Abstract
With the advent of powerful computers, simulation studies are becoming an important tool in statistical methodology research. However, computer simulations of a specific process are only as good as our understanding of the underlying mechanisms. An attractive supplement to simulations is the use of plasmode datasets. Plasmodes are data sets that are generated by natural biologic processes, under experimental conditions that allow some aspect of the truth to be known. The benefit of the plasmode approach is that the data are generated through completely natural processes, thus circumventing the common concern of the realism and accuracy of computer simulated data. The estimation of admixture, or the proportion of an individual’s genome that originates from different founding populations, is a particularly difficult research endeavor that is well suited to the use of plasmodes. Current methods have been tested with simulations of complex populations where the underlying mechanisms such as the rate and distribution of recombination are not well understood. To demonstrate the utility of this method data derived from mouse crosses is used to evaluate the effectiveness of several admixture estimation methodologies. Each cross shares a common founding population so that the ancestry proportion for each individual is known, allowing for the comparison of true and estimated individual admixture values. Analysis shows that the different estimation methodologies (Structure, AdmixMap and FRAPPE) examined all perform well with simple datasets. However, the performance of the estimation methodologies varied greatly when applied to a plasmode consisting of three founding populations. The results of these examples illustrate the utility of plasmodes in the evaluation of statistical genetics methodologies.
Keywords: Admixture, ancestry, simulation, plasmode, population structure
1. INTRODUCTION
Admixture refers to the process in which individuals from populations with different allele frequencies begin to mate and form a new, mixed or ‘hybrid’ population. In subsequent generations disequilibrium among linked loci (points in the genome that tend to be inherited together) in this admixed population may span a greater genetic distance than generally found in populations that have been randomly mating for many generations. This extended linkage disequilibrium (LD, non-random association of loci) has the potential to facilitate the detection of regions of the genome that contain phenotype-influencing loci by reducing the required number of marker loci needed for mapping when compared to disequilibrium mapping in randomly mating populations [1–6]. However, not only can the admixture process produce LD, it can also produce disequilibrium between pairs of unlinked loci. This admixture induced disequilibrium between unlinked loci can create confounding, or spurious associations, in genetic association studies and therefore needs to be accommodated in association studies [3,6,7].
There are several methods that have been proposed to deal with confounding due to admixture-induced disequilibrium and variations in individual ancestry [8]. These methods can be grouped into two fundamentally different categories: Genomic Control (GC) and Structured Association Testing (SAT). GC corrects for stratification in association studies by adjusting for a uniform overall inflation statistic estimated from random null markers [9]. Structured Association Testing methods can be divided into two subcategories: those based on individual ancestry estimates and those based on a measure of genetic background obtained through principal component analysis (PCA). In the first category of SAT methods, individual ancestry estimates are used to cluster individuals into subpopulations and control for population structure during association testing. [10–15]. The second SAT subgroup employs PCA to estimate a genetic background score for each individual and in an attempt to control for stratification by accounting for variation in the data associated with the differences in allele frequencies [16–18]. Methods based on individual admixture estimations are commonly used for controlling for population stratification and will be used for the example presented here.
Several computational methods have been developed that estimate individual admixture from genetic data [3,8]. Within this paper, we utilize plasmodes to evaluate three often cited approaches to admixture estimation; Structure, AdmixMap, and FRAPPE [12,14,15,19,20]. Subsequent to testing via simulations, illustration of most of these methods, with the exception of Structure, has been limited to data from human populations such as African-Americans and Hispanic-Americans [13,14,21]. Such validation is also problematic (we use the word validation here loosely, in actuality “field-test” or “illustrate” would be more accurate). Limiting the testing to these populations for method evaluation implicitly assumes that they are representative of all admixed populations, and that the answers, in this case the individual admixture values, are known a priori.
Despite recent interest in applying admixture methods to diverse populations ranging from rice to tiger salamanders and buffalo to humans [22–26], most methods were developed and tested via simulation of genotypes for individuals in an admixed human population. There are two major issues with using simulated datasets in this manner. First, simulations may not accurately capture the complexity of biological data [27]. Biologists are sometimes suspicious of computer generated simulated biological data, doubting that current simulation methods can effectively emulate the complex processes involved in such things as non-random mating, recombination, hot spots, and interference. Second, population genetic models used to simulate the test populations are often limited to the island or continuous gene flow models, neither of which are likely to accurately represent the complexity of the process by which the majority of admixed human, or non-human, populations are formed [28–30].
