Abstract
The influenza A virus M2 proton channel equilibrates pH across the viral membrane during entry and across the trans-Golgi membrane of infected cells during viral maturation. It is an important target of adamantane-family antiviral drugs, but drug resistance has become a critical problem. Two different sites for drug interaction have been proposed. One is a lipid-facing pocket between 2 adjacent transmembrane helices (around Asp-44), at which the drug binds and inhibits proton conductance allosterically. The other is inside the pore (around Ser-31), at which the drug directly blocks proton passage. Here, we describe structural and functional experiments on the mechanism of drug inhibition and resistance. The solution structure of the S31N drug-resistant mutant of M2, a mutant of the highly pathogenic avian influenza subtype H5N1, shows that replacing Ser-31 with Asn has little effect on the structure of the channel pore, but dramatically reduces drug binding to the allosteric site. Mutagenesis and liposomal proton flux assays show that replacing the key residue (Asp-44) in the lipid-facing binding pocket with Ala has a dramatic effect on drug sensitivity, but that the channel remains fully drug sensitive when replacing Ser-31 with Ala. Chemical cross-linking studies indicate an inverse correlation between channel stability and drug resistance. The lipid-facing pocket contains residues from 2 adjacent channel-forming helices. Therefore, it is present only when the helices are tightly packed in the closed conformation. Thus, drug-resistant mutants impair drug binding by destabilizing helix–helix assembly.
Keywords: S31N, structure, NMR
The M2 proton channel of influenza A virus equilibrates pH across the viral membrane during cell entry and across the trans-Golgi membrane of infected cells during viral maturation. It is the target of adamantane compounds such as amantadine and rimantadine, which were the first effective drugs licensed for influenza treatment. Emergence of adamantane-resistant M2 has now greatly compromised the effectiveness of these drugs, with resistance >90% (1). In earlier work, we determined the solution structure of M2 with bound rimantadine (2). The channel has a minimalist architecture (Fig. 1). The polypeptide chain is a single-span membrane protein, which tetramerizes in the viral membrane to form a functional proton conductor. A left-handed 4-helix bundle forms the channel pore (2–5). Tetramerization is further stabilized by intermolecular contacts between C-terminal amphipathic helices (2). The packing of Trp-41 indole rings creates a channel gate (6), which closes off the C-terminal end of the pore. The imidazole rings of His-37 are inside the pore and serve as pH sensors (7). To lock the Trp-41 gate, a network of intersubunit hydrogen bonds and salt bridges link the side chains of Trp-41, Asp-44, and Arg-45, forming a gating triad (2).
Fig. 1.
Proposed adamantane binding sites of the M2 channel. (A) The lipid-facing adamantane binding pocket composed of critical channel gating elements from 2 adjacent TM helices (2RLF). (B) The proposed drug-binding site inside the channel pore (3C9J).
The location of the functional adamantane binding site has been a source of controversy. The solution NMR structure shows 4 equivalent rimantadine sites, accessible from the lipid bilayer at the subunit interface. Trp-41, Ile-42, and Arg-45 from one transmembrane (TM) helix, and Leu-40, Leu-43, and Asp-44 from the adjacent TM helix create this lipid-facing pocket (Fig. 1A) (2). It contains residues of the gating triad, consistent with a model in which the drug inhibits the channel allosterically by stabilizing the closed conformation of the gate. An alternative channel pore binding site (Fig. 1B) has been proposed based on electron density in a crystallographic study of a TM segment of M2 (5); its location would suggest a direct pore-blocking mechanism. In this model, the hydrophobic adamantyl cage is coordinated to the hydroxyls of Ser-31 (Fig. 1B), making the model difficult to reconcile with the chemical properties of the drug (8, 9).
The mutation S31N accounts for the vast majority of all resistant viruses (1). Other mutations known to confer drug resistance (L26F, V27A, A30T, G34E, and L38F) are spread over 3 helical turns of the TM helix (≈17 Å), and cover a region much larger than the dimensions of the drugs (Fig. 1), suggesting that the mechanism of resistance is more complicated than alteration of a localized binding site. Lack of an accurate picture of drug inhibition and resistance has severely limited progress in improving the adamantane inhibitors. To provide this picture, we report a combined structural and functional analysis of M2. Our results show that the lipid-facing pocket is a site of rimantadine inhibition, and that the mechanisms of drug resistance are allosteric.
Results
Functional Relevance of the 2 Proposed Drug Binding Sites.
