Abstract
Cell elongation in plants is controlled by environmental cues such as light and internal growth regulators including plant steroid hormones, brassinosteroids (BRs). In this study, we found that 3 related receptor-like kinases (RLKs), HERCULES1, THESEUS1, and FERONIA, are transcriptionally induced by BRs and are down-regulated in the loss-of-function BR mutant bri1 and up-regulated in the constitutive BR-response mutant bes1-D. These RLKs belong to the CrRLK family that has 17 members in Arabidopsis. We hypothesize that these RLKs are involved in BR-regulated processes. Although 2 of the RLKs were recently found to mediate male-female interaction during pollen tube reception (FERONIA) and to sense cell wall integrity (THESEUS1), our genetic studies demonstrated that they are required for cell elongation during vegetative growth as herk1 the1 double and fer RNAi mutants displayed striking dwarf phenotypes. The herk1 the1 double mutant enhances the dwarf phenotype of bri1 and partially suppresses bes1-D phenotype, supporting a role of HERK1/THE1 in BR-mediated cell elongation. Microarray experiments demonstrated that these RLKs control the expression of a unique set of genes including those implicated in cell elongation and 16% of the genes affected in herk1 the1 are regulated by BRs. Our results, therefore, identify a previously unknown pathway that functions cooperatively with, but largely independent of the BR pathway to regulate cell elongation. The work establishes a platform to identify other signaling components in this important pathway for plant growth and provides a paradigm to study the coordination of independent pathways in the regulation of a common biological process.
Keywords: brassinosteroid, FERONIA, HERCULES1, THESEUS1
Understanding signaling mechanisms and gene networks regulating growth and development is a central issue in developmental biology. Plant growth and development are controlled by intrinsic growth regulators or hormones and environmental cues through interconnected signal transduction pathways (1). A single hormone can regulate many different processes and likewise different hormones can cooperate to control the same cellular process. Mechanisms underlying the cross-talk between different signaling pathways have been described recently (2, 3). Plant steroid hormones, brassinosteroids (BRs), regulate many growth and developmental processes such as cell elongation, senescence, vascular development, reproduction, and various stress responses (4–7). BRs signal through a membrane-localized, leucine-rich repeat (LRR) receptor kinase BRI1 and coreceptor BAK1 to regulate several intermediate signaling components (4, 8–10). BR signaling eventually controls BES1 and BZR1 family transcription factors, which mediate the expression of many genes for various BR responses (11–14). Consistent with the main function of BRs in cell elongation, BRs promote the expression of a number of cell wall remodeling enzymes required for cell elongation (15–17). In addition, BRs also regulate a variety of other genes implicated in signal transduction including several receptor-like kinases (RLKs), which may be involved in communicating BR signals to other pathways. In this study, we report on the characterization of 3 related genes, HERCULES Receptor Kinase 1 (HERK1, At3g46290), THESEUS1 (THE1, At5g54380) and FERONIA (FER, At3g51550), all of which are induced by BRs.
These 3 RLKs belong to CrRLK family that has 17 members in Arabidopsis (18, 19). During the course of characterizing these genes, THE1 and FER were reported to repress cell elongation in various systems (18). Loss-of-function mutations in THE1 were shown to suppress the hypocotyl elongation defect of a cellulose-deficient mutant cesA6 in dark-growing seedlings. Therefore, THE1 was proposed to be a sensor of cell wall integrity - inhibiting cell elongation when the cell wall is damaged (20). However, FER was found to be involved in male-female interaction during pollen tube reception in Arabidopsis (21). More specifically, FER, which is expressed in synergid cells of female gametophytes, was proposed to receive a signal from incoming pollen tubes and to inhibit pollen tube elongation, leading to eruption of the pollen tip and release of sperm cells to fertilize the egg cell (21, 22). In complete loss-of-function mutants of FER, feronia (fer) and sirene (sir), pollen tubes continued to grow without fertilizing the egg cell (21, 23, 24).
The broad expression patterns of FER, THE1, and HERK1, especially during vegetative growth (18), imply additional functions for these RLKs. Our genetic studies demonstrated that the HERK1/THE1/FER pathway is required for optimal cell elongation and interacts with the BR pathway. Microarray experiments indicated that most of the genes affected in the mutants are not BR target genes. We therefore conclude that HERK1/THE1/FER are components of a newly recognized pathway that functions cooperatively, but largely independent of the BR pathway to promote cell elongation.
Results
HERK1, THE1, and FER Are Induced by BRs and Modulated in BR mutants.
