Summary
The eukaryotic DNA replication machinery must traverse every nucleosome in the genome during S phase. As nucleosomes are generally inhibitory to DNA-dependent processes, chromatin structure must undergo extensive reorganization to facilitate DNA synthesis. However, the identity of chromatin-remodeling factors involved in replication and how they affect DNA synthesis is largely unknown. Here we show that two highly conserved ATP-dependent chromatin-remodeling complexes in Saccharomyces cerevisiae, Isw2 and Ino80, function in parallel to promote replication fork progression. As a result, Isw2 and Ino80 play especially important roles for replication of late-replicating regions during periods of replication stress. Both Isw2 and Ino80 complexes are enriched at sites of replication, suggesting that these complexes act directly to promote fork progression. These findings identify ATP-dependent chromatin-remodeling complexes promoting DNA replication, and define a specific stage of replication that requires remodeling for normal function.
The compaction of DNA into chromatin is essential for organization and transmission of the eukaryotic genome. The fundamental repeating unit of chromatin is the nucleosome, a structure composed of 147 base pairs of DNA wrapped around an octamer of histone proteins. Since a majority of DNA in the eukaryotic genome is occupied in nucleosomes, every process requiring a DNA template is strongly influenced by the positioning and structural integrity of nucleosomes. Therefore, mechanisms controlling chromatin structure can positively or negatively affect such nuclear processes.
ATP-dependent chromatin-remodeling complexes regulate many DNA-dependent processes by disrupting histone-DNA contacts. The enzymatic activity of these complexes results in the repositioning of nucleosomes along DNA, increased availability of DNA on the nucleosome surface, and/or exchange of histone proteins within the nucleosome1,2. Although their ability to regulate transcription is best characterized1,3, the involvement of ATP-dependent chromatin-remodeling factors in DNA replication4, repair5, and recombination6,7 has recently been revealed.
Eukaryotes encode a variety of ATP-dependent chromatin-remodeling factors with unique subunit compositions, suggesting individualized functions. These complexes are classified into families based on the amino acid sequence of the ATPase subunit of the complex8,9. The presence of multiple types of remodeling factors within an organism emphasizes the importance and diversity of chromatin regulation mechanisms, but potentially complicates identification of the biological functions of these chromatin regulators. For example, the budding yeast S. cerevisiae encodes members of 13 major subfamilies of remodeling enzymes9, but mutations in most of these enzymes cause relatively mild phenotypes. One explanation for this phenomenon is that other chromatin regulators can account for the loss of an individual ATP-dependent chromatin-remodeling factor by performing similar or compensating functions.
In order to uncover previously unknown functions of an ATP-dependent chromatin-remodeling factor, we conducted a genetic screen designed to identify genes required for normal growth in the absence of the S.cerevisiae Isw2 complex. The Isw2 ATP-dependent chromatin-remodeling complex is well characterized with respect to its mechanism of action in vivo10,11, and thus provides a powerful model for defining the biological roles of chromatin remodeling. Our genetic analyses revealed that subunits of the Ino80 ATP-dependent chromatin-remodeling complex are required for normal growth in the absence of Isw2. Since Isw2 remodels chromatin by sliding nucleosomes along DNA in cis10, and Ino80 has been proposed to facilitate histone exchange12, the above result suggests that two chromatin remodeling complexes with distinct biochemical activities have compensatory functions. To identify these functions, we investigated the roles of Isw2 and Ino80 complexes in DNA replication by whole-genome replication profiling, and found that these complexes promote DNA replication specifically in the late-replicating regions. Furthermore, we found that mutations of both chromatin-remodeling complexes decrease the rate of replication-fork progression, especially during periods of replication stress. These results identify Isw2 and Ino80 chromatin-remodeling complexes as factors able to promote DNA replication, and identified replication fork progression as a step in DNA synthesis facilitated by these chromatin regulators.
Results
S phase is extended in MMS-treated isw2 nhp10
To identify the genetic backgrounds in which ISW2 is required for normal cell growth, we performed systematic synthetic genetic-interaction screens13. We found that deletions of NHP10, IES2, IES3, and IES5, which encode subunits of Ino80 chromatin remodeling complex, cause a growth defect in strains bearing an isw2 mutation. Although the growth conditions used in the genetic screens revealed synthetic growth defects, the growth defect of each double mutant under optimal conditions (YPD media, 30 °C) is mild, resulting in a 33% increase in doubling time over that of wild type (data not shown). The ies2, ies3, ies5, and nhp10 deletion mutations likely compromise a subset of Ino80 complex function as the ino80 deletion mutation, a likely null mutation for Ino80 complex function, has a severe growth defect12,14. For simplicity, we will refer to the isw2 nhp10 double mutant as a representative of this genetic interaction as it behaved identically to isw2 ies2, isw2 ies3, and isw2 ies5 double mutants in growth rate assays.
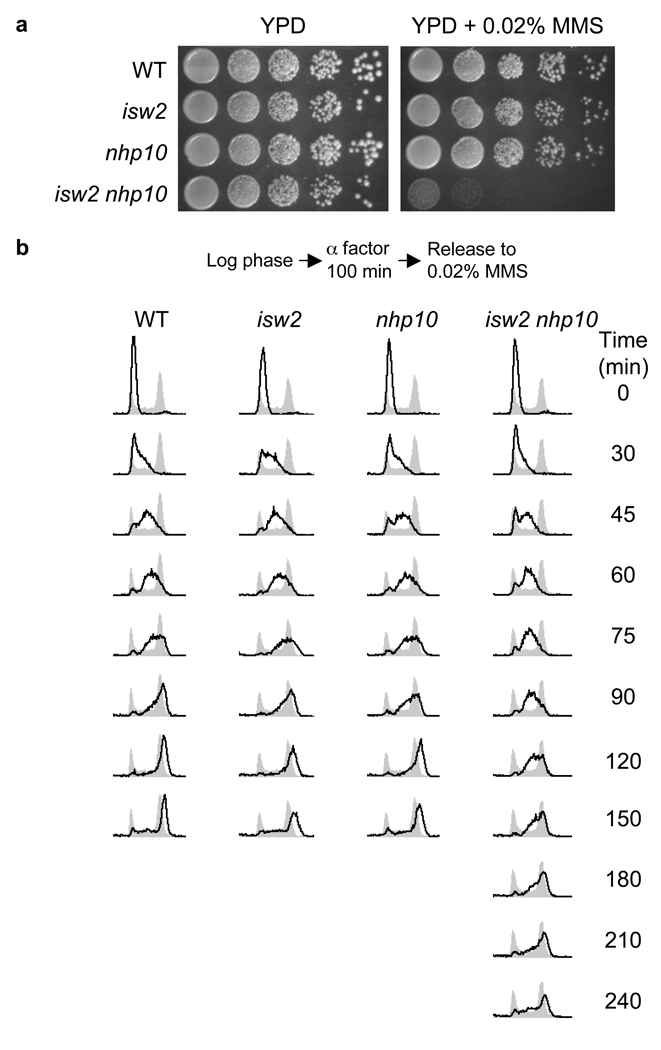
We tested the response of isw2 nhp10 double mutants to various environmental stresses to gain insight into the biological functions of the Isw2 and Ino80 complexes. Of the stress conditions tested, exposure to the DNA alkylating agent methyl methanesulfonate (MMS) caused the most striking growth defect (Fig. 1a). Importantly, the sensitivity to MMS is unique to the double mutant; both single mutants show the same MMS sensitivity as wild type (Fig. 1a, Supplementary Fig. 1a), showing that ISW2 and NHP10 function in a parallel and partially compensatory pathway that plays an important role(s) in the cellular response to MMS. We believe that the genetic interactions between ISW2 and NHP10 represent parallel functions of Isw2 and Ino80 complexes for the following reasons: (1) Mutations in multiple Ino80 complex subunits, the ies2, ies3, ies5, and nhp10 deletion mutations, cause identical growth defects in combination with an isw2 deletion mutation under all conditions tested (data not shown); (2) the Nhp10 protein is present exclusively in the Ino80 complex in vivo15; and (3) a deletion of approximately 900 bp from the 5′ end of INO80 gene (ino80Δ900), which encodes the ATPase subunit of Ino80 complex and partially inactivates Ino80 complex function12, causes a strong synthetic MMS sensitivity in combination with an isw2 mutation (Supplementary Fig. 1a).