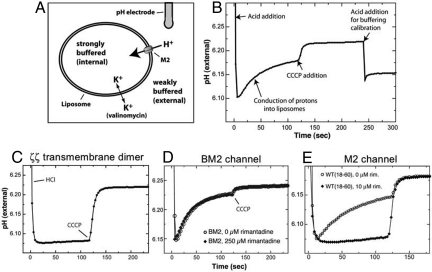
To investigate the functional relevance of the 2 different drug sites proposed, we have established a liposomal proton flux assay (10–13), and performed systematic analyses of various M2 mutants. In the activity assay, the M2 channel is reconstituted into liposomes from a known quantity of purified peptide under strong buffering conditions, and then exchanged into weakly buffered solution. Proton conduction is initiated under conditions of rapid solution mixing by the addition of concentrated acid to the external solution, and then observed as an increase in pH of the external solution as protons move down the pH gradient into the liposomes (Fig. 2 A and B). This experimental setup mimics what occurs after endocytosis of the virus, as the host cell acidifies the endosomal compartment (14). Initial experiments verified that (i) little or no proton translocation is observed in the presence of an arbitrary membrane-embedded protein, in particular, the TM domain of the ζζ dimeric chain of the T cell receptor complex (Fig. 2C) (15); (ii) no inhibition of conductance is observed for a channel known to be adamantane resistant, here, a channel-forming segment of the B/M2 protein from influenza B virus (Fig. 2D); and (iii) the WT M2 construct containing residues 18 to 60 [WT(18–60)] has a proton conductance of ≈6 H+/sec/channel that is completely inhibited by rimantadine at concentrations ≥10 μM rimantadine (Fig. 2E).
Fig. 2.
The liposomal proton flux assay. (A) Schematic representation of the M2 activity assay set-up. For additional details, see Methods. (B) A typical proton flux assay. The assay is initiated by the addition of protons (as HCl) to the external solution. M2 channel activity results in recovery of the pH as protons are conducted into the liposome interior. The experiment is terminated by addition of the proton uncoupler CCCP. The buffering capacity of the system is then measured by adding a known quantity of protons. (C) Proton permeability of the membrane is minimal in the presence of a nonchannel TM protein, the ζζ TM dimer of the T cell receptor complex. (D) The drug-resistant proton channel BM2 from influenza B was tested for inhibition by rimantadine. No inhibition was observed, consistent with its known insensitivity to the adamantanes. (E) The proton conductance of WT(18–60) is almost completely inhibited by 10 μM rimantadine.
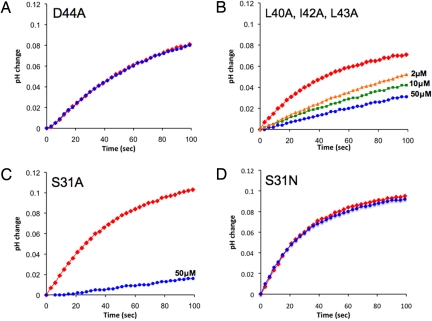
The most direct approach to test the functional relevance of a drug pocket is to introduce pocket-modifying mutations. The lipid-facing pocket has a unique amphipathic property, in which the amine of rimantadine is in contact with the polar sidechain of Asp-44, probably through a hydrogen bond or salt bridge, whereas the methyl groups and adamantyl cage of rimantadine interact with the hydrophobic part of the pocket (Leu-40, Ile-42, and Leu-43; Fig. 1A). Because the rimantadine amine group is essential for adamantane inhibition (8), and because it interacts directly with the carboxyl group of Asp-44, we tested the drug resistance of the D44A mutation. We found that D44A(18–60) is tetrameric, as indicated by chemical cross-linking (Fig. S1), but that its proton conductance is ≈4-fold lower than WT (Table 1). This residual proton conductance is completely insensitive to rimantadine, even at concentrations as high as 50 μM (Fig. 3A). The impaired conductance of the D44A mutant probably explains why this mutation is not observed naturally. In rare isolates (e.g., A/Chicken/Germany/34), the Asp-44 is replaced by Asn. Asn can form enthalpically favorable contacts with the rimantadine amine group, and the D44N mutant has WT-like conductance and retains drug sensitivity (16, 17). Along the proton conduction pathway, the aspartate or Asn at position 44 is the first hydrophilic residue after His-37, and it may be required to relay protons or hydronium ions. A recent study using whole-cell electrophysiology reported that M2 having the D44A mutation showed drug inhibition comparable with WT (18). We are unable to reproduce this result in our assay (for more detailed discussion of this discrepancy, see Discussion).
Table 1.