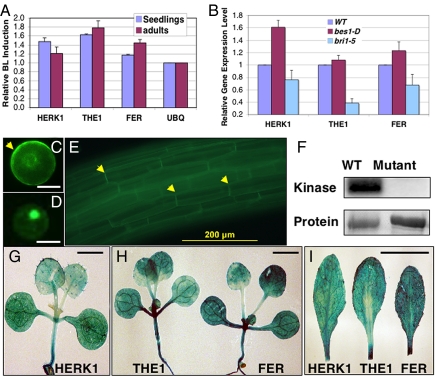
In our effort to identify and characterize BR regulated genes, we found that HERK1, THE1 and FER are induced by BRs as revealed by both published (15) and publicly available microarray data (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). HERK1, THE1, and FER are the most highly induced genes by BR and, therefore, were chosen for further study. Our quantitative RT-PCR experiments confirmed that brassinolide (BL), the most active BR, up-regulated these RLKs by ≈20–80% in seedlings and/or adult plants (Fig. 1A). The expression changes are significant as demonstrated by student's t test (P < 0.01). In addition, the expression levels of the RLK genes were reduced in the loss-of-function mutant bri1 and increased in the constitutive BR-response mutant bes1-D (Fig. 1B). The close relationship of these 3 RLKs in the phylogenetic tree, especially between HERK1 and THE1 (Fig. S1) and their induction by BRs suggest an involvement in BR-regulated processes.
Fig. 1.
HERK1, THE1, and FER are related receptor-like kinases induced by BRs. (A) Expression of HERK1, THE1, and FER is induced by BL treatment in 10-day-old seedlings and 4-week-old adult plants. Triplicate biological samples were used to prepare RNA for quantitative RT PCR. The relative expression levels compared with UBQ5 gene were used to determine the BL-induction levels of each gene. The averages and standard deviations from 3 biological repeats are shown. (B) HERK1, THE1, and FER are up-regulated in bes1-D and down-regulated in bri1. Ten-day-old seedlings were used to prepare RNA for qRT-PCR as described in A. (C–E) HERK1 cellular localization: A HERK1-GFP (C) or BES1-D-GFP (D) construct was introduced into protoplasts or transgenic plants (E). HERK1 is mostly localized at the plasma membrane (C and E), whereas BES1-D is primarily in the nucleus (D). (F) HERK1 displayed kinase activity. Recombinant proteins fused with MBP representing the WT HERK1 kinase domain (WT), and a mutant version with disrupted kinase activity (mutant), were used in kinase assay. Autophosphorylation was detected by phosphorimaging, and the proteins were detected by SYPRO RUBY staining. (G–I) Expression patterns of HERK1, THE1, and FER in 10-day-old seedlings (G and H) or 3-week-old adult leaves (I) as revealed by GUS reporter gene. (Scale bars: C and D, 10 μm; G and H, 2.5 mm; I, 10 mm.)
We first determined the cellular localization of HERK1 by fluorescence microscopy. Similar to FER (21) and THE1 (20), the HERK1-GFP fusion localized to the plasma membrane when expressed in Arabidopsis protoplasts and plants (Fig. 1 C and E). The kinase activity of HERK1 was tested with recombinant proteins expressed and purified from Escherichia coli. The WT kinase domain of HERK1 was autophosphorylated, whereas a mutant form, in which a conserved lysine was mutated to arginine (K513R), lost its kinase activity in vitro (Fig. 1F). Taken together, these results demonstrate that HERK1, like its homologs THE1 and FER, is a plasma membrane-localized receptor kinase.
HERK1, THE1 and FER are universally expressed in most vegetative tissues, including leaves, stems and roots as revealed by our promoter-GUS (β-glucuronidase) reporter gene studies (Fig. 1 G–I) and by studies carried out by others (20, 21). The expression is particularly strong in the regions undergoing cell elongation, such as in hypocotyls and leaf petioles. Because HERK1, THE1, and FER are regulated by BRs and modulated in BR mutants, we hypothesize that HERK1, THE1, and FER have a more general role in cell elongation processes, in addition to the function of THE1 in sensing cell wall integrity and FER in pollen tube and female gametophyte interactions.
HERK1, THE1, and FER Are Required for Cell Elongation During Vegetative Growth.
T-DNA insertion mutants were identified for both HERK1 and THE1, designated as herk1-1 and the1-4, which had the T-DNA insertions in the kinase domains in both genes. Both mutants are null because transcripts were not detected in mutant plants (see Fig. 4). The close homology and overlapping expression patterns of HERK1 and THE1 prompted us to test whether these two genes function redundantly. Toward that end, we constructed a herk1-1 the1-4 double mutant, referred to hereafter as herk1 the1. Because there are no T-DNA insertion mutants available for FER and the null allele of fer was embryonic lethal due to failed fertilization, we used an artificial microRNA, or amiRNA (25) to knockdown the FER gene to determine its function during vegetative growth. An amiRNA specifically targeting FER was cloned into an expression vector driven by the strong, constitutive BRI1 gene promoter (26). More than 80% of the transgenic lines displayed a reduced growth phenotype (Fig. 2B). Two transgenic lines, designated as fer1 and fer2, with different degrees of FER reduction, were chosen for further studies.
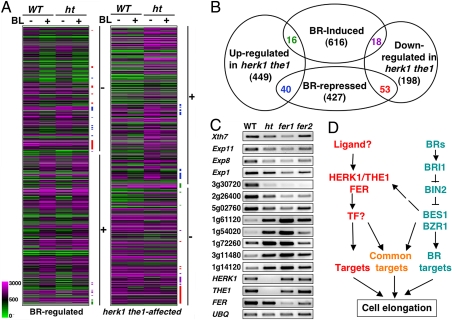
Fig. 4.