Figure 1.
MMS sensitivity of isw2 nhp10 mutants is due to a prolonged S phase. (a) isw2 nhp10 double mutants grow slowly in the presence of MMS. Wild type (W1588-4c), isw2 (YTT1080), nhp10 (YTT2060), isw2 nhp10 (YTT2109) strains were grown to saturation then 10-fold serial dilutions were plated onto YPD plates with or without 0.02% MMS. (b) S phase is prolonged in MMS-treated isw2 nhp10 mutants. The above strains were arrested in G1 and then released into S phase in the presence of 0.02% MMS as diagrammed. Cells were collected at the indicated times after release and DNA content was determined by flow cytometry (black lines). Gray profiles are from asynchronous cells collected prior to G1 arrest.
Sensitivity to MMS can be due to defects in DNA repair, DNA damage checkpoint pathways, or DNA replication. Multiple independent lines of evidence indicate that the MMS sensitivity an isw2 nhp10 mutant is not a result of a deficiency in DNA repair or DNA damage checkpoint pathways (Supplementary Fig. 1b–c, Supplementary Table 1, Supplemental Discussion). Since the isw2 nhp10 mutant grows very slowly in the presence of MMS without showing a detectable increase in cell death (Supplementary Fig. 1b), we tested whether the mutant exhibits specific defects in cell cycle progression in the presence of MMS. To this end, we arrested cells in G1, released them into S phase in the presence of MMS, and monitored their DNA content by flow cytometry (Fig. 1b). Wild type cells, as well as isw2 and nhp10 single mutants all exhibit similar kinetics of replication. The isw2 nhp10 double mutant displays similar S phase kinetics to the above strains until 60 minutes post release, after which there is a pronounced delay in S phase progression. The delay in S phase progression is the primary cause of the MMS induced growth defect in isw2 nhp10 mutants as arrest and release from the G2/M boundary, in the presence of MMS, similarly resulted in an S phase specific delay (data not shown). Together, these results suggest that the MMS-induced growth defect of the isw2 nhp10 mutant is due to a delay in S phase progression. This conclusion is consistent with the observation that the isw2 nhp10 mutants are sensitive to other inhibitors of DNA replication (Supplementary Fig. 1d).
Isw2 and Ino80 facilitate replication in late-regions
A delay in S phase in the isw2 nhp10 mutants can either be due to a uniform delay of DNA replication kinetics throughout the genome or due to replication defects at specific loci. Conventional replication analyses such as FACS analysis or immuno-fluorescence staining after BrdU incorporation cannot distinguish between these possibilities. To address this key question, we utilized DNA microarrays to directly determine the kinetics of DNA replication of MMS-treated cells on a genome-wide scale16 (see Supplementary Fig. 2a for the schematic drawing of the procedure). Briefly, wild type and isw2 nhp10 strains were arrested in G1, treated with MMS, released into S phase in the presence of MMS, and collected at intervals during S phase (Supplementary Fig. 2b). Newly synthesized DNA was separated from unreplicated DNA by dense isotope transfer in a variation17 of the Meselson/Stahl experiment. Unreplicated DNA and newly replicated DNA were then alternatively labelled and hybridized to yeast ORF microarrays. A value for percent replication was determined for each spot on the microarray and the normalized data were plotted against the chromosomal position to generate a replication profile 18.