Inhibition of proton conduction
| Conductance/tetramer | % inhibition at 50 μM rimantadine | % inhibition at 10 μM rimantadine | % inhibition at 2 μM rimantadine | |
|---|---|---|---|---|
| WT(18–60) | 6.5 (0.5; 5) | 99 (5; 4) | 94 (1) | — |
| WT(20–46) | 2.7 (0.4; 3) | 75 (4; 3) | — | — |
| V27A | 9.7 (0.9; 4) | 15 (8; 3) | — | — |
| S31N | 4.2 (0.7; 5) | 5 (8; 3) | — | — |
| S31A | 6.1 (0.5; 5) | 91 (4; 3) | — | 84 (1) |
| D44A | 2.1 (0.8; 7) | 2.5 (9; 5) | — | — |
| L40A, I42A, L43A | 2.5 (0.4; 5) | 74 (5; 3) | 64 (6; 2) | 57 (3; 2) |
Proton conduction rates, in protons/sec/channel, were measured as the initial rate of change on dropping the external pH to 6.05 (internal pH 7.7). The two numbers in parentheses are SD and number of samples, respectively.
Fig. 3.
Investigation of 2 proposed drug-binding sites by functional mutagenesis. Red represents traces in the absence of drug; blue, 50 μM rimantadine; green, 10 μM rimantadine; and orange, 2 μM rimantadine. (A) The D44A mutation abolishes drug inhibition at rimantadine concentration of 50 μM. (B) The triple mutant (L40A, I42A, and L43A) shows substantially decreased inhibition at 3 different concentration of rimantadine. (C) The S31A mutant is very similar to the WT, both in proton conductance and in drug inhibition. (D) The natural drug resistant mutant S31N. The channel concentration is 2.5 μM in A, 2.5 μM in B, 1.25 μM in C, and 2.5 μM in D.
In further experiments, we mutated to Ala the hydrophobic residues Leu-40, Ile-42, and Leu-43, which form the hydrophobic wall of the lipid-facing pocket. These changes decrease the depth and amphipathic character of the lipid-facing pocket. The triple mutant forms functional tetramer (Fig. S1), and is less sensitive to inhibition by rimantadine (Fig. 3B). We also tested channels formed by the TM-only segment, WT(20–46), which preserves the pore-binding site proposed in the X-ray study (5), but exposes the lipid-facing pocket to end effects such as helix fraying. Consistent with disruption of the drug active site by the L40A/I42A/L43A triple mutant or D44A mutation, inhibition of WT(20–46) is substantially reduced with respect to the longer construct (Table 1). A rimantadine concentration of 50 μM was required to obtain ≈75% inhibition, whereas WT(18–60) is almost completely inhibited at ≈10 μM rimantadine.
Last, to test the proposed pore binding site, in which the drug interacts with Ser-31, we mutated Ser-31 to Ala. Both Ala and Ser are frequently found at helix–helix interfaces in membrane proteins (19). Thus, changing Ser-31 to Ala should continue to satisfy the requirements for interhelical stability while changing the chemical nature of the proposed pore binding site. The channel behavior of the S31A mutant is nearly identical to that of WT, both in proton conductance and inhibition of conductance by drug (Fig. 3C), in contrast to S31N, which almost abolishes drug inhibition (Fig. 3D). These results suggest that Ser-31 does not interact directly with the drug, and that the effect of the S31N mutation is allosteric.
Mechanism of Drug Resistance.
Structural analysis of the S31N mutant: the lipid-facing pocket no longer “sees” the drug.
To allow direct comparison between the S31N mutant and WT structure, we examined the S31N(18–60) construct under the same conditions as those used in an NMR study of WT(18–60): 0.75 mM peptide (monomer), ≈300 mM DHPC, 40 mM rimantadine, and 30 mM glutamate (NMR spectrum shown in Fig. S2) (2). The high concentration of rimantadine (40 mM) used in the previous WT study facilitated structure determination by stabilizing the tetramer, but we have found that as little as 4 mM rimantadine specifically perturbs the NMR resonances of the lipid-facing pocket of WT channel.
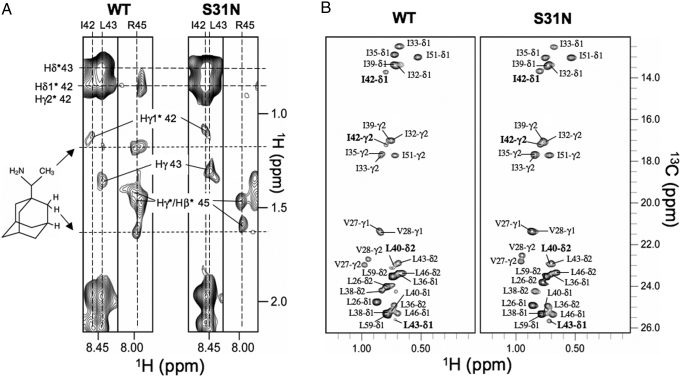
In the S31N mutant, there are no protein-drug NOEs for residues in the lipid-facing pocket (Fig. 4A), indicating that the mutation in the N-terminal portion of the TM affected drug binding to the lipid-facing pocket in the C-terminal TM region. Also, the mutation significantly reduces chemical exchange broadening of the drug pocket methyl resonances (Fig. 4B), indicating that S31N changes the dynamic properties of the lipid-facing binding site.