HERK1/THE1/FER and BR pathways affect independent genes with some overlap. (A) Cluster analysis of BR-regulated genes in WT and mutant plants. The majority of BL-repressed genes (marked by −) and BL-induced genes (marked by +) are not affected in herk1 the1 double mutant (ht). Likewise, the majority of genes down-regulated (−) or up-regulated (+) in herk1 the1 double mutant are not regulated by BRs. The overlap between BR- and HERK1/THE1-affected genes are indicated by colored bars corresponding to the colored numbers in the overlaps shown in Fig. 4B. (B) A diagram showing the overlap of BR- and HERK1/THE1-regulated genes. Lists of the genes represented in this diagram are presented in Table S1, Table S2, Table S3, and Table S4. (C) Semiquantitative PCR with primers from indicated genes to confirm the microarray data in the herk1 the1 double mutant (ht) and to examine the expression levels in fer1 and fer2 mutants. Genes either down- or up-regulated in the herk1 the1 double mutant and the 3 receptor genes were tested. (D) A model for the BR- and HERK1/THE1-pathways in the regulation of cell elongation. See Discussion for details. TF stands for putative transcription factor.
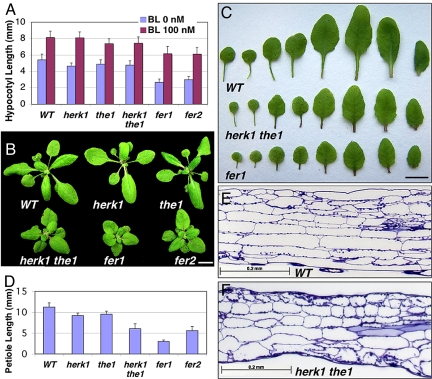
Fig. 2.
HERK1, THE1 and FER are required for cell elongation. (A) BR responses of the herk1, the1, and fer mutants at the seedling stage. The seeds were germinated in 1/2MS media containing the indicated concentrations of brassinolide (BL) and grown under the light for 10 days. Twenty to 30 hypocotyls were measured, and the average hypocotyl lengths and standard deviations are presented. (B) Shoot phenotypes of 24-day-old adult plants. (Scale bar: 10 mm.) (C) The leaves of the WT, herk1 the1 double mutants, and fer1 mutants, showing the reduced lengths in leaf blades and leaf petioles. (Scale bar: 10 mm.) (D) Quantification of petiole lengths of the 6th leaf in WT and mutants. Averages and standard deviations from 10 plants are shown. (E and F) The herk1 the1 double mutant plants have reduced cell elongation. The petioles of the WT (E) and herk1 the1 double mutant (F) plants were fixed, stained with toluidine blue, and embedded. Longitudinal sections were examined under a bright-field light microscope and photographed. The sections shown are midway along the length of the petioles and cut through the center (traces of vascular tissues can be seen on both WT, and mutant sections can be seen on the right side).
We first determined the BR response of the mutants by measuring hypocotyl lengths in the absence or presence of BL. BRs stimulate cell elongation in the light-grown seedlings, and hypocotyl elongation has been widely used as a measure BR activity. herk1, the1, and herk1 the1 double mutants have somewhat shorter hypocotyls compared with WT without BL treatment, whereas fer1 and fer2 have shorter hypocotyls than WT with or without BL treatment (Fig. 2A). Interestingly, the BR response was largely unchanged in the mutants (Fig. 2A). We therefore concluded that HERK1, THE1 and particularly, FER, are required for hypocotyl elongation mostly in a BR-independent manner at the seedling stage.
At the adult stage, although herk1 and the1 single mutants are indistinguishable from WT, herk1 the1 double mutants were clearly stunted in growth (Fig. 2B). The elongation of both leaves and leaf petioles was affected (Fig. 2 C and D). We focused on leaf petioles that were most severely affected in the mutants. Although the elongation of petioles was slightly affected in herk1 and the1 single mutants, the petioles were reduced to half the length of the WT in herk1 the1 double mutants (Fig. 2 C and D). To determine the basis for the stunted growth phenotypes, we examined the cells in the leaf petioles. Most cells in the mutant leaf petioles were shorter than the corresponding cell types in WT (Fig. 2 E and F).
To confirm that the mutant phenotype is caused by T-DNA knockout, a genomic clone of HERK1 under the control of its native promoter was transformed back into the herk1 the1 double mutant. From >10 transgenic lines recovered, approximately half of them showed clear rescue of the mutant phenotype (Fig. S2 A–C). The level of the HERK1 transcript was higher in the transgenic plants with rescued phenotype (Fig. S2D). We conclude that HERK1 and THE1 function redundantly to promote cell elongation, especially in adult plants.
Both fer1 and fer2 adult plants have almost the same phenotypes as the herk1 the1 double mutant, with a more severe cell elongation defect in fer1 (Fig. 2 B–D). The fertility in both lines was also reduced, presumably because of impaired FER function in pollen tube-female gametophyte interaction. The extent of both vegetative and reproductive phenotypes correlated well with the reduction in expression of the FER gene (Fig. 2 B–D, Fig. 4C).