To overcome some of the limitations of testing via simulations and human populations, we propose the use of plasmode datasets to evaluate individual admixture estimation methodologies. A plasmode is defined as a collection of data that: 1) is the result of a real biological process; 2) is not merely the result of a computer simulation and; 3) has been constructed so that at least some aspect of the ‘truth’ of the data generating process is known [31]. A commonly used type of plasmode is a ‘spike in’ experiment in microarray expression analysis, where a known amount of transcript is added to serve as a positive control. One of the primary advantages is that, unlike what is generally observed with computer simulations, distributions and correlations are realistic because they are taken directly from real data [27,31,32].
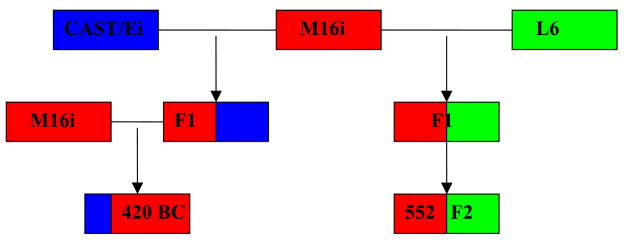
The plasmodes used here are formed from a collection of two experimental mouse crosses. The three founding populations are M16i, L6 and CAST/Ei. The first cross is the result of a mating between M16i and L6 mice, to obtain an F1 (first generation). These F1 mice were then mated together to obtain a F2 (second generation) population [33,34]. Hence, all mice within the first plasmode will have ancestry of 0.5 from M16i and L6 (Figure 1). The second is a backcross (BC) resulting from the mating of M16i mice with CAST/Ei to obtain F1 individuals which were then mated with M16i mice to obtain the BC ((CAST/EixM16i)xM16i) [35,36]. By this design, all mice within the second cross will have ancestry of 0.75 from M16i and 0.25 from the CAST/Ei. All mice were genotyped with 100% ancestry informative microsatellite markers and phenotype data was gathered for a number of traits. Since both crosses share M16i as a common ancestor, we will use the fraction of markers inherited from the M16i strain as the reference to evaluate the different admixture estimation methodologies. Because individual admixture estimates provided by the different estimation techniques are a function of true ancestry, random variation due to recombination, and measurement error, the individual admixture estimates provided by the estimation approaches should vary around the true ancestry values. [37,38]. Although the use of this plasmode does not allow for complex models such as non-random mating or extended time since the admixture event to be tested, it does intrinsically incorporate processes such as recombination, hotspots and interference not accounted for in current simulation studies, and provides a simple example to illustrate the utility of the concept.
Figure 1. Design of mouse plasmodes.
Illustration of the breeding scheme and proportion of ancestry for the mouse plasmodes. Corresponding with Figures 2–4 M16i, the shared founder is indicated in red, CAST/Ei in blue and L6 in red.
In this manuscript we provide an example of the application of plasmode datasets as a supplement to simulation in the evaluation of individual admixture estimation software. In particular, we will use three popular methods: Structure, AdmixMap and FRAPPE [12,14,15,19,20]. We then present a comparison of these three methods using a mouse plasmode with known ancestry to evaluate each algorithm’s performance.
2. APPLICATION OF A PLASMODE, AN ILLUSTRATIVE EXAMPLE
Current methods for estimating individual admixture can be divided into two classes: Bayesian and maximum likelihood (ML). Programs such as AdmixMap, AncestryMap and Structure are software that utilize data from the founding populations, in the form of individuals for Structure or allele frequencies for AdmixMap, to provide an informative prior to calculate the posterior distribution of admixture estimates using Markov Chain Monte Carlo (MCMC) in the Bayesian based framework [11–15]. ML estimation methods, such as IBGA, PSMIX and FRAPPE, fall under the frequentist framework [20,21,39]. Individual admixture estimates are obtained by maximizing the likelihood of the individual ancestry proportion of each individual given the available data. Although the program AncestryMap has been successfully used for admixture based mapping of disease genes [40–42], we do not include it here since it is restricted to two founding populations and diallelic markers.