Fig. 4.
NMR investigation of rimantadine binding to the WT and S31N mutant. (A) Strips from 3D 15N-edited NOESY spectra (110 NOE mixing time) recorded for WT(18–60) and S31N(18–60) in the presence of rimantadine, indicating the absence of drug binding to the S31N mutant at the lipid-facing pocket. (B) WT(18–60) and S31N(18–60) methyl spectra show that the S31N mutation greatly reduced exchange broadening of the methyl-baring residues in the lipid-facing pocket.
From a complete NOE analysis, we derived a structural model of the mutant. The overall structure of S31N(18–60) is similar to that of WT(18–60) (Fig. 5A; Fig. S3), except for minor local changes due to the mutation at residue 31. In the WT structure, the Ser-31 side chain points to the helix–helix packing interface in the closed channel. The Asn-31 side chain in the mutant is also on average positioned at the helical packing interface (Fig. 5B). We suggest that the bulkier Asn side chains at position 31 perturb packing of the TM helices and, therefore, indirectly disrupt the lipid-facing pocket. This observation is not consistent with the proposed drug resistance mechanism for S31N derived from the pore-blocking model, in which carboxamides point into the channel, and form a hydrogen-bonded carbonyl ring in the center of the pore to prevent amantadine from blocking at that position (5).
Fig. 5.
Structural analysis of the S31N drug resistant mutant. (A) Ribbon representation of a structural model of the S31N mutant, illustrating the 2 lysines used in cross-linking. (B) Results of structural refinement of the helix–helix interface of S31N(18–60) based on experimental NOE data. The dashed lines represent intersubunit NOEs involving the side-chain NH2 of Asn-31. For comparison, a similar view of WT(18–60) (PDB, 2RLF) is shown. The side chains of both Ser-31 in the WT and Asn-31 in the mutant are positioned at the helix–helix interface. (C) SDS/PAGE of M2(18–60) variants after being treated with 75 μM DSP for 15 min (for details, see Methods). (D) Same as in C except 2.5 mM DSP and 60 min of reaction time were applied.
Resistance-conferring mutations destabilize helical packing in the tetramers.
How does a mutation near the N-terminal end of the channel affect drug binding near the C-terminal end if the overall structures of WT and S31N mutant are similar? Because the lipid-facing pocket is composed of residues from 2 adjacent TM helices, it is only present when the helices are closely packed. We suggest that energetically unfavorable interhelical packing of Asn side chains at position 31 decreases channel stability and, therefore, indirectly disrupts the lipid-facing pocket. We carried out quantitative chemical cross-linking experiments using dithiobis[succinimidylpropionate] (DSP) to investigate the effect of mutations on the stability of the oligomeric assembly. According to the structure, there are only 2 places where cross-linking can occur, one between Lys-49 of one subunit and Lys-60 of the adjacent subunit, and the other between the N-terminal alpha-amino groups. The conformation-specific cross-linking is, thus, a direct way of reporting dynamics of M2 tetramers, because the rate of cross-linking has a simple dependence on the probability of the reactive groups being close to each other. At a suitable concentration of DSP cross-linkers (75 μM), nearly all of WT could be cross-linked to a tetramer, whereas under the same conditions, the S31N and V27A mutants showed a distribution of monomer, dimer, and tetramer (Fig. 5C). Less efficient cross-linking indicates weaker tetrameric assembly for some regions of the protein; it does not suggest dissociation of the tetramer, because the mutants could all be cross-linked to nearly homogeneous tetramers under a stronger cross-linking condition (Fig. 5D). V27A is apparently more stable than S31N (compare Fig. 5C, lanes 4 and 5), and inhibition of the mutant V27A at 50 μM rimantadine was greater than that of S31N (Table 1). On the contrary, the S31A mutant, which remains fully drug sensitive, forms as stable a tetramer as WT (Fig. 5C). Together, these results indicate that resistance mutations destabilize channel assembly, and that the instability correlates with loss of drug inhibition.