To determine the gain-of-function phenotype for the RLKs, we identified several transgenic plant lines overexpressing the HERK1 gene under its own promoter, aided by CAMV 35S enhancer elements. Of these, we characterized 1 representative line that accumulated ≈10 times more HERK1 RNA than WT (Fig. S3). Overexpression of HERK1 appeared to increase petiole length by ≈15–20% (Fig. S3A). Consistent with the phenotype, several HERK1/THE1 regulated genes (see next) were up-regulated by ≈20–70% (Fig. S3B). Taken together, our results demonstrate that, in addition to their established roles in sensing cell wall-integrity (20) and pollen and female gametophyte interaction (21), HERK1/THE1/FER RLKs play a major role in promoting cell elongation during vegetative growth.
Genetic Interactions Between BR and HERK Pathways.
Triple mutants of bri1-5 herk1 the1 and bes1-D herk1 the1 were constructed to investigate the genetic interactions between BR and HERK signaling (Fig. 3). bri1-5 is a weak loss-of-function allele of the BR receptor gene, BRI1, that displays a semidwarf phenotype (27). The herk1 the1 double mutant enhanced the bri1-5 dwarf phenotype (Fig. 3 A–C). However, bes1-D is a gain-of-function mutant that displays constitutive BR responses including excessive cell elongation (13). The herk1 the1 double mutant partially suppressed the cell elongation phenotype of bes1-D (Fig. 3 D and E). These genetic studies suggest that HERK1 and THE1 cooperate with the BR pathway and mediate part of BR-regulated cell elongation.
Fig. 3.
herk1 the1 double mutant enhances bri1-5 and suppresses bes1-D mutant phenotypes. (A) Two-week-old seedlings. (B) Adult plants (≈35-day-old). (C) Fifth and 6th leaves. (D) Nine-day-old seedlings. (E) Adult plants (≈24-day-old). Note that one of the leaves from the bes1-D herk1 the1 plant was removed for genotyping.
Most of the HERK1/THE1/FER Affected Genes Are Not Regulated By BRs.
Despite similar defects in cell elongation, the herk1 the1 double or fer mutants differ from BR mutants. BR mutants have epinastic and dark green leaves that were not observed in herk1 the1 double or fer mutants (Fig. 2). We asked whether the cell elongation phenotype of herk1 the1 double mutant was due to changes in BR-regulated gene expression. To that end, the global gene expression patterns of herk1 the1 double mutant, in the absence and presence of BL during the adult stages, were first determined by microarray analysis (Fig. 4 A and B; Table 1; and Table S1, Table S2, Table S3, and Table S4). In adult plants, although BL induces and represses the expression of 650 and 520 genes respectively, 269 and 505 genes were down- or up-regulated, respectively, in herk1 the1 double mutant compared with WT control. Using GeneSpring (www.chem.agilent.com/enUS/Products/software/lifesciencesinformatics/genespringgx/Pages/default.aspx), we performed cluster analysis of BR- and HERK1/THE1-regulated genes and found that the majority of BR-regulated genes were not affected in the mutant, and likewise, the majority of HERK1/THE1 regulated genes were not clearly regulated by BL (Fig. 4 A and B). It appears that only ≈10% of BR-regulated genes were affected in the herk1 the1 double mutant or 16% genes affected in herk1 the1 double mutant are regulated by BRs (Fig. 4B). These results suggest that the HERK pathway is largely independent of the BR pathway with a subset of overlapping genes.
Table 1.
The regulation of genes implicated in cell elongation by BRs and HERK1/THE1
| Treatment/genotype | Gene no. | WT-BL | WT + BL | herk1 the1-BL | herk1 the1 + BL | Annotation |
|---|---|---|---|---|---|---|
| BL | AT5G57560 | 2277 | 7344 | 2728 | 7941 | XTH22 |
| AT1G10550 | 47 | 223 | 48 | 191 | XTH33 | |
| AT1G11545 | 453 | 1178 | 357 | 1249 | XTH8 | |
| At2g06850 | 7247 | 13455 | 5096 | 11194 | XTH4 | |
| At4g30290 | 153 | 297 | 162 | 259 | XTH19 | |
| At1g65310 | 137 | 214 | 95 | 302 | XTH17 | |
| At3g23730 | 559 | 819 | 636 | 1069 | XTH16 | |
| At3g07010 | 427 | 939 | 325 | 829 | PLL20 | |
| At1g04680 | 2466 | 3402 | 1982 | 2932 | PLL26 | |
| At4g38400 | 188 | 433 | 151 | 381 | EXPL2 | |
| At3g45970 | 581 | 1339 | 529 | 970 | EXPL1 | |
| At2g37640 | 384 | 641 | 340 | 594 | EXP3 | |
| At3g45960 | 24 | 92 | 18 | 44 | EXPL3 | |
| At1g20190 | 141 | 395 | 35 | 225 | EXP11 | |
| BL/herk1 the1 | At2g40610 | 1006 | 2407 | 437 | 1586 | EXP8 |
| At1g69530 | 6798 | 8465 | 3595 | 6462 | EXP1 | |
| herk1 the1 | At2g20750 | 295 | 198 | 161 | 157 | EXPB1 |
| At2g28950 | 3287 | 2524 | 2074 | 1919 | EXP6 | |
| At3g29030 | 2323 | 2112 | 1523 | 1848 | EXP5 | |
| At4g37800 | 2899 | 3419 | 1831 | 2864 | XTH7 |
Affymetrix Arabidopsis genomic arrays were used to detect gene expression. BL-regulated or herk1 the1-affected genes implicated in cell elongation are included. Average of the gene expression level from 3 biological replicates are shown. Note that several BL-induced genes appeared to be reduced in herk1 the1 double mutant, but did not pass the statistical test.