Several authors reported correlations of the admixture estimates obtained with their methods with Structure estimates, but they do not report the correlation of their estimates with true admixture, as determined by simulation. Tang et al. provide the only direct evaluation of individual admixture estimates [20]. They show via simulation that with informative markers and well represented parental populations, both Structure and FRAPPE estimation work well. With less informative markers, or only a few members of the parental populations, the FRAPPE estimation is unbiased while Structure estimates can be highly biased. Here, to demonstrate the feasibility and utility of plasmodes to evaluate methods and software, we present a systematic evaluation of three individual admixture estimation methods, Structure, AdmixMap, and FRAPPE through the use of the mouse plasmodes. The use of plasmodes allows us to compare the individual admixture estimates from each algorithm with known individual ancestry.
2.1 Evaluation of Ancestry Estimates in the BC and F2 plasmodes
We gauge the performance of Structure, AdmixMap and FRAPPE with the simple and straightforward individual BC and F2 datasets. Results with all markers are presented in Table 1. Root mean square error (RMSE) was used to measure the variation from estimated individual admixture from true ancestry [20].
Table 1.
Average individual admixture proportions estimated with Structure, AdmixMap, and FRAPPE for the individual BC and F2 plasmodes.
| A | Structure | FRAPPE |
|---|---|---|
|
F2 Expected M16i proportion 0.5000 | ||
| Average M16i | 0.4970 | 0.4953 |
| STDEV | 0.0534 | 0.0625 |
| RMSE | 0.0423 | 0.0047a |
|
| ||
|
BC Expected M16i proportion 0.7500 | ||
| Average M16i | 0.7410 | 0.7498 |
| STDEV | 0.0552 | 0.0572 |
| RMSE | 0.0453 | 0.0020a |
| B | Structure | AdmixMap |
|
F2 Expected M16i proportion 0.5000 | ||
| Average M16i | 0.4959 | 0.4948 |
| STDEV | 0.0543 | 0.0529 |
| RMSE | 0.0435 | 0.0429 |
|
| ||
|
BC Expected M16i proportion 0.7500 | ||
| Average M16i | 0.7304 | 0.7417 |
| STDEV | 0.0472 | 0.0388 |
| RMSE | 0.0416 | 0.0325 |
Admixture estimates, standard deviation, and RMSE estimates for Structure, FRAPPE, and AdmixMap. A. Structure Linkage model compared to FRAPPE. B. Structure PopAdx model compared to AdmixMap.
FRAPPE gives identical estimates on repeated runs.
Results from two models for Structure are presented in order to provide a direct comparison with both AdmixMap and FRAPPE. Structure results obtained under the “Linkage model” are most directly comparable to AdmixMap, whereas Structure results derived under the “PopAdx model” are most directly comparable to the FRAPPE results [11,12,14,20,43,44]. The linkage model incorporates genetic distance information for each marker into the calculation of the admixture estimates. The PopAdx model, on the other hand, does not utilize linkage information, but instead uses prior information on the population of origin of the founders. The linkage model allows for admixture in all individuals, while the PopAdx model only allows for admixture in the offspring. Examples for the Structure output for the BC and F2 plasmodes are provided in Figures 2 & 3. Although FRAPPE is not designed to utilize linkage information, the estimates did not seem to be affected when linked markers are used, in accordance to Tang et al.’s observations [20].
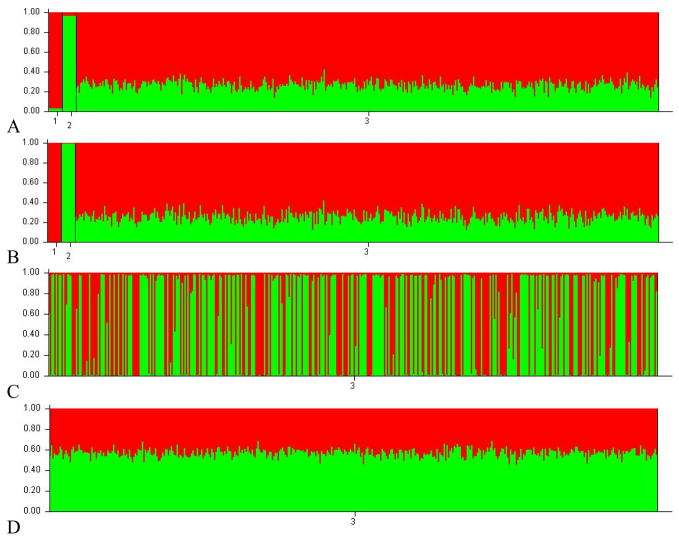
Figure 2.