Discussion
The functional and structural data we have presented are all consistent with the proposal that rimantadine inhibition of the M2 channel comes from binding at the lipid-facing pocket; they are not consistent with the alternative site within the pore. First, the structural data show that the channel pore of the S31N mutant is very similar to that of WT, but that the lipid-facing pocket no longer binds the drug (indicated by the absence of drug NOEs). Second, functional experiments show that mutating residues of the lipid-facing pocket to Alanines dramatically lowers sensitivity to drug, whereas mutating Ser-31 inside the pore to Ala has almost no effect. Third, although the prevalence of the S31N resistance mutation might suggest that a drug can bind at this position, neither the known structure-activity relationship (SAR) of adamantane drugs (8), nor the properties of the S31A mutant, nor the structure of S31N itself support this view. Indeed, naturally-occurring drug-resistance mutations have been observed in many positions, which together span >3 alpha-helical turns, a region much larger than the dimensions of the drug. We show here that the S31N mutation results in a more loosely assembled tetrameric channel. Because the lipid-facing binding site is composed of residues from neighboring subunits and can only exist when the 4-helix bundle is tightly assembled (Fig. 6), destabilization of channel assembly disrupts the binding pocket and lowers drug affinity
Fig. 6.
Schematic representation of drug inhibition and drug resistance. In the WT channel, 2 adjacent TM helices form the lipid-facing pocket. The drug binds to the tightly assembled pocket in the closed state, making the channel harder to open. The S31N mutation weakens TM helical packing and thereby disrupts the drug pocket. Consequently, drug affinity is dramatically reduced while channel activity is preserved. The fuzzy lines represent increased conformational heterogeneity caused by weaker helical packing. Channel activation requires His-37 protonation at low pH; thus, destabilization of channel assembly is not sufficient for proton flux.
Destabilization of the channel by mutations, as indicated by less efficient cross-linking, does not indicate that the tetramers fall apart. Results from stronger cross-linking experiments clearly indicate that the mutants are intact tetramers in the closed state (Fig. 5D). We suggest that weaker packing of TM helices in the mutants leads to less efficient cross-linking, but intermolecular contacts between the C-terminal amphipathic helices hold the tetramers together (2). That is, resistance-conferring mutations weaken TM helical packing and increase structural heterogeneity of the closed state (represented by the fuzzy drawings in Fig. 6). We emphasize that, although proton conductance requires loosening of the channel assembly to unlock the Trp-41 gates, destabilization of assembly does not necessarily lead to greater proton conductance, because conductance is dominated by other factors such as protonation of His-37, and arrangement of proton relay groups in the pore. This point is illustrated by our observation that the S31N mutant is not a better conductor than the WT.
Why have resistance mutations closer to the lipid-facing pocket not evolved? Residues 37–46 in the C-terminal portion of the TM domain, which include those that form the drug pocket, are highly conserved (20). This conservation is presumably due to the functional constraints on His-37, Trp-41, Asp-44, and Arg-45 for pH sensing and channel gating. However, there is one observed mutation in the C terminus, L38F, which, together with V27I and D44N, contributes to drug resistance (21). Leu-38 is the only residue in the C-terminal half of the channel, besides the critical residues of the gating triad, that makes intersubunit contacts for channel assembly, and we interpret its contribution to resistance as a weakening of helical packing. The long-range communication between the 3 sites is only understandable in the proposed allosteric mechanism, because it is not possible for a drug-binding site to span this distance.
How can we explain the potentially contradictory data supporting a pore binding site? The strongest support for such a site is the crystallographic study of the TM segment of M2 in the presence of amantadine. Because the TM construct does not include the amphipathic helix that follows the pore segment, the tetramer splays apart at its base, and the lipid-facing pocket is not present. Density on the axis of the pore adjacent to Ser-31 was interpreted as a bound amantadine molecule. This interpretation results in an unlikely pairing of hydrophobic and hydrophilic groups, in which the hydroxyl groups of Ser-31 coordinate the nonpolar adamantyl cage (PDB code 3C9J; Fig. S4a) (5, 9). The structure in the presence of amantadine was determined at relatively low resolution (3.5 Å), making it impossible to identify unambiguously small molecules such as amantadine (the maximum diameter of the approximately spherical adamantyl cage is ≈3.4 Å). A 2.0-Å crystal structure determined in the absence of drug in the same study also showed density near the Ser-31 side chain. This feature comes from a cluster of water molecules (PDB code 3BKD; Fig. S4b), consistent with the hydrophilicity of the hydroxyls. Because the protein conformation and the position of the Ser-31 hydroxyls are essentially identical in the 2 crystal structures, it is difficult to explain how a hydrophobic drug could expel water from the hydrophilic binding site. Until the proposed amantadine density is verified by further experiments, it could be interpreted as water or headgroup of β-octyl glucoside used in the crystallization buffer.