Consistent with the similar phenotypes between fer1, fer2 and herk1 the1 double mutant, 12 of 16 randomly chosen genes affected in herk1 the1 double mutant are similarly affected in fer1 and fer2 mutants (Fig. 4C). The gene expression changes correlate well with the mutant phenotypes, i.e., the genes are reduced or increased more in fer1 than in fer2 and herk1 the1 double mutant. The results support the conclusion that HERK1/THE1 and FER likely affect a similar set of genes and, therefore, function in a common pathway.
To gain a better understanding of how the BR and HERK/THE pathways function to regulate similar processes, we further examined genes implicated in cell elongation (Table 1). It is well established that BRs promote cell elongation by inducing the expression of the genes involved in cell wall-loosening enzymes, such as xyloglucan endotransglycosylase/hydrolase (XTH) and Pectin Lyase-like (PLL) as well as expansins (EXP) (28–30). Consistent with its major function in promoting cell elongation, BRs induce the expression of 7 XTHs, 2 PLLs and 7 EXPs in adult plants. Similarly, HERK1/THE1 is also required for the expression of 6 cell elongation genes, including 1 XTH, and 5 EXPs. Interestingly, 2 genes (EXP1 and EXP8) are induced by both pathways and are likely to be common targets. These results indicated that although BR and HERK induce different sets of genes for cell elongation, there are some common target genes that may integrate signals from both pathways. Because only 2 of 16 BR-induced genes implicated in cell elongation are reduced in herk1 the1 double mutant, it seems unlikely that the mutant phenotype is solely due to the changes in BR target gene expression.
Discussion
Based on our results, we propose a working model for the HERK1/THE1/FER pathway and its relationship with the BR pathway in the regulation of cell elongation (Fig. 4D). Although BRs function through BRI1 and BIN2 to regulate the BES1/BZR1 family transcription factors and, therefore, modulate the expression of target genes including those required for cell elongation, HERK1, THE1, and FER function in an independent pathway to regulate different genes including those implicated in cell elongation. The regulation of common targets may represent cross-talk between these 2 pathways.
Our study, therefore, identified an important pathway required for plant growth. Plant cell elongation is regulated by environmental cues such as light and several plant hormones, including BRs, auxin and gibberellin (31, 32). Our qRT-PCR information and the public microarray data revealed that although BRs induce the expression of all 3 RLKs (Fig. 1 and Fig. S4A), other hormones, including auxin, gibberellin, ethylene, and cytokinin, which are known to regulate cell elongation or division, modulate individual RLKs (http://bbc.botany.utoronto.ca/efp/cgi-bin/efpWeb.cgi). For example, our qRT-PCR analysis indicated that GA3 up-regulates the expression of HERK1, THE1, and FER whereas IAA appears to induce HERK1 (Fig. S4B). However, the herk1 the1 double mutant responds normally to gibberellin in hypocotyl elongation assays and to auxin in root elongation assays (Fig. S4 C and D), implying that HERK/THE1/FER RLKs are unlikely involved in gibberellin or auxin perception. It is more likely that different hormones can modulate the expression of HERK1/THE1/FER to regulate plant growth. By comparison to other hormone-regulated genes (33), we found that among 774 herk1 the1 affected genes, 61, 7, 41, and 37 genes are known to be regulated by auxin (789 genes), gibberellin (122 genes), ethylene (532 genes), and cytokinin (491 genes), respectively. The observation further supports the notion that HERK1/THE1 and FER represent another pathway required for optimal cell elongation. The dwarf mutant phenotypes of HERK1/THE1 and FER at adult stage should greatly facilitate the identification of other signaling components for this important family of RLKs.
What is the relationship of HERK1, THE1 and FER? The fact that fer1 and fer2 showed almost identical phenotypes and affected similar genes to the herk1 the1 double mutant suggests that HERK1/THE1 and FER function in the same pathway. For example, FER could serve as a coreceptor for HERK1/THE1 such that a knockdown of FER has the same effect as a knockout of both HERK1 and THE1. It's worth noting that several other members in the family, such as At2g39360, At5g24010, At1g30570, At5g38990/At5g39000, and At5g39020 are slightly induced by BL (Fig. S4A). They may function in other processes including cell elongation. It's possible that different combinations of members regulate growth in different tissues or organs at different developmental stages.
HERK1/THE1/FER signaling apparently interacts with the BR pathway. First, BRs induce the expression of HERK1, THE1 and FER through BRI1 and BES1 (Fig. 1 A and B). Second, herk1 the1 double mutants enhance bri1 and suppress bes1-D mutant phenotypes (Fig. 3). Third, the 2 pathways regulate a common set of 127 target genes, including those involved in cell elongation (Fig. 4 and Table 1). It's possible that functional cooperation between effectors in each pathway (i.e., BES1 and its counterpart in the RLK pathway) accounts for the expression of the common target genes. Identification of downstream effector protein(s) in the HERK1/THE1/FER pathway can help test the hypothesis.