Structure results for the BC plasmode. In A & B M16i (red) and CAST/Ei (green) founders are represented by number 1 and 2, respectively, on the x notations of the graph. Admixed individuals are identified as 3 in A, B, C, and D. Individuals are represented by bars on the x axis and population proportion on the y. The proportion of M16i and CAST/Ei for each individual are illustrated by the amount of red and green for each individual. A) Linkage analysis with all markers and 10 founders, B) PopAdx analysis with all markers and 10 founders, C) PopAdx with all markers and no founders D) Linkage with all markers and no founders.
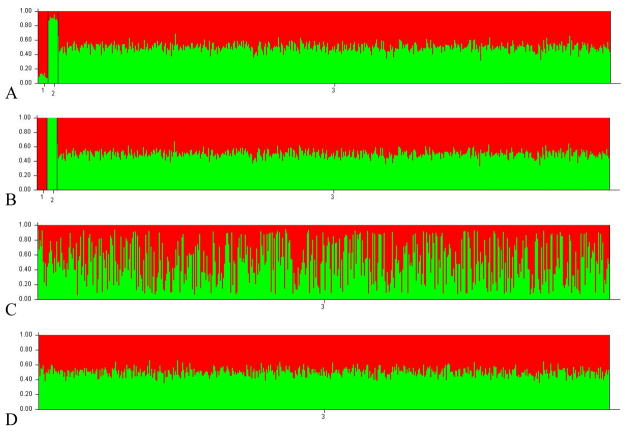
Figure 3.
Structure results for the F2 plasmode M16i proportions are indicated in red. A) Linkage analysis with all markers and 10 founders, B) PopAdx analysis with all markers and 10 founders, C) PopAdx with all markers and no founders D) Linkage with all markers and no founders.
All the programs performed equally well when founders were included in the analysis, and all identified the correct the number of founding populations (K). However, when no founders were included in the analysis of the two plasmodes, the Structure and FRAPPE estimates no longer accurately reflect the true structure of the data with M16i estimates of approximately 50% for both methods in both populations (e.g. Figures 2 & 3). For both estimation approaches, RMSE greatly increases in the absence of founders. AdmixMap does provide accurate estimates without founders included in the analysis (0.4948 and 0.7460 M16i for the F2 and BC respectively). This difference is to be expected since, as noted in the software documentation, these programs utilize founders included in the analysis to estimate the founding allele frequencies, whereas AdmixMap utilizes pre-supplied founding allele frequencies.
2.2 Evaluation of Ancestry Estimates in the combined plasmode
For the relatively simple BC or F2 analysis using the recommended conditions, including founders in Structure and FRAPPE analysis, all the methods perform well. However, it is unrealistic to expect that in typical association studies all subjects will have uniform ancestry. For a more complex analysis we pooled the data from the BC and F2 plasmodes to create a combined plasmode. The combined plasmode has 3 founding populations, M16i, which is the common founder for both the BC and F2 plasmodes, L6 from the BC, and CAST/Ei from the F2. The original F2 and BC datasets had only 11 markers in common, which is a potential limitation. However, the markers we have are 100% ancestry informative and our purpose here is to estimate individual admixture, for which a lesser number of markers is required than for association studies [45–48]. Results from the combined plasmode are presented in Table 2 and illustrated in Figure 4.
Table 2. Structure.