Another potentially conflicting result concerns the functional properties of the D44A mutant. We showed, using the liposomal proton flux assay, that the D44A mutant is insensitive to rimantadine (Fig. 3A), but Jing et al. (18) showed, using whole-cell electrophysiology, that the mutant is as sensitive to the drug as the WT. The discrepancy could come from the fundamental differences between the 2 techniques used. The whole cell is a complex system, containing many endogenous ion channels in addition to the expressed M2 channels. It has been reported that M2, along with several other single-pass membrane proteins, can modulate the behavior of endogenous ion channels in Xenopus oocytes, and that minor mutations in the M2 TM domain alter such modulation (22). Also, in the whole-cell voltage-clamp setup, all ions contribute to the current, which could complicate the result if a mutation alters ion selectivity of the channel. In fact, it was reported that, although the H37E mutant maintains conductance and drug sensitivity, it acquires new selectivity for Cl−/Na+ in addition to H+ (7). It is unclear from Jing et al. (18) whether the ion selectivity of the D44A mutant is different from that of WT. Therefore, we believe that the whole-cell setup is less suitable than the liposome assay for quantifying the effect of mutations on M2 activity and drug inhibition. The latter assay is a simple system, in which only proton conductance is recorded, and in which the conductance must come from the M2 channels. A potential weakness of the liposome assay is the use of a truncated version of M2 (residues 18–60), but its proton conductance is very similar to that observed for the full-length M2 reconstituted into liposomes (11).
In conclusion, we have shown that the lipid-facing pocket near the Trp-41 gate is relevant to adamantane inhibition. The binding site is formed by residues from TM helices of neighboring subunits. Therefore, it can only exist when the 4-helix bundle is tightly assembled, as in the closed channel conformation. Drug binding to the lipid-facing pocket stabilizes the closed conformation, making the channel harder to open. This finding explains the lower affinity of drug for the open channel (21), because channel activation is coupled to destabilization of the 4-helix bundle (2). In principle, the virus could escape drug inhibition through mutations in the lipid-facing pocket. However, most residues in the pocket are highly conserved, and thus, presumably important for function. The reduction in proton conductance by the D44A mutation illustrates the tradeoff required for changing residues in this region of the channel. Full channel activity is preserved in resistance-conferring mutations located far from the drug binding site, while simply destabilizing the TM helical packing. Weaker channel assembly preserves WT proton conductance, as shown for the S31N(18–60) and V27A(18–60) constructs, while at the same time, disrupting the lipid-facing pocket to resist drug binding.
Methods
Liposomal Proton Flux Assay.
An activity assay for M2 channels was established based on works from the Schroeder, Miller, and Busath laboratories (10, 12, 13). In our M2 assays, a proton gradient was used to drive proton conduction, because this most directly mimics what is thought to happen in the cell. Briefly, liposomes were made with identical pH and ion concentrations inside and outside, but highly buffered inside, and only weakly buffered outside. Protein-mediated conductance of protons from the external bath into the liposome interior was initiated by adding hydrochloric acid under continuous rapid mixing. Proton flux was monitored as an increase in pH of the external bath (Fig. 2).
M2 channels were reconstituted into liposomes by mixing 10 mg of Escherichia coli polar lipid extract (Avanti Polar Lipids), 5, 10, or 20 nmol of M2 peptide, and 0.2 nmol of the potassium ionophore valinomycin in 1.1 mL of a 2:1 mixture of chloroform and methanol. The solution was dried down to thin films under nitrogen gas. The films were redissolved in 750 μL of chloroform and dried down a second time under nitrogen, resulting in high-quality, transparent thin films. Liposomes were then formed by resuspending the thin films in strongly-buffered internal liposome buffer (50 mM phosphate/50 mM citrate/122 mM KCl/122 mM NaCl/0.01% NaN3, pH 7.7), and extruding 21 times through 0.2 μM polycarbonate membranes. The external buffer was exchanged by running 750 μL of the liposome solution over a PD-10 column (GE Health Sciences) preequilibrated with weakly-buffered external vesicle buffer (EVB; 2 mM phosphate/2 mM citrate/122 mM KCl/122 mM NaCl/0.01% NaN3, pH 7.8). Final eluted volume was 1.5 mL, containing 5 mg/mL lipid, 3, 6, or 12 μM M2 peptide, and 0.1 μM valinomycin. Valinomycin in small quantities was required to allow potassium ions to flow across the membrane in the opposite direction of protons to avoid generating a charge potential. No proton conductance was observed in the absence of valinomycin. The liposomes had diameters of ≈160 nm, as determined by dynamic light scattering. We estimate that, we estimate that there are ≈30, 60, or 90 channels per liposome, 50% of which, it is assumed, have the correct orientation to conduct protons into the liposome (11). Initial pH inside and outside of liposomes was identical. Protein-mediated conductance of protons from the bath into the liposomes was initiated lowering the external pH to 6.05 by addition of 3 μL of 1 M HCl with continuous rapid mixing with a microstir bar. Proton flux was monitored as an increase in pH of the external bath with a pH microelectrode (InLab). Reported flux rates were taken as the average rate observed over the period from 15 to 45 sec after the addition of HCl. The assay was terminated by the addition of 5 μM of the proton ionophore carbonyl cyanide m-chlorophenylhydrazone (CCCP). The effects of buffering from citrate, phosphate, rimantadine, and lipids was evaluated by addition of 5 μL of 50 mM HCl. Because the proton fluxes are taken as the initial change in pH, a correction factor was required to account for the increased buffering capacity of the solution after the system was uncoupled by CCCP. The excess buffering from the uncoupled liposome interiors was determined in a series of peptide-free controls containing 4.7, 5.0, or 5.3 mg/mL of lipid by adding 5 μL of 50 mM HCl before and after addition of CCCP. The excess buffering capacity of the external buffer was found to be 12 ± 2% that of the CCCP uncoupled system. To assay channel inhibition, rimantadine was added from concentrated stock solutions in anhydrous ethanol (Sigma) 5 minutes before initiation of proton flux. The maximum volume of ethanol added for drug titrations was 7.5 μL. No change in pH on addition of up to 50 μL ethanol was observed.