Our study provides significant insight into the function of CrRLK family of RLKs. HERK1, THE1, and FER are members of the CrRLK family RLKs in Arabidopsis. Two members of the family, FER and THE, were found recently to inhibit cell elongation in two different contexts (18). In contrast, we observed that knockout of both the HERK1 and THE1 genes or the reduction of FER led to reduced cell elongation. Meanwhile, overexpression of HERK1 resulted in up-regulation of some HERK target genes and slightly enhanced petiole elongations (Fig. S3). Therefore, our results suggest a role of the RLK family in promoting cell elongation. Several possibilities can explain the seemingly opposite functions. First, the pathways may have different outputs in different tissues/organs at different developmental stages. Second, like many signaling pathways, different signaling intensities can lead to opposite biological effects. For example, although exogenously applied BRs promote hypocotyl elongation in light-grown seedlings, they inhibit hypocotyl growth in the dark where cell elongation is already very active.
Finally, our results extend the role of FER from female: male interaction to vegetative growth. Interestingly, BRs, which are well established for their role in vegetative growth, function in reproduction as well. BRs are required for pollen tube growth, as demonstrated by the fact that BR mutants are defective in pollen tube elongation and have reduced fertility (17, 34). Because FER functions to block further pollen tube growth once the tube reaches synergid cells, it is apparent that although BRs and FER have similar functions in vegetative tissue, they have opposite roles in pollen tube growth. The identification of other signaling components is needed to fully understand the functional divergence of the HERK1/THE1/FER pathway.
In summary, this study has identified signaling components that regulate an important plant growth process, cell elongation. The established phenotypes and target genes will help identify the ligand(s) and other signaling components. In addition, the apparent interactions between BR and HERK pathways provide a paradigm for the way in which different signaling pathways contribute to the regulation of a similar process, a general theme for growth and development.
Materials and Methods
Plant Materials and Growth Conditions.
Arabidopsis thaliana ecotype Col-0 was the WT. Seeds were geminated and grown on ½ MS medium with 1% sucrose under 15 h light/9 h dark cycle at 22 °C. Two-week-old seedlings were usually transferred to soil and grown in growth rooms under same conditions.
HERK1 Localization and Kinase Activity.
A HERK1-GFP or BES1-GFP construct was introduced to protoplasts derived from 5-day-old seedlings grown in liquid culture by PEG-mediated transfection and culture for 16 h (35). The BRI1::HERK1-GFP was also introduced into Arabidopsis plants by floral dip method (36). Transgenic plants were identified by screening T1 seeds in ½MS containing 50 mg/L Kanamycin. Protoplasts or hypocotyls cells from 3-day-old transgenic lines (T2) grown in the dark were observed under an Olympus fluorescence microscope with a filter for GFP visualization. For the in vitro kinase assay, WT HERK1 kinase domain (WT), and a mutant version (K513R), were cloned into a MBP (Maltose Binding Protein) fusion vector and expressed in E. coli. Recombinant proteins were purified using amylase resin (New England Biolabs) and used in kinase assay in 50 mM Tris·HCl, 150 mM NaCl, 1 mM MaCl2, 0.1 mM ATP (pH 7.5), plus 2 μL of 32P-γ-ATP (Perkin Emer, 3,000 Ci/mmol), room temperature, for 0.5 h. Autophosphorylation was detected after protein gel electrophoresis and phosphorimaging with a Typhoon 9400 (GE Healthcare). Protein was visualized by SYPRO RUBY protein gel stain (Invitrogen).
Transgenic Studies.
The promoters of HERK1 (-2114/-19 relative to the translational start site) and FER (-1193/-27) were amplified from BAC DNA, and THE1 (-2222/-43) was amplified from genomic DNA, and cloned to the pBI101 with the β-glucuronidae (GUS) gene (Clontech). A 5.3-kb HERK1 genomic fragment was cloned into pMN20 that harbors 4 copies of CaMV 35S enhancer (37). The constructs were transfected into Agrobacterium strain GV3101, which were used to transform Arabidopsis. T2 or T3 transgenic plants were used to detect reporter gene expression by GUS assay (38) or phenotypic analysis.
Identification of T-DNA Knockout Mutants and Creation of Artificial microRNA (amiRNA) Mutants.
The T-DNA knockout seeds, herk1 (SALK_008043) and the1-4 (CS829966), were obtained from the Arabidopsis Biological Resource Center (www.arabidopsis.org). Homozygous plants were identified by PCR genotyping with primers listed in Table S5. Double mutants were constructed by cross single homozygous mutants and F2 plants were genotyped for double homozygous plants. The amiRNA system was used to knockdown FER. A set of 4 primers (Table S5) were designed using the tools found at http://wmd.weigelworld.org/cgi-bin/mirnatools.pl and used to amplify an amiRNA fragment (25), which was cloned into a binary vector under the control of BRI1 promoter.
Characterization of Growth Phenotype.