, AdmixMap, and FRAPPE average individual admixture estimates for the combined plasmode.
| A | Structure | FRAPPE | |||
|---|---|---|---|---|---|
| founders | 10 | 30 | 10 | 30 | |
|
| |||||
| F2 | Expected M16i proportion 0.5000 | ||||
|
| |||||
| Average M16i | 0.0219 | 0.0321 | 0.3976 | 0.3741 | |
| STDEV | 0.0606 | 0.0800 | 0.1462 | 0.1484 | |
| RMSE | 0.4854 | 0.4701 | 0.1024 | 0.1259 | |
|
|
|||||
| BC | Expected M16i proportion 0.7500 | ||||
|
|
|||||
| Average M16i | 0.4361 | 0.4523 | 0.5187 | 0.4306 | |
| STDEV | 0.4651 | 0.4602 | 0.1729 | 0.1932 | |
| RMSE | 0.5942 | 0.4912 | 0.2313 | 0.3194 | |
| B | Structure | AdmixMap | |||
|
| |||||
| founders | 10 | 30 | 0a | 0b | 30 |
|
| |||||
| F2 | Expected M16i proportion 0.5000 | ||||
| Average M16i | 0.0062 | 0.3534 | 0.4280 | 0.4606* | 0.4815* |
| STDEV | 0.0077 | 0.1477 | 0.1188 | 0.1146* | 0.1149* |
| RMSE | 0.4970 | 0.1700 | 0.1112 | 0.0968* | 0.0930* |
|
| |||||
| BC | Expected M16i proportion 0.7500 | ||||
|
| |||||
| Average M16i | 0.9741 | 0.7230* | 0.6743 | 0.7004* | 0.7261* |
| STDEV | 0.0032 | 0.0914* | 0.0990 | 0.0902* | 0.0854* |
| RMSE | 0.5537 | 0.0779* | 0.1013 | 0.0825* | 0.0728* |
Admixture estimates, standard deviations and RMSE for the F2 and BC individuals in the combined plasmode with 0, 10 or 30 founders included in the analysis. Starred (*) values indicate those that have a RMSE less than 0.1000. A. Structure Linkage model compared to FRAPPE. B. Structure PopAdx model compared to AdmixMap.
AdmixMap analysis conducted with no founders or prior allele frequency information (K=3).
AdmixMap analysis conducted with no founders but with prior allele frequencies for the founding populations.
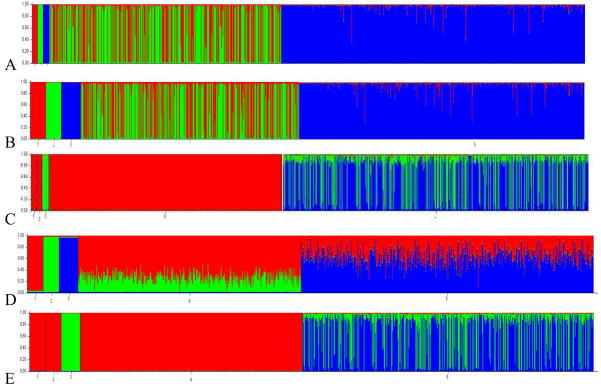
Figure 4.
Graphical output for Structure analysis of combined plasmode with the first 3 bars (groups) representing the founders 1) M16i - red 2) CAST/Ei - green 3) L6 - blue with the BC ((CAST/Ei × M16i) × M16i) sample in the middle and the F2 (M16i × L6) on the far right: A) PopAdx 11 markers 10 founders, B) PopAdx 11 markers 30 founders C) Linkage 11 markers 10 founders- note that Structure does not differentiate between the M16i and CAST/Ei founders, D) Linkage 11 markers 30 founders, E) Linkage 11 markers 30 founders illustrating lack of convergence between two successive runs.
Despite having completely ancestry informative markers, neither Structure nor FRAPPE returned accurate estimate of the population structure or individual admixture values in the combined plasmode (Table 2, Figure 4). Structure did not provide accurate results under the PopAdx model. This model is comparable to the model used in FRAPPE that produced better estimates and lower RMSE than the Structure PopAdx model. For both 10 and 30 founders, Structure clearly identified the three founding populations, but indicated that the individual mice in the F2 sample were of pure L6 ancestry and that the BC sample was an equal mix of CAST/Ei and M16i (Figure 4). This is particularly surprising since the PopAdx model allows for the user to specify which individuals belong to which population, and that information is used in the assignment of individuals to clusters. As with FRAPPE, there was not a difference in the estimates or RMSE when 10 or 30 founders were included.