NMR Spectroscopy.
NMR experiments were conducted at 30 °C on spectrometers equipped with cryogenic probes (Bruker). Sequence specific assignment of backbone 1HN, 15N, and 13Cα chemical shifts were accomplished using a combination of the available WT(18–60) resonance assignments and a pair of HNCA and HNCOCA experiments (23, 24), recorded with a 15N-, 13C-, and 85% 2H-labeled protein. NOEs involving both backbone and side chain protons were assigned using the 3D 15N-edited and 13C-edited NOESYs recorded with NOE mixing times of 110 and 150 ms, respectively, on a sample containing 15N-, 13C-labeled protein, rimantadine, and deuterated DHPC (D35-DHPC) (Avanti Polar Lipids).
Structural Model of the S31N Mutant.
Because S31N(18–60) does not form a rigid tetramer, as indicated by the cross-linking data shown in Fig. 5C, it is not meaningful to calibrate NOE-derived distances to calculate a structure with high precision. Nevertheless, for the purpose of qualitative analysis of the mutant structure, we constructed a model of the S31N(18–60) tetramer based on the observed intra and intersubunit NOEs. Based on the similarities in both chemical shifts and NOE patterns between S31N(18–60) and WT(18–60), the S31N(18–60) structural model was obtained by refining a homology model derived from the WT(18–60) structure against the S31N(18–60) NMR restraints (including 1004 intra and 68 intersubunit NOEs). A low-temperature simulated annealing protocol was implemented in the program X-PLOR-NIH (25), in which the bath is cooled from 500 to 20 K with a temperature step of 20 K, and 6.7 ps of Verlet dynamics at each temperature step, using a time step of 3 fsec. The force constants for NOE restraints were ramped from 25 to 50 kcal·mol−1·Å−2, whereas those for dihedral angles were ramped from 10 to 30 kcal·mol−1·rad−2. The structural restraints yielded an ensemble of 10 structures with a backbone rmsd of 0.87 Å for the TM helix and 1.01 Å for the amphipathic helix (Fig. S3).
Chemical Cross-Linking.
Cross-linking was performed using DSP at different time and cross-linker concentrations. DSP was dissolved in DMSO at 50 or 6 mM, and added to reconstituted protein at 0.13 mM (monomer) in ≈100 mM DHPC to final DSP concentration of 2.5 mM or 75 μM. Samples were incubated at 25 °C for 1 h (for higher DSP concentration) and 15 min (for lower DSP concentration), followed by quenching with 200 mM Tris buffer (pH 7.5) and SDS/PAGE.
Supplementary Material
Acknowledgments.
We thank Stephen Harrison, Kirill Oxenoid, Matthew Call, and Donald Coen for many discussions; and Chris Miller for suggesting the liposomal proton flux assays. This work was supported by National Institutes of Health (NIH) Grant AI067438 and by The Pew Scholars Program in the Biomedical Sciences (J.J.C.). J.R.S was supported by an NIH F32 Postdoctoral Fellowship.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0902548106/DCSupplemental.