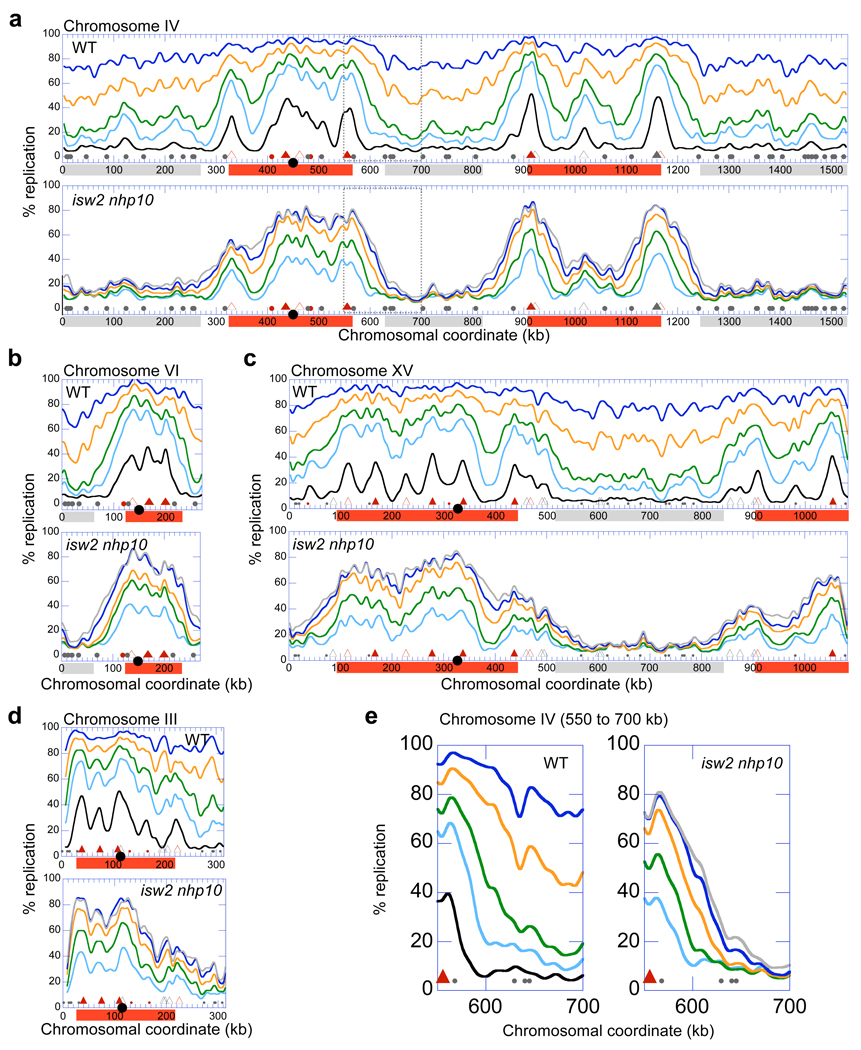
Replication profiles of wild type cells show initiation of replication (termed firing) occurring early in S phase in broad chromosomal domains containing early-firing origins of replication (hereafter origins) (Fig. 2a–d; red boxes). Remarkably, virtually every peak in the profiles detected early in S phase corresponds to known16,19,20 or predicted21,22 origins, showing that our analysis accurately reflects the kinetics of DNA replication throughout the genome. Our results are also consistent with previous reports that early-firing origins generally fire efficiently in the presence of replication stress 23,24. Replication of the regions that contain only inefficient and/or late-firing origins (late-replicating regions: Fig. 2a–c; grey boxes) proceeds slowly compared to early-replicating regions. The difference in replication timing between early and late-replicating regions in this analysis is likely enhanced due to MMS-induced activation of the S phase DNA damage checkpoint, which delays the firing of late-firing origins18,20,23,24.
Figure 2.
Isw2 and Ino80 complexes are required for efficient replication of late-replicating regions in the presence of MMS. (a–d) Replication profiles of chromosomes IV (a), VI (b), XV (c) and III (d) from WT (YTT1831) and isw2 nhp10 (YTT3306) strains undergoing S phase in the presence of MMS. (e) Close view of the replication profile at the interface between early and late-replicating regions on chromosome IV at chromosomal coordinates 550–700 kb (dotted box in a). Profiles were generated from cells collected at 30 (black), 45 (light blue), 60 (green), 90 (orange), 120 (dark blue), and 150 (grey) minutes after release from G1 arrest. Collection of isw2 nhp10 samples was initiated at 45 minutes due to a less efficient release from α-factor arrest characteristic of this strain (See Supplementary Fig. 2b, 4b). Positions of confirmed and likely ARSs43 are indicated at the bottom of each graph. Triangles correspond to positions of origins that are replicated early in a normal S phase (filled triangles represent origins that are amongst the first 25% replicated in two studies of replication timing16,19; open triangles represent origins that are amongst the earliest 25% in only one of the two studies). The filled circles correspond to remaining origins. Origins that are fired in a wild type strain during DNA damage checkpoint activation by 200mM Hydroxyurea20 (early firing origins) are indicated in red. The black circle on the x-axis indicates position of the centromere. The red and grey boxes represent approximations of early and late-replicating regions respectively.
By analyzing the replication profiles of the isw2 nhp10 mutant, we are able to identify specific regions where Isw2 and Ino80 complexes are required for facilitation of DNA replication (Fig. 2a–c). During early time points in S phase, replication initiation in the double mutant occurs in the same domains as in the wild type strain. This result shows that the initiation of replication from early-firing origins is not strongly affected in an isw2 nhp10 mutant. Two-dimensional (2-D) gel electrophoresis analysis at early-firing origins further supports this conclusion (Supplementary Fig. 3a). In sharp contrast to early-replicating regions, late-replicating regions in an MMS treated isw2 nhp10 mutant exhibit a strong defect in replication (Fig. 2a–c, grey boxes). Indeed, very limited amounts of DNA synthesis are detected within late-replicating regions in the first 150 minutes after release from G1. These specific replication defects in late-replicating regions occur throughout the genome in the isw2 nhp10 mutant; severe defects in DNA synthesis within late-replicating regions are observed on every single chromosome (Supplementary Fig. 2c) except for chromosome III, the only chromosome that lacks extended stretches of late-replicating regions (Fig. 2d). Consistent with the fact that the survival of the isw2 nhp10 mutant in MMS is equivalent to that of wild type (Supplementary Fig. 1b), DNA synthesis in late-replicating regions is eventually completed during a dramatically extended period in S phase (Supplementary Fig. 4a).
There are two prominent trends in the replication profiles. First, the replication defect in isw2 nhp10 mutants worsens as the distance from early-replicating regions increases (e.g. Fig. 2a; e.g. 570–700 kb). Second, the regions where DNA replication is most dramatically affected in isw2 nhp10 mutants extensively overlap with those most dependent on Clb5 for normal rates of replication, a S-phase cyclin required for efficient initiation from late-firing origins (Fig. 2a–c, note positions of early and late origins; H. McCune, M.K. Raghuraman, and B. Brewer, University of Washington, personal communication). This result confirms our conclusion that Isw2 and Ino80 are required for efficient replication of late-replicating regions. Together, the genome-wide analysis of replication kinetics revealed that the requirement for Isw2 and Ino80 complexes in DNA replication in the presence of MMS is not uniform, and that these chromatin-remodelling factors play particularly crucial roles in promoting DNA synthesis in the regions distant from efficient origins of replication.
Isw2 and Ino80 facilitate replication fork progression
After monitoring the replication dynamics for MMS treated wild type and isw2 nhp10 mutants, we analyzed the respective replication profiles for clues as to how DNA replication is affected in late-replicating regions. Replication profiles at the interface between early and late-replicating regions suggest defects in replication-fork progression in the isw2 nhp10 double mutant (Fig. 2e). In these areas, the slope of wild type replication profiles flattens as S phase progresses, indicating that fork progression from early into late-replicating regions contributes significantly to DNA synthesis in these regions. In contrast, the slope of replication profiles in the isw2 nhp10 mutant is slow to change during the first 150 minutes into S phase, suggesting that fork progression is less efficient in this strain.