When Structure was run with the Linkage model, which is comparable to AdmixMap, the RMSE did decrease with the inclusion of additional founders (Table 2, Figure 4). However, Structure did not provide accurate results with 10 founders and could not differentiate between M16i and CAST/Ei mice in the founder or the BC samples. When 30 founders are included, Structure does provide the correct population structure, but only provided accurate individual admixture estimates for the BC sample and not the F2 portion.
Structure Linkage model results also provide an example of a non-identifiability issue that can occur with Structure. Structure’s MCMC sampler can become ‘stuck’ in a mode, or fail to converge to the same answer over multiple iterations (Figure 4D & E), which often occurs when Structure is unsure of the true structure present in the data [20]. When founders are not included in the analysis, it becomes impossible to identify founding populations in multiple runs of algorithm. This emphasizes the need to run multiple replications with the same number of populations (K).
The program AdmixMap is the only one that gave accurate estimates for the combined plasmode (Table 2). In agreement with Tang’s [20] observation, inclusion of founders in the analysis slightly improved the individual admixture estimates, although the algorithm is designed to run without founders in the dataset. The improved performance of AdmixMap when compared to Structure is not entirely unexpected since AdmixMap utilizes pre-supplied founding allele frequencies, and Structure must estimate the frequencies from the data. When AdmixMap is run on the combined plasmode without the pre-supplied allele frequencies (with the number of populations set to K=3 and no founders) the estimates are more similar to those obtained via Structure or FRAPPE (Table 2).
3. DISCUSSION
In this work we introduce the concept of plasmode datasets as a supplement to simulation. We then use a mouse plasmode to provide an illustration of the application with the comparison of three individual admixture estimation methodologies, Structure, AdmixMap, and FRAPPE.
Plasmodes offer a unique and important addition to the tool box of methodologists. Because they are created through a natural process, the complexities of a biological dataset are intrinsically incorporated. Simulations are a staple of statistical methodologic research since they allow the investigator to specify and control every aspect of the study. However, biological data sets are by nature very intricate. While simulation methods are continually improving, they are limited by the researchers knowledge and understanding of a complex system. Recombination location and rates provides a telling example. In simulating admixed populations recombination rates are typically assumed to be constant across the genome. It is further assumed that recombination events occur independently, and have a Poisson distribution. It is well know that recombination occurs more frequently in certain regions of the genome (hotspots) and that a phenomenon known as interference prevents recombination from occurring near each other in the same generation. It has recently been shown that recombination rates are in fact variable across the genome and for different populations [49–51]. Because the plasmodes used here were created from breeding populations, the distribution and rate of recombination are inherently incorporated, without requiring prior specific knowledge.
Concerns have been raised in the past about the properties of the methods when few markers are used. The use of plasmodes allows for the examination of these issues in a dataset generated by biological process. The poor performance of Structure and FRAPPE may in fact be the result of the low number of markers. However, all the markers are 100% ancestry informative and visual inspection of the data clearly differentiates between the BC, F2 and founding populations. Although we purposely avoided including in this manuscript computer matings, or simulations, simulations were conducted that mimic the combined plasmode. These simulations did show that both Structure and FRAPPE required both more markers and additional founders to provide accurate estimates.
The importance of including founders in the analysis has also been a matter of contention. It is not surprising that Structure performs better with an increased number of founders, since it is recommended in the user’s guide that at least half of the sample should consist of individuals from the founder populations. Tsai et al. [8] report that Structure requires a minimal ratio of founders to admixed samples in the analysis. Tang et al. [20] also report the need for founders, or pseudo-founders, in the analysis for FRAPPE, Structure, and AdmixMap. This is in direct opposition of the literature describing AdmixMap which claims that founders should not be included in the data. McKeigue claims that with the assumption of a unimodal distribution of individual admixture proportions in AdmixMap’s model, inclusion of founders in the analysis will cause the distribution to shift, which results in the model having a poor fit [44,52]. Regardless of potential poor model fit, it is apparent that based on the analysis of these plasmodes all the methods return more accurate estimations when founders are included in the analysis.