References
- 1.Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. J Am Med Assoc. 2006;295:891–894. doi: 10.1001/jama.295.8.joc60020. [DOI] [PubMed] [Google Scholar]
- 2.Schnell JR, Chou JJ. Structure and mechanism of the M2 proton channel of influenza A virus. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinto LH, et al. A functionally defined model for the M2 proton channel of influenza A virus suggests a mechanism for its ion selectivity. Proc Natl Acad Sci USA. 1997;94:11301–11306. doi: 10.1073/pnas.94.21.11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovacs FA, Denny JK, Song Z, Quine JR, Cross TA. Helix tilt of the M2 transmembrane peptide from influenza A virus: An intrinsic property. J Mol Biol. 2000;295:117–125. doi: 10.1006/jmbi.1999.3322. [DOI] [PubMed] [Google Scholar]
- 5.Stouffer AL, et al. Structural basis for the function and inhibition of an influenza virus proton channel. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tang Y, Zaitseva F, Lamb RA, Pinto LH. The gate of the influenza virus M2 proton channel is formed by a single tryptophan residue. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 7.Wang C, Lamb RA, Pinto LH. Activation of the M2 ion channel of influenza virus: A role for the transmembrane domain histidine residue. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aldrich PE, et al. Antiviral agents. 2. Structure-activity relationships of compounds related to 1-adamantanamine. J Med Chem. 1971;14:535–543. doi: 10.1021/jm00288a019. [DOI] [PubMed] [Google Scholar]
- 9.Huang RB, Du QS, Wang CH, Chou KC. An in-depth analysis of the biological functional studies based on the NMR M2 channel structure of influenza A virus. Biochem Biophys Res Commun. 2008;377:1243–1247. doi: 10.1016/j.bbrc.2008.10.148. [DOI] [PubMed] [Google Scholar]
- 10.Nguitragool W, Miller C. Uncoupling of a CLC Cl-/H+ exchange transporter by polyatomic anions. J Mol Biol. 2006;362:682–690. doi: 10.1016/j.jmb.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 11.Lin TI, Schroeder C. Definitive assignment of proton selectivity and attoampere unitary current to the M2 ion channel protein of influenza A virus. J Virol. 2001;75:3647–3656. doi: 10.1128/JVI.75.8.3647-3656.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moffat JC, et al. Proton transport through influenza A virus M2 protein reconstituted in vesicles. Biophys J. 2008;94:434–445. doi: 10.1529/biophysj.107.109082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schroeder C, Ford CM, Wharton SA, Hay AJ. Functional reconstitution in lipid vesicles of influenza virus M2 protein expressed by baculovirus: Evidence for proton transfer activity. J Gen Virol. 1994;75:3477–3484. doi: 10.1099/0022-1317-75-12-3477. [DOI] [PubMed] [Google Scholar]
- 14.Helenius A. Unpacking the incoming influenza virus. Cell. 1992;69:577–578. doi: 10.1016/0092-8674(92)90219-3. [DOI] [PubMed] [Google Scholar]
- 15.Call ME, et al. The structure of the zetazeta transmembrane dimer reveals features essential for its assembly with the T cell receptor. Cell. 2006;127:355–368. doi: 10.1016/j.cell.2006.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chizhmakov IV, et al. Differences in conductance of M2 proton channels of two influenza viruses at low and high pH. J Physiol. 2003;546:427–438. doi: 10.1113/jphysiol.2002.028910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Betakova T, Ciampor F, Hay AJ. Influence of residue 44 on the activity of the M2 proton channel of influenza A virus. J Gen Virol. 2005;86:181–184. doi: 10.1099/vir.0.80358-0. [DOI] [PubMed] [Google Scholar]
- 18.Jing X, et al. Functional studies indicate amantadine binds to the pore of the influenza A virus M2 proton-selective ion channel. Proc Natl Acad Sci USA. 2008;105:10967–10972. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walters RF, DeGrado WF. Helix-packing motifs in membrane proteins. Proc Natl Acad Sci USA. 2006;103:13658–13663. doi: 10.1073/pnas.0605878103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito T, Gorman OT, Kawaoka Y, Bean WJ, Webster RG. Evolutionary analysis of the influenza A virus M gene with comparison of the M1 and M2 proteins. J Virol. 1991;65:5491–5498. doi: 10.1128/jvi.65.10.5491-5498.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang C, Takeuchi K, Pinto LH, Lamb RA. Ion channel activity of influenza A virus M2 protein: Characterization of the amantadine block. J Virol. 1993;67:5585–5594. doi: 10.1128/jvi.67.9.5585-5594.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shimbo K, Brassard DL, Lamb RA, Pinto LH. Viral and cellular small integral membrane proteins can modify ion channels endogenous to Xenopus oocytes. Biophys J. 1995;69:1819–1829. doi: 10.1016/S0006-3495(95)80052-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salzmann M, Wider G, Pervushin K, Wuthrich K. Improved sensitivity and coherence selection for [N-15,H-1]-TROSY elements in triple resonance experiments. J Biomol NMR. 1999;15:181–184. doi: 10.1023/a:1008358030477. [DOI] [PubMed] [Google Scholar]
- 24.Kay LE, Ikura M, Tschudin R, Bax A. Three-dimensional triple resonance NMR spectroscopy of isotopically enriched proteins. J Magn Reson. 1990;89:496–514. doi: 10.1016/j.jmr.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 25.Schwieters CD, Kuszewski J, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2002;160:66–74. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.