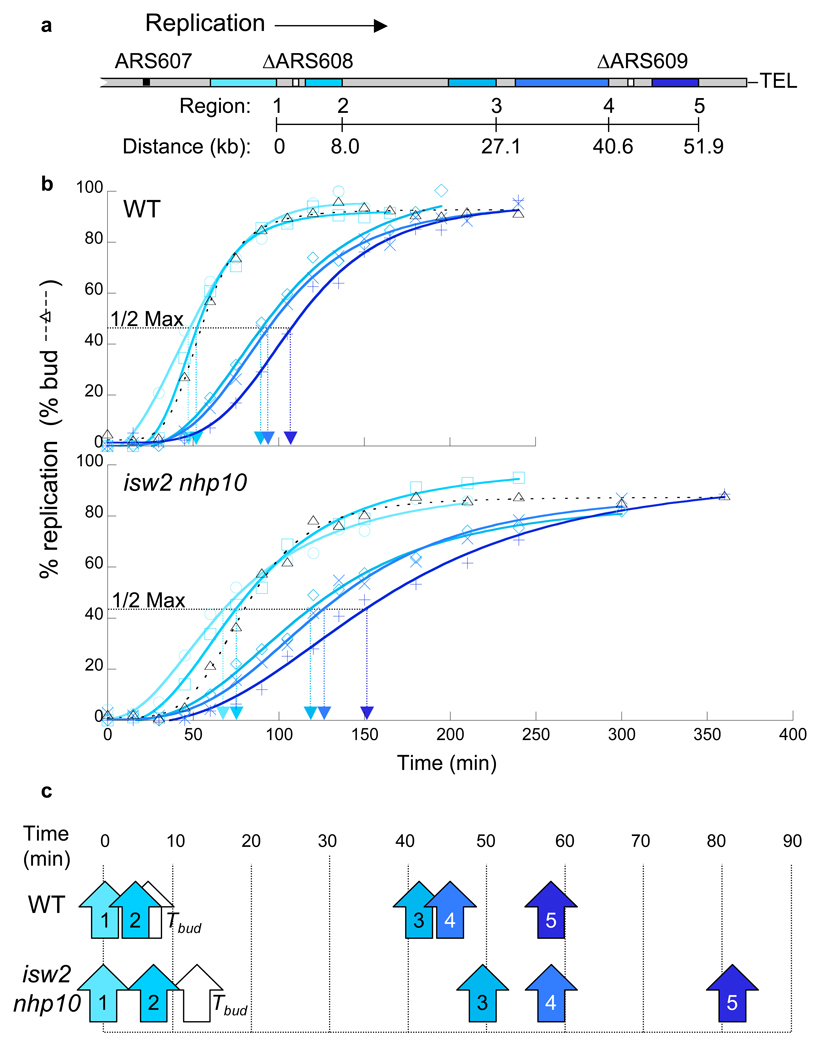
To directly test whether Isw2 and Ino80 complexes facilitate replication fork progression during replication stress, we determined the rate of replication fork progression in wild type and isw2 nhp10 strains using dense isotope transfer experiments. To this end, we employed strains bearing deletions of two inefficient origins on the right arm of chromosome VI, ARS608 and ARS609 (Fig. 3a), such that replication in this region proceeds in one direction from the efficient, and early-firing, ARS607 origin25 to the right telomere. We then determined the kinetics of replication at 5 loci interspersed over 52 kilobases (Fig. 3b, Supplementary Fig. 4b). Measurements of replication timing (Trep; time of half-maximal replication17) during MMS treatment showed that isw2 nhp10 mutants require more time than wild type to replicate each of the 5 loci between ARS607 and the right telomere (Fig. 3c). We estimate that forks in isw2 nhp10 mutants progress at approximately 75% the rate of wild type in this region (621 bp/min and 837 bp/min respectively, see Supplementary Table 2 for raw data). Similar decreases in replication fork rates were detected in the isw2 nhp10 mutants in two independent experiments (data not shown).
Figure 3.
Replication-fork progression is slowed in isw2 nhp10 mutants. (a) Diagram of the right arm of chromosome VI in the strains used for replication-fork progression measurements. The locations of the five regions (Regions 1–5) monitored for replication are shown in shades of blue. ARS608 and ARS609 (white boxes) are deleted to promote unidirectional replication from ARS607 to the telomere25. (b) Replication kinetics along chromosome VI. Percent replication of Regions 1–5 (colors correspond to diagram in a) was determined throughout S phase in the presence of MMS. The dashed line corresponds to the percentage of budded cells. The horizontal dotted line corresponds to the percent replication value one-half of the maximum obtained for that experiment. The Trep for that region is indicated on the x-axis by the arrowhead of the same color. Wild type (YTT3528) and isw2 nhp10 (YTT3531) strains were treated as in Supplementary Figure 2a. The kinetic curves of replication in the double mutant are more flat than those of wild type cells because the mutant is released from G1 arrest in a less synchronous fashion (see the budding index and Supplementary Fig. 4b). Importantly, the difference in the release kinetics only affects the slope of kinetic curve at each time point, not the distance between each line that reflects replication fork rate. (c) Replication times relative to Region 1. The values for Trep determined in (b) were plotted relative to the value for Region 1 and are indicated by arrows (colors correspond to diagram in a). The white arrows indicate the time of half-maximal budding in the population (Tbud).
We believe the differences in replication fork rates between wild type and isw2 nhp10 mutants are underestimated by this assay, especially given the substantial difference in the replication profiles observed in the whole-genome analysis (Fig. 2a–c). One key difference between these two replication analyses is that the fork-rate measurement (Fig. 3) only monitors fork progression over a small portion of the genome. The genome-wide replication kinetics clearly indicate that the replication defect in isw2 nhp10 mutant is the most severe in late-replicating regions at distances greater than 50 kb from early-firing origins (Fig. 2). Because we monitored fork progression of a 50 kb region of chromosome VI that does not encompass an extended late-replicating region (Fig. 2b), it is highly likely that the difference in the replication fork rate we observed in this region is an underestimate of the actual difference in fork rate. Nonetheless, it is important to note that our measurement of replication fork rates provides the first direct evidence that Isw2 and Ino80 complexes facilitate replication fork progression in the presence of replication stress.
How do Isw2 and Ino80 function to promote DNA replication?
We considered two possible mechanisms by which Isw2 and Ino80 promote replication; these factors may act (1) indirectly by maintaining a transcriptional program required to promote replication, or (2) directly by promoting fork progression at the sites of replication. To address the first possibility, we tested whether isw2 nhp10 mutants are deficient in expression of genes that contribute to replication in the presence of MMS. Given that the MMS sensitivity is specific to the double mutant (Fig. 1a, Supplementary Fig. 1a), we sought to identify genes specifically mis-regulated in isw2 nhp10 mutants. To this end, we directly compared transcript profiles of isw2 and nhp10 single mutants to those of an isw2 nhp10 double mutant (Supplementary Fig. 5a–b). In a previous study, our laboratory found that the direct comparison of transcript profiles between single and double mutants is more effective for determining defects in transcription specific to the double mutant than comparing transcript profiles of mutants to those of wild type cells26. Our analysis revealed minor differences in transcription profiles between single and double mutants: 93 genes showed more than 1.5-fold reduction in RNA levels in the double mutant as compared to each single mutant. Notably, none of these genes has been shown to be involved in replication, DNA repair, or S phase checkpoint functions (Fig. 4a, Supplementary Fig. 5c). Furthermore, none of the 93 genes causes MMS sensitivity when deleted27. Based on these results, we conclude that it is unlikely that Isw2 and Ino80 facilitate replication fork progression through transcriptional induction of genes involved in replication, DNA repair, or DNA damage checkpoint.