For BR response experiments, brassinolide (BL, from Wako Chemicals USA Inc, Richmond VA) was added to the medium after autoclaving and cooling to 50 °C. The seeds were treated at 4 °C for 4 days and germinated and grown for 10 days before measuring hypocotyl lengths. For adult phenotypes, WT and mutant plants were grown side-by-side in a growth flat. The 24-day-old mature plants were examined for the growth phenotype and photographed. The petiole lengths of the 6th leaves, from 10 independent plants, were measured. The average and standard deviations were calculated. To examine the cell lengths, leaf petioles were fixed with 2% glutaraldehyde (wt/vol) and 2% paraformaldehyde (wt/vol) in 0.1M cacodylate buffer (pH 7.2) for 48 h at 4 °C. Samples were rinsed in 0.1M cacodylate buffer and dehydrated in a graded ethanol series, infiltrated and embedded using LR White resin (Electron Microscopy Sciences). Resin blocks were polymerized for 48 h at 60 °C. Thick sections were made using a Reichert UC6 ultramicrotome (Leeds Precision Instruments). Sections were stained with 1% toluidine blue in 1% borax and images were taken using a Zeiss Axioplan II compound microscope with a MRC digital camera and Axiovision software (Carl Zeiss).
Gene Expression Studies.
Microarray experiments were performed with 24-day-old adult plants. The whole plants were sprayed with either water or 1 μM BL and incubate for 2.5 h. The plants were pooled into 3 groups as triplicates (8 plants per group). Total RNA was prepared with RNeasy Plant Mini Kit (QIAGEN) and used to make probes for microarray experiments with Affymetrix Arabidopsis Genomic arrays. The probe labeling, hybridization, and scanning were performed according to manufacture's instructions. Microarray data were normalized by the MAS 5.0 method implemented in the R package affy and the linear model was applied to 2 sample comparison (samples with and without BL treatment or WT compared with mutant), using the limma package (39, 40). Genes with an adjusted P ≤ 0.01 were considered to be differentially expressed. The BR-induced genes, HERK1/THE regulated genes were used for clustering analysis with the GENESPRING program (Silicon Genetics), using Pearson correlation.
For reverse transcription-PCR (RT-PCR), 2 μg of total RNA was reverse-transcribed to cDNA by SuperScript II Reverse Transcriptase (Invitrogen). For real-time quantitative RT-PCR, the primers were designed so that the products are between 200 and 300 bp. The SYBR Green PCR Master mix (Applied Biosystems) was used and the PCRs were run on the Mx4000 multiplex quantitative PCR system (Stratagene). Two or 3 biological replicates were each analyzed with 2–3 RT-PCRs. For semiquantitative RT-PCR, the same amount of cDNA was used for each PCR. The primers used are listed in Table S5, and the UBQ5 gene was the control. PCRs were stopped in the linear range of the amplification and were repeated for 2 times. Very similar results were obtained with both sets of samples.
Supplementary Material
Acknowledgments.
We thank Tracey Pepper at the Bessey Microscopy Facility (Ames, IA) for tissue processing and sectioning, Jiqing Peng at Iowa State University Microarray Facility (Ames, IA) for microarray experiments, Martijn van de Mortel for help with the GENESPRING program, and Drs. Steve Howell and Tom Peterson for editing the manuscript. The research was supported by National Science Foundation Grant IOS0546503 and a faculty start-up fund from Iowa State University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0812346106/DCSupplemental.
References
- 1.Chory J, Wu D. Weaving the complex web of signal transduction. Plant Physiol. 2001;125:77–80. doi: 10.1104/pp.125.1.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Muller B, Sheen J. Cytokinin and auxin interaction in root stem-cell specification during early embryogenesis. Nature. 2008;453:1094–1097. doi: 10.1038/nature06943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hardtke CS. Transcriptional auxin-brassinosteroid crosstalk: Who's talking? Bioessays. 2007;29:1115–1123. doi: 10.1002/bies.20653. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Jin H. Regulation of brassinosteroid signaling. Trends Plants Sci. 2007;12:37–41. doi: 10.1016/j.tplants.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 5.Gendron JM, Wang ZY. Multiple mechanisms modulate brassinosteroid signaling. Curr Opin Plant Biol. 2007;10:436–441. doi: 10.1016/j.pbi.2007.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Belkhadir Y, Chory J. Brassinosteroid signaling: A paradigm for steroid hormone signaling from the cell surface. Science. 2006;314:1410–1411. doi: 10.1126/science.1134040. [DOI] [PubMed] [Google Scholar]
- 7.Clouse S, Sasse J. Brassinosteroids: Essential regulators of plant growth and development. Annu Rev Plant Physiol Plant Mol Biol. 1998;49:427–451. doi: 10.1146/annurev.arplant.49.1.427. [DOI] [PubMed] [Google Scholar]
- 8.Wang X, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Chory J. Brassinosteroids regulate dissociation of BKI1, a negative regulator of BRI1 signaling, from the plasma membrane. Science. 2006;313:1118–1122. doi: 10.1126/science.1127593. [DOI] [PubMed] [Google Scholar]