This manuscript is not meant to serve as comparison of results from simulation studies and plasmode analysis, but as a proof of principle for the utility of plasmodes in the evaluation of statistical genetic methodologies. As we have illustrated here, plasmodes offer an attractive addition to the use of simulations and real populations for the testing of such methodologies. It is important to note that we do not support the use of plasmodes at the complete abandonment of simulation studies. Instead we believe that by using both methods, the limitations of each will be overcome. The lack of appropriate data sets for the creation of plasmodes is one of the largest hurdles in its application. The plasmodes here are limited by the number of markers in common between the two crosses. As use of plasmodes gains acceptance, we hope that plasmode datasets which more appropriately represent complex human data will also become widely available. Future research will include the examination of more complex plasmodes.
4. PLASMODE CONSTRUCTION, ADMIXTURE ESTIMATION & STATISTICAL ANALYSIS
4.1 Plasmode Construction
Mouse data for the creation of the plasmode have been described in detail elsewhere [33,36,53]. Briefly, the data consist of 420 ((CAST/Ei × M16i) × M16i) backcrossed mice (BC) genotyped with 88 microsatellite markers and 552 (M16i × L6 F2) intercross mice (F2) genotyped with 61 microsatellite markers. Because crosses are derived from three independent lines, each locus is completely ancestry informative (i.e. M16i individuals have only the A allele, L6 only the B allele, and CAST/Ei only the C allele). Although marker information was not provided for individual founders, because of the completely ancestry informative markers genotypes could be unambiguously assigned to founders. Thus, the genotype data for founders can be derived from the cross genotypes. Thirty, ten, or zero founders were included in the analysis as indicated. The two datasets were compiled to form the combined plasmode based on 11 common microsatellite markers: D2MIT133 (60), D2MIT224 (64), D2MIT22 (73), D2MIT49 (84), D4MIT27 (36), D7MIT55 (15), D9MIT2 (17), D12MIT5 (41), D13MIT53 (50), D18MIT19 (2), and D18MIT51 (27). Marker names indicate the chromosome on which the marker is located, (e.g. D2 markers are on chromosome 2) with the chromosomal location from the MGD database in Haldane (cM) units indicated in parenthesis. In this dataset we know the true M16i ancestry for each individual, 75% for each BC mouse and 50% for the F2 individuals. As discussed in the introduction, true admixture is true ancestry plus biological ‘error’ from recombination and measurement error (for a more detailed discussion refer to Redden et al. 2006 [37]). Therefore, we expect that the individual admixture estimates provided by the different programs will vary around true ancestry.
4.2 Structure version 2.1
The Structure software takes a Bayesian approach developed by Pritchard and colleagues [12,15] available at http://pritch.bsd.uchicago.edu/structure.html. It employs a model-based clustering approach to infer population structure, to estimate the proportion of the genome derived from each founding population for each individual, and to estimate population allele frequencies via a Markov chain Monte Carlo (MCMC) procedure. Genotype data for founders and admixed individuals, from linked or unlinked markers are used to probabilistically assign individuals to populations, or mixtures of populations. Hardy-Weinberg Equilibrium and Linkage Equilibrium are assumed within populations, and no particular mutation process or model for the formation of the admixed population are specified. Several models are allowed such as no admixture, admixture, or linkage are described in detail in [12].
Structure analysis was conducted via the windows front end interface using the following models: 1) Linkage - where the admixture model is used, but admixture estimates are calculated while taking into account linkage information; and 2) PopAdx - Prior information with admixture. The two models which correspond to the models employed by AdmixMap and FRAPPE were run with correlated and independent allele frequencies for the founding populations, and three iterations for each number of populations (three iterations of each K at K=1–3 for the BC and F2 and K=1–4 for the combined). The ‘correct’ number of populations, or K, was selected based on prior knowledge or estimated from the data [12,15,28,43]. Analyses were conducted with 20,000 burn-in and 50,000 iterations.
4.3 AdmixMap version 3.1
AdmixMap was developed by Hoggart et al. [11,14] building on McKeigue 1998 and 2000 papers [1,52] and available at http://www.ucd.ie/genepi/software.html. It is a hybrid of Bayesian and classical approaches that utilizes MCMC methods to jointly estimate the admixture proportions for individuals and control for admixture induced confounding in association studies. Briefly, a Bayesian model is used to calculate the admixture values for individuals, based on the supplied prior distribution of allele frequencies in the founding populations. The posterior distribution of admixture and the ancestry specific allele frequencies at each locus, given the observed genotype and trait data, is generated by MCMC simulation. The