Figure 4.
Isw2 and Ino80 may directly facilitate DNA replication. (a) Venn diagram showing the lack of overlap of genes deactivated 1.5 fold in isw2 nhp10 (gray) with genes involved in replication, repair, checkpoint (yellow), and MMS resistance (blue). The 93 genes represented are a collection of genes that are deactivated in isw2 nhp10 mutants in comparison to isw2 and nhp10 single mutants under normal or MMS growth conditions. The Venn diagram encompasses data from 6144 genes. (b) Chromatin immunoprecipitation (ChIP) of Pol1, Nhp10, and Isw2 during S phase. YTT3735 and YTT3736 cells (Pol1-3Flag, Nhp10-13Myc, Isw2-Avi) were arrested in G1 and released into S phase in the presence of 200 mM HU. Cells were harvested at indicated time points in S phase and utilized for ChIP analysis. DNA from immunoprecipitated fractions was analyzed by radioactive PCR with primers corresponding to an early origin (ARS607) and a late origin (ARS1502). ARS1502 does not initiate replication under this experimental condition. The mean and standard deviation of the signals at ARS607 relative to ARS1502 from three biological replicates are presented.
To test the possibility that Isw2 and Ino80 directly facilitate replication, we performed chromatin immunoprecipitation (ChIP) to compare the localization of Isw2, Nhp10, and Pol1 during S phase. Pol1 is the catalytic subunit of the DNA polymerase α primase complex required for leading and lagging strand synthesis28,29, and is thus localized to replication forks. If Isw2 and Ino80 chromatin-remodeling complexes function directly to promote DNA synthesis, we expect Isw2, Nhp10, and Pol1 to be enriched at the sites of active replication. To facilitate synchronous progression of replication forks, we performed the ChIP experiments in the presence of hydroxyurea (HU), which slows down replication forks in a more controlled fashion than MMS. As expected, the Pol1 signal peaks early in S phase at an efficient and early-firing origin ARS607 (Fig. 4b). The ChIP signals for Nhp10 and Isw2 show modest, but reproducible, increases during S phase at the same locus; we saw similar results at another early-firing origin ARS305 (data not shown). The degree of enrichment of Isw2 and Nhp10 at origins during S phase that we observe is very similar to the degree of enrichment of the Ino80 ATPase at origins in a separate study under the same condition (Papamichos-Chronakis and Peterson, in press). These results are consistent with the possibility that Isw2 and Ino80 complexes are enriched at the sites of active replication.
Isw2 and Ino80 facilitate replication in the absence of MMS
Our findings that Isw2 and Ino80 facilitate replication fork progression during MMS treatment led us to examine their roles during growth in the absence of MMS. We considered the possibility that MMS facilitates the detection of a pre-existing replication defect in isw2 nhp10 mutants. Because MMS delays firing of late-firing origins through activation of S phase checkpoint, fewer origins fire during later stages in S phase in the presence of MMS. As a result, each replication fork must travel a longer distance than normal under this condition, providing a more sensitive environment to detect defects in replication fork progression. Consistent with our model, we observed modest but reproducible decreases in replication fork rate in isw2 nhp10 mutants in the absence of MMS at both 30°C and 16°C (Supplementary Fig. 4a–b, Supplementary Table 2), but no detectable differences in the efficiency of firing from late origins (Supplementary Fig. 3b). If our model is correct, one can predict that the efficiency of late-origin firing dictates whether cells depend on Isw2 and Ino80 for efficient S phase progression.
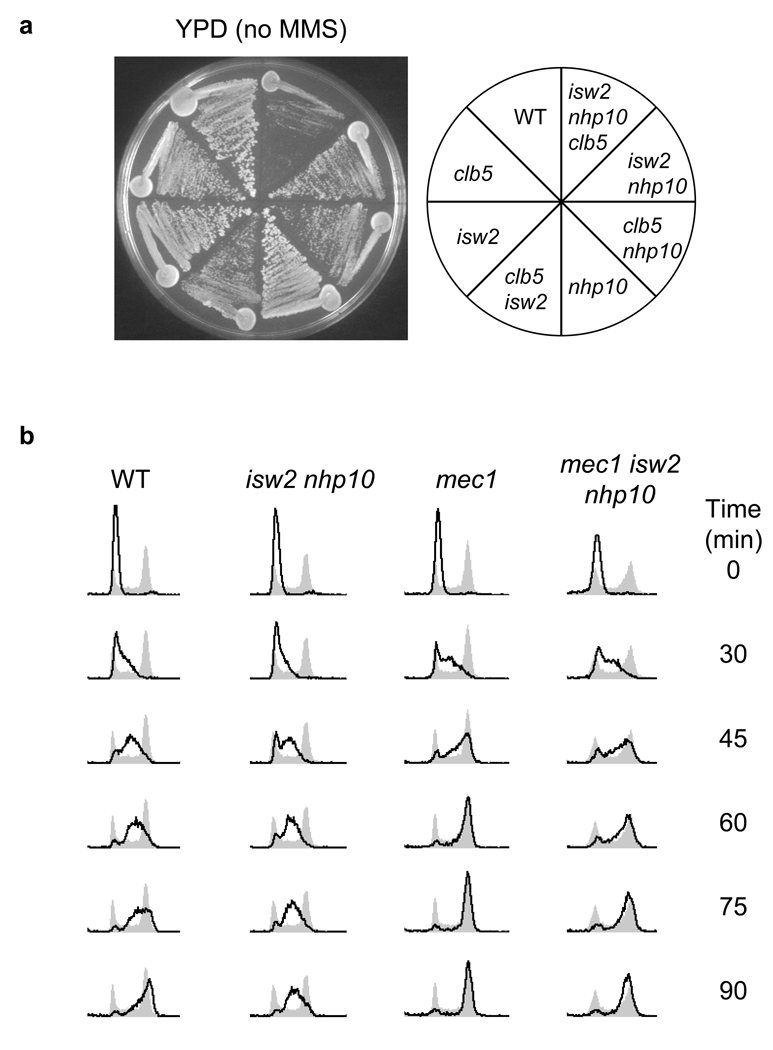
To test our model, we took two complementary approaches to manipulate late-origin activity in isw2 nhp10 mutants. First, we reduced the efficiency of late-origins in the isw2 nhp10 mutant independently of MMS. Deletion of CLB5, a gene encoding an S phase cyclin, results in a decreased frequency of late-origin firing without affecting early-origin efficiency30. As shown in Figure 5a, the isw2 nhp10 clb5 triple mutant has a severe growth defect compared to corresponding double and single mutants in the absence of MMS. This result shows that a clb5 mutation, which suppresses late-firing origins, increases the cell’s dependence on Isw2 and Ino80 complexes for normal growth in the absence of MMS. In a complementary approach, we tested whether an increase in late-origin activity can relieve the S phase delay in MMS-treated isw2 nhp10 mutants. We found that deletion of MEC1, which abrogates the S phase checkpoint and permits efficient firing of late origins during DNA damage23,24, results in a shortened S phase in isw2 nhp10 mutants during MMS treatment (Fig. 5b). Importantly, the difference in the S phase progression rate in the presence and absence of both Isw2 and Nhp10 is largely diminished in a mec1 background (Fig. 5b). This result indicates that the efficient firing of late-origins can effectively mask the replication fork progression defect in isw2 nhp10 mutants even in the presence of MMS. These results collectively support our model that Isw2 and Ino80 facilitate replication fork progression in the absence of MMS, and that treatment of cells with MMS or deletion of CLB5 reveals the function of these remodelling factors during periods of normal DNA replication by suppressing late-firing origins.