- 11.Yin Y, et al. A new class of transcription factors mediate brassinosteroid-regulated gene expression in Arabidopsis. Cell. 2005;120:249–259. doi: 10.1016/j.cell.2004.11.044. [DOI] [PubMed] [Google Scholar]
- 12.He JX, et al. BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005;307:1634–1638. doi: 10.1126/science.1107580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yin Y, et al. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002;109:181–191. doi: 10.1016/s0092-8674(02)00721-3. [DOI] [PubMed] [Google Scholar]
- 14.Wang ZY, et al. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002;2:505–513. doi: 10.1016/s1534-5807(02)00153-3. [DOI] [PubMed] [Google Scholar]
- 15.Nemhauser JL, Mockler TC, Chory J. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. 2004;2:E258. doi: 10.1371/journal.pbio.0020258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goda H, et al. Comprehensive comparison of auxin-regulated and brassinosteroid regulated genes in Arabidopsis. Plant Physiol. 2004;134:1555–1573. doi: 10.1104/pp.103.034736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clouse SD. Molecular genetic studies confirm the role of brassinosteroids in plant growth and development. Plant J. 1996;10:1–8. doi: 10.1046/j.1365-313x.1996.10010001.x. [DOI] [PubMed] [Google Scholar]
- 18.Hematy K, Hofte H. Novel receptor kinases involved in growth regulation. Curr Opin Plant Biol. 2008;11:321–328. doi: 10.1016/j.pbi.2008.02.008. [DOI] [PubMed] [Google Scholar]
- 19.Shiu SH, Bleecker AB. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol. 2003;132:530–543. doi: 10.1104/pp.103.021964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hematy K, et al. A receptor-like kinase mediates the response of Arabidopsis cells to the inhibition of cellulose synthesis. Curr Biol. 2007;17:922–931. doi: 10.1016/j.cub.2007.05.018. [DOI] [PubMed] [Google Scholar]
- 21.Escobar-Restrepo JM, et al. The FERONIA receptor-like kinase mediates male-female interactions during pollen tube reception. Science. 2007;317:656–660. doi: 10.1126/science.1143562. [DOI] [PubMed] [Google Scholar]
- 22.McCormick S. Plant science. Reproductive dialog. Science. 2007;317:606–607. doi: 10.1126/science.1146655. [DOI] [PubMed] [Google Scholar]
- 23.Rotman N, et al. Female control of male gamete delivery during fertilization in Arabidopsis thaliana. Curr Biol. 2003;13:432–436. doi: 10.1016/s0960-9822(03)00093-9. [DOI] [PubMed] [Google Scholar]
- 24.Huck N, Moore JM, Federer M, Grossniklaus U. The Arabidopsis mutant feronia disrupts the female gametophytic control of pollen tube reception. Development. 2003;130:2149–2159. doi: 10.1242/dev.00458. [DOI] [PubMed] [Google Scholar]
- 25.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Chory J. A putative leucine-rich repeat receptor kinase involved in brassinosteroid signal transduction. Cell. 1997;90:929–938. doi: 10.1016/s0092-8674(00)80357-8. [DOI] [PubMed] [Google Scholar]
- 27.Noguchi T, et al. Brassinosteroid-insensitive dwarf mutants of Arabidopsis accumulate brassinosteroids. Plant Physiol. 1999;121:743–752. doi: 10.1104/pp.121.3.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palusa SG, Golovkin M, Shin SB, Richardson DN, Reddy AS. Organ-specific, developmental, hormonal and stress regulation of expression of putative pectate lyase genes in Arabidopsis. New Phytol. 2007;174:537–550. doi: 10.1111/j.1469-8137.2007.02033.x. [DOI] [PubMed] [Google Scholar]
- 29.Becnel J, Natarajan M, Kipp A, Braam J. Developmental expression patterns of Arabidopsis XTH genes reported by transgenes and Genevestigator. Plant Mol Biol. 2006;61:451–467. doi: 10.1007/s11103-006-0021-z. [DOI] [PubMed] [Google Scholar]
- 30.Darley CP, Forrester AM, McQueen-Mason SJ. The molecular basis of plant cell wall extension. Plant Mol Biol. 2001;47:179–195. [PubMed] [Google Scholar]
- 31.Chory J, et al. From seed germination to flowering, light controls plant development via the pigment phytochrome. Proc Natl Acad Sci USA. 1996;93:12066–12071. doi: 10.1073/pnas.93.22.12066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Grauwe L, Vandenbussche F, Tietz O, Palme K, Van Der Straeten D. Auxin, ethylene and brassinosteroids: Tripartite control of growth in the Arabidopsis hypocotyl. Plant Cell Physiol. 2005;46:827–836. doi: 10.1093/pcp/pci111. [DOI] [PubMed] [Google Scholar]
- 33.Nemhauser JL, Hong F, Chory J. Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell. 2006;126:467–475. doi: 10.1016/j.cell.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 34.Szekeres M, et al. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996;85:171–182. doi: 10.1016/s0092-8674(00)81094-6. [DOI] [PubMed] [Google Scholar]
- 35.Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat Protoc. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- 36.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 37.Weigel D, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jefferson RA, Kavanagh TA, Bevan MW. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smyth GK, Michaud J, Scott HS. Use of within-array replicate spots for assessing differential expression in microarray experiments. Bioinformatics. 2005;21:2067–2075. doi: 10.1093/bioinformatics/bti270. [DOI] [PubMed] [Google Scholar]
- 40.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.