol Chem. 2005;280:11599–11607. doi: 10.1074/jbc.M410933200. [DOI] [PubMed] [Google Scholar]
- 19.Healy FG, Krasnoff SB, Wach M, Gibson DM, Loria R. Involvement of a cytochrome P450 monooxygenase in thaxtomin A biosynthesis by Streptomyces acidiscabies. J Bacteriol. 2002;184:2019–2029. doi: 10.1128/JB.184.7.2019-2029.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schultz AW, et al. Biosynthesis and structures of cyclomarins and cyclomarazines, prenylated cyclic peptides of marine actinobacterial origin. J Am Chem Soc. 2008;130:4507–4516. doi: 10.1021/ja711188x. [DOI] [PubMed] [Google Scholar]
- 21.Tang MR, Sternberg D, Behr RK, Sloma A, Berka RM. Use of transciptional profiling and bioinformatics to solve production problems. Ind Biotechnol. 2006;2:66–74. [Google Scholar]
- 22.Denisov IG, Makris TM, Sligar SG, Schlichting I. Structure and chemistry of cytochrome P450. Chem Rev. 2005;105:2253–2277. doi: 10.1021/cr0307143. [DOI] [PubMed] [Google Scholar]
- 23.McLean KJ, et al. Expression, purification and spectroscopic characterization of the cytochrome P450 CYP121 from Mycobacterium tuberculosis. J Inorg Biochem. 2002;91:527–541. doi: 10.1016/s0162-0134(02)00479-8. [DOI] [PubMed] [Google Scholar]
- 24.Bischoff D, et al. The Biosynthesis of Vancomycin-Type Glycopeptide Antibiotics-The Order of the Cyclization Steps. Angew Chem Int Ed Engl. 2001;40:4688–4691. doi: 10.1002/1521-3773(20011217)40:24<4688::aid-anie4688>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 25.Bischoff D, et al. The Biosynthesis of Vancomycin-Type Glycopeptide Antibiotics-New Insights into the Cyclization Steps. Angew Chem Int Ed Engl. 2001;40:1693–1696. doi: 10.1002/1521-3773(20010504)40:9<1693::aid-anie16930>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 26.Funa N, Funabashi M, Ohnishi Y, Horinouchi S. Biosynthesis of hexahydroxyperylenequinone melanin via oxidative aryl coupling by cytochrome P-450 in Streptomyces griseus. J Bacteriol. 2005;187:8149–8155. doi: 10.1128/JB.187.23.8149-8155.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Howard-Jones AR, Walsh CT. Staurosporine and rebeccamycin aglycones are assembled by the oxidative action of StaP, StaC, and RebC on chromopyrrolic acid. J Am Chem Soc. 2006;128:12289–12298. doi: 10.1021/ja063898m. [DOI] [PubMed] [Google Scholar]
- 28.Ikezawa N, Iwasa K, Sato F. Molecular Cloning and Characterization of CYP80G2, a Cytochrome P450 That Catalyzes an Intramolecular C-C Phenol Coupling of (S)-Reticuline in Magnoflorine Biosynthesis, from Cultured Coptis japonica Cells. J Biol Chem. 2008;283:8810–8821. doi: 10.1074/jbc.M705082200. [DOI] [PubMed] [Google Scholar]
- 29.Makino M, et al. Crystal structures and catalytic mechanism of cytochrome P450 StaP that produces the indolocarbazole skeleton. Proc Natl Acad Sci USA. 2007;104:11591–11596. doi: 10.1073/pnas.0702946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ralston I, Dunford HB. Horseradish peroxidase. XXXII. pH dependence of the oxidation of L-(-)-tyrosine by compound I. Can J Biochem. 1978;56:1115–1119. doi: 10.1139/o78-175. [DOI] [PubMed] [Google Scholar]
- 31.Ralston IM, Dunford HB. Horseradish peroxidase. XLII. Oxidations of L-tyrosine and 3,5-diiodo-L-tyrosine by compound II. Can J Biochem. 1980;58:1270–1276. doi: 10.1139/o80-170. [DOI] [PubMed] [Google Scholar]
- 32.Eickhoff H, Jung G, Rieker A. Oxidative phenol coupling-tyrosine dimers and libraries containing tyrosyl peptide dimers. Tetrahedron. 2001;57:353–364. [Google Scholar]
- 33.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 34.Gao Q, Kripke K, Arinc Z, Voskuil M, Small P. Comparative expression studies of a complex phenotype: Cord formation in Mycobacterium tuberculosis. Tuberculosis. 2004;84:188–196. doi: 10.1016/j.tube.2003.12.013. [DOI] [PubMed] [Google Scholar]
- 35.Gao Q, et al. Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology. 2005;151:5–14. doi: 10.1099/mic.0.27539-0. [DOI] [PubMed] [Google Scholar]
- 36.Prasad C. Bioactive cyclic dipeptides. Peptides. 1995;16:151–164. doi: 10.1016/0196-9781(94)00017-z. [DOI] [PubMed] [Google Scholar]
- 37.Schuster I, Bernhardt R. Inhibition of cytochromes P450: Existing and new promising therapeutic targets. Drug Met Rev. 2007;39:481–499. doi: 10.1080/03602530701498455. [DOI] [PubMed] [Google Scholar]
- 38.Mowat CG, et al. Crystallization and preliminary crystallographic analysis of a novel cytochrome P450 from Mycobacterium tuberculosis. Acta Crystallogr D Biol Crystallogr. 2002;58:704–705. doi: 10.1107/s0907444902002676. [DOI] [PubMed] [Google Scholar]
- 39.CCP4 Collaborative Computational Project N. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 40.Vagin A, Teplyakov A. MOLREP: An automated program for molecular replacement. J Appl Cryst. 1997;30:1022–1025. [Google Scholar]
- 41.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 42.Roussel A, Cambillau C. Silicon Graphics Geometry Partner Directory. Mountain View, CA: Silicon Graphics; 1989. pp. 77–78. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.