Figure 5.
Isw2 and Ino80 promote replication in the absence of MMS. (a) A clb5 deletion causes a severe growth defect in the absence of Isw2 and Nhp10. Wild type (W1588-4c), isw2 (YTT1080), nhp10 (YTT2060), isw2 nhp10 (YTT2109), clb5 (YTT3402),isw2 clb5 (YTT3434) nhp10 clb5 (YTT3437), and isw2 nhp10 clb5(YTT3441) were grown on YPD media at 30°C for 2 days. (b) A mec1 deletion suppresses the extended S phase of isw2 nhp10 mutants in the presence of 0.02% MMS. mec1 (YTT3003) and mec1 isw2 nhp10 (YTT3048) mutants were treated and their DNA content determined as in Figure 1b. WT and isw2 nhp10 FACS profiles are reproduced from Fig.1b for comparison. Both strains contain an sml1 deletion to suppress lethality of a mec1 deletion38.
Discussion
It has been proposed that the regulators of chromatin structure can profoundly affect DNA replication4,31, but how chromatin-mediated regulation of replication takes place is unknown. Although some differences in the activity of late-firing origins may also contribute to the overall defect in replication of late-replicating regions in the presence of MMS (Supplementary Fig. 3a), our replication profiling experiments revealed that the Isw2 and Ino80 complexes facilitate replication fork progression. As a consequence, these complexes play particularly important roles in the replication of late-replicating regions during replication stress, which occurs in the presence of MMS or in a clb5 mutant. We propose that defects in replication are evident under such conditions because each replication fork has to travel a longer distance than normal, thus providing a more sensitive environment to detect replication fork progression defects. This is the first time in which ATP-dependent chromatin-remodeling factors are shown to facilitate a specific step in DNA replication. Based on transcriptional analyses in the isw2 nhp10 mutant cells as well as co-enrichment of Isw2 and Ino80 complexes with DNA polymerase α during S phase, we propose that both complexes function directly at replication forks to facilitate fork progression through chromatin templates. A direct role for Ino80 complex in DNA replication was also suggested by recent independent studies (Papamichos-Chronakis and Peterson, in press). These observations lead to a number of interesting questions; how does altering chromatin structure promote fork-progression, and how do two complexes, each with distinct subunit composition and biochemical activities, function in a partially compensatory manner? Isw2 slides nucleosomes along DNA in vivo10 and Ino80 has been proposed to facilitate histone exchange12. It is possible that these complexes either remodel chromatin ahead of the replication fork, facilitating fork passage, or behind the fork, where re-establishing proper chromatin structure may be important for fork-progression32. How and where these complexes remodel chromatin in relation to a replication fork remains to be determined. Our studies have defined a system to begin investigating these fundamentally important questions.
Previous work has implicated the ISWI class of ATP dependent chromatin-remodeling factors in promoting replication in metazoans33–36. These studies generally used cytological methods to show that ISWI-family remodeling factors co-localize with regions containing newly replicated DNA throughout S phase in both mammalian33,35,36 and Xenopus34 cells. Depletion of ISWI remodeling complexes, ACF or NoRC, resulted in a delay in replication of heterochromatic regions33,36. It remains to be seen whether these ISWI complexes function specifically at the replication fork, and which step in replication is affected by their activity. However, together with our results, these studies suggest that ATP-dependent chromatin remodeling may have conserved roles in directly facilitating replication throughout eukarya.
Methods
Yeast Strains
Yeast strains contain a correction of a weak rad5 mutant allele present in the W303-1a parental strain37,38 (see Supplementary Table 3). We carried out single -step gene disruptions using KanMX, NatMX, and HphMX drug-resistance markers as described39,40. Strains with deletions of ARS608 and ARS609 were derived from a cross between YJT8025 and YTT3109. YTT1080 and YTT3306 are both ADE+ due to pRS402 integration at the ade2-1 locus.
Yeast Culture
Cells were grown at 30°C unless otherwise noted. Log-phase cells for arrest and release experiments were grown to an optical density from 0.25 – 0.30 at 600 nm (OD600). Arrest in G1 was accomplished by treatment with 5 µg ml−1 α–factor. Cells were released from α–factor arrest via filtration on 0.45 µm nitrocellulose membranes, washed, then suspended in pre-warmed media without α–factor. For experiments involving MMS treatment of cells in liquid culture, MMS (Sigma) was added to 0.02% (v/v) when cells were cultured in YPD, or 0.015% (v/v) when synthetic media was required. Cells grown on agar plates were incubated at 30°C for 2 days before imaging. Plates containing drugs were used within 24 hours of preparation.
Flow cytometry
Cells were collected and fixed overnight at 4°C in a final concentration of 66.7% (v/v) ethanol (0.5 ml culture: 1 ml ethanol). Samples were washed in water, resuspended in 2 mg ml−1 RNaseA in 50mM Tris-HCl pH 8.0, and incubated at 37°C for 4 hours. Cells were then centrifuged, resuspended in 2 mg ml−1 Proteinase K in 50mM Tris-HCl pH 7.5, and incubated at 50°C for 45 minutes. After resuspension in 50mM Tris-HCl pH 7.5 and brief sonication at low power, SYTOX Green nucleic acid stain (Invitrogen) was added. DNA content analysis was performed using a BD FACScan flow cytometer and Cell Quest software (BD Biosciences).
Dense isotope transfer
Density transfer experiments were performed essentially as described17 (http://fangman-brewer.genetics.washington.edu/density_transfer.html). Cells were grown a minimum of 7 generations in minimal medium containing 13C and 15N as the sole carbon and nitrogen sources (dense media). Cells were synchronized with α–factor for 105 minutes. Cells were then filtered and transferred to complete media containing 12C and 14N (light media) in the continued presence of α–factor for 75 minutes prior to release. This “conditioning” phase promoted a more synchronous release into S phase. For drug treatment, MMS was added to 0.015% (v/v) 15 minutes into the conditioning phase resulting in a 60-minute exposure during G1 arrest. After the conditioning phase, cells were filtered, washed, and released in light media in the presence or absence of 0.015% MMS (v/v). Samples were collected at the indicated times and genomic DNA was isolated. Genomic DNA was digested with EcoRV (New England BioLabs) for microarray experiments, or with a combination of ClaI and SalI (NEB) for fork-progression analysis. DNA comprised of two heavy strands (HH) was separated from DNA comprised of a heavy and light strand (HL) by ultracentrifugation in a cesium chloride gradient.
Replication profiling with Microarrays
HH and HL fractions were pooled separately for each collection, alternatively labeled with Cy3-dUTP and Cy5-dUTP (GE Healthcare), and co-hybridized to yeast ORF arrays (GEO accession number GPL1914, Fred Hutchinson CRC genomics facility). Image analyses of microarrays were performed with Gene Pix Pro v6.0 (Molecular Devices). Data were normalized and smoothed as described18. Normalized and smoothed data were averaged for each dye swap pair and plotted with KaleidaGraph v 4.0 (Synergy Software).
Fork progression rate
Percent replication of ClaI / SalI restriction fragments between ARS607 and right telomere of chromosome VI was determined by slot-blotting of HH and HL fractions as described (http://fangman-brewer.genetics.washington.edu/density_transfer.html). Probes to Regions 1–5 are identical to probes 2–6 described by Tercero and Diffley25. Kinetic curves of replication end either at the last collection time, or at the last time prior to detection of light-light (LL) DNA at that region (indicative of a new round of replication in cells that have completed S phase and Mitosis since the beginning of the experiment). The maximum percentage of budded cells provided a good estimate of the maximum percent replication of each restriction fragment and was therefore used to determine the Trep. The distance relative to Region 1 was measured from the right most point of each probed restriction fragment. An estimate of average fork rate from Region 1 to 5 was provided by the inverse of the slope of the best-fit line accounting for all 5 data points25. Generation of graphs, determination of Trep values, and curve fitting were done using KaleidaGraph. For determination of fork rate at a low temperature, cells were synchronized in G1 at 30°C, then released at 16°C.
Expression analysis
Logarithmically dividing cells were grown to an OD600 of 0.35 in YPD media then divided in two. One culture was treated with 0.02% (v/v) MMS. Cells were then incubated for two hours then harvested for RNA extraction by acid-phenol method. Transcript-microarray analysis of was performed as described41. The “deletion causes MMS sensitivity” gene list is as described27. The “replication, repair, and checkpoint” gene list was compiled based on descriptions provided at the Saccharomyces Genome Database. The complete transcript array data set (MIAME compliant) is available at http://www.fhcrc.org/science/labs/tsukiyama/.
Chromatin immunoprecipation (ChIP)
ChIP was done essentially as described41,42 with the following modifications. 300 mL of culture was collected at indicated time points after release from G1 arrest to 200mM HU and fixed at room temp with 1% formaldehyde for 20 min. Immunoprecipitation performed with 20 µL of protein G dynabeads (Dynal) prebound overnight at 4°C with either 1 µL of Flag (Sigma) or 2 µL of 9E10 (Covance) antibodies, or 100 µL of M-280 streptavidin dynabeads (Dynal Biotech). A detailed protocol is available at http://www.fhcrc.org/science/labs/tsukiyama/.
PCR analysis
28 cycles of PCR analysis was performed using 0.5 µL of [α-32P] dCTP per 100µl reaction. Serial dilution of input DNA confirmed that PCR was within the linear range. Radioactive PCR was done in duplicate for ChIP samples from three independently prepared samples. PCR products were separated on 6% (w/v) polyacrylamide gels, and visualized on a phosphorimager (Molecular Dynamics). Primer sequences are listed in Supplementary Table 4.
Supplementary Material
Acknowledgements
We thank G.M. Alvino, S. Biggins, B.J. Brewer, D. Collingwood, H.S. Malik, M.K. Raghuraman, and members of the Tsukiyama lab for critical reading of the manuscript; J.F.X. Diffley (Cancer Research UK) for the YJT80 strain; members of the Biggins and Brewer/Raghuraman labs for helpful discussions; D. Collingwood for help with analysis of replication data; H.S. Malik for the manuscript title suggestion; C.L. Peterson for sharing data prior to publication; and G.M. Alvino for a DNA labeling protocol for microarrays, technical advice on density transfer experiments, and help with replication data analysis. This work was supported by grants from the National Institutes of Health and the Leukemia and Lymphoma Society to T.T. J.A.V. and T.J. K. were supported in part by training grants from the National Institutes of Health.
References
- 1.Becker PB, Horz W. ATP-dependent nucleosome remodeling. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 2.Smith CL, Peterson CL. ATP-dependent chromatin remodeling. Curr Top Dev Biol. 2005;65:115–148. doi: 10.1016/S0070-2153(04)65004-6. [DOI] [PubMed] [Google Scholar]
- 3.Narlikar GJ, Fan HY, Kingston RE. Cooperation between complexes that regulate chromatin structure and transcription. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 4.Varga-Weisz P. Chromatin remodeling factors and DNA replication. Prog Mol Subcell Biol. 2005;38:1–30. doi: 10.1007/3-540-27310-7_1. [DOI] [PubMed] [Google Scholar]
- 5.Ataian Y, Krebs JE. Five repair pathways in one context: chromatin modification during DNA repair. Biochem Cell Biol. 2006;84:490–504. doi: 10.1139/o06-075. [DOI] [PubMed] [Google Scholar]
- 6.Jaskelioff M, Van Komen S, Krebs JE, Sung P, Peterson CL. Rad54p is a chromatin remodeling enzyme required for heteroduplex DNA joint formation with chromatin. J Biol Chem. 2003;278:9212–9218. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 7.van Attikum H, Gasser SM. ATP-dependent chromatin remodeling and DNA double-strand break repair. Cell Cycle. 2005;4:1011–1014. doi: 10.4161/cc.4.8.1887. [DOI] [PubMed] [Google Scholar]
- 8.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucl. Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fazzio TG, Tsukiyama T. Chromatin Remodeling In Vivo: Evidence for a Nucleosome Sliding Mechanism. Molecular Cell. 2003;12:1333–1340. doi: 10.1016/s1097-2765(03)00436-2. [DOI] [PubMed] [Google Scholar]
- 11.Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat Struct Mol Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- 12.Papamichos-Chronakis M, Krebs JE, Peterson CL. Interplay between Ino80 and Swr1 chromatin remodeling enzymes regulates cell cycle checkpoint adaptation in response to DNA damage. Genes Dev. 2006;20:2437–2449. doi: 10.1101/gad.1440206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tong AH, et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364–2368. doi: 10.1126/science.1065810. [DOI] [PubMed] [Google Scholar]
- 14.Shen X, Mizuguchi G, Hamiche A, Wu C. A chromatin remodelling complex involved in transcription and DNA processing. Nature. 2000;406:541–544. doi: 10.1038/35020123. [DOI] [PubMed] [Google Scholar]
- 15.Morrison AJ, et al. INO80 and gamma-H2AX interaction links ATP-dependent chromatin remodeling to DNA damage repair. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 16.Raghuraman MK, et al. Replication dynamics of the yeast genome. Science. 2001;294:115–121. doi: 10.1126/science.294.5540.115. [DOI] [PubMed] [Google Scholar]
- 17.McCarroll RM, Fangman WL. Time of replication of yeast centromeres and telomeres. Cell. 1988;54:505–513. doi: 10.1016/0092-8674(88)90072-4. [DOI] [PubMed] [Google Scholar]
- 18.Alvino GM, et al. Replication in Hydroxyurea: It's a matter of time. Mol Cell Biol. 2007 doi: 10.1128/MCB.00719-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yabuki N, Terashima H, Kitada K. Mapping of early firing origins on a replication profile of budding yeast. Genes Cells. 2002;7:781–789. doi: 10.1046/j.1365-2443.2002.00559.x. [DOI] [PubMed] [Google Scholar]
- 20.Feng W, et al. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nat Cell Biol. 2006;8:148–155. doi: 10.1038/ncb1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wyrick JJ, et al. Genome-wide distribution of ORC and MCM proteins in S. cerevisiae: high-resolution mapping of replication origins. Science. 2001;294:2357–2360. doi: 10.1126/science.1066101. [DOI] [PubMed] [Google Scholar]
- 22.Nieduszynski CA, Knox Y, Donaldson AD. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev. 2006;20:1874–1879. doi: 10.1101/gad.385306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Santocanale C, Diffley JF. A Mec1- and Rad53-dependent checkpoint controls late-firing origins of DNA replication. Nature. 1998;395:615–618. doi: 10.1038/27001. [DOI] [PubMed] [Google Scholar]
- 24.Shirahige K, et al. Regulation of DNA-replication origins during cell-cycle progression. Nature. 1998;395:618–621. doi: 10.1038/27007. [DOI] [PubMed] [Google Scholar]
- 25.Tercero JA, Diffley JF. Regulation of DNA replication fork progression through damaged DNA by the Mec1/Rad53 checkpoint. Nature. 2001;412:553–557. doi: 10.1038/35087607. [DOI] [PubMed] [Google Scholar]
- 26.Fazzio TG, et al. Widespread collaboration of Isw2 and Sin3-Rpd3 chromatin remodeling complexes in transcriptional repression. Mol Cell Biol. 2001;21:6450–6460. doi: 10.1128/MCB.21.19.6450-6460.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang M, Bellaoui M, Boone C, Brown GW. A genome-wide screen for methyl methanesulfonate-sensitive mutants reveals genes required for S phase progression in the presence of DNA damage. Proc Natl Acad Sci U S A. 2002;99:16934–16939. doi: 10.1073/pnas.262669299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sugino A. Yeast DNA polymerases and their role at the replication fork. Trends Biochem Sci. 1995;20:319–323. doi: 10.1016/s0968-0004(00)89059-3. [DOI] [PubMed] [Google Scholar]
- 29.Burgers PM. Eukaryotic DNA polymerases in DNA replication and DNA repair. Chromosoma. 1998;107:218–227. doi: 10.1007/s004120050300. [DOI] [PubMed] [Google Scholar]
- 30.Donaldson AD, et al. CLB5-dependent activation of late replication origins in S. cerevisiae. Mol Cell. 1998;2:173–182. doi: 10.1016/s1097-2765(00)80127-6. [DOI] [PubMed] [Google Scholar]
- 31.Falbo KB, Shen X. Chromatin remodeling in DNA replication. J Cell Biochem. 2006;97:684–689. doi: 10.1002/jcb.20752. [DOI] [PubMed] [Google Scholar]
- 32.Ye X, et al. Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell. 2003;11:341–351. doi: 10.1016/s1097-2765(03)00037-6. [DOI] [PubMed] [Google Scholar]
- 33.Collins N, et al. An ACF1-ISWI chromatin-remodeling complex is required for DNA replication through heterochromatin. Nat Genet. 2002;32:627–632. doi: 10.1038/ng1046. [DOI] [PubMed] [Google Scholar]
- 34.Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. Embo J. 2002;21:2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Poot RA, et al. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol. 2004;6:1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- 36.Li J, Santoro R, Koberna K, Grummt I. The chromatin remodeling complex NoRC controls replication timing of rRNA genes. Embo J. 2005;24:120–127. doi: 10.1038/sj.emboj.7600492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thomas BJ, Rothstein R. The genetic control of direct-repeat recombination in Saccharomyces: the effect of rad52 and rad1 on mitotic recombination at GAL10, a transcriptionally regulated gene. Genetics. 1989;123:725–738. doi: 10.1093/genetics/123.4.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- 39.Guldener U, Heck S, Fielder T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldstein AL, McCusker JH. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi: 10.1002/(SICI)1097-0061(199910)15:14<1541::AID-YEA476>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 41.Lindstrom KC, Vary JC, Jr, Parthun MR, Delrow J, Tsukiyama T. Isw1 functions in parallel with the NuA4 and Swr1 complexes in stress-induced gene repression. Mol Cell Biol. 2006;26:6117–6129. doi: 10.1128/MCB.00642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gelbart ME, Bachman N, Delrow J, Boeke JD, Tsukiyama T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 2005;19:942–954. doi: 10.1101/gad.1298905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nieduszynski CA, Hiraga S, Ak P, Benham CJ, Donaldson AD. OriDB: a DNA replication origin database. Nucleic Acids Res. 2007;35:D40–D46. doi: 10.1093/nar/gkl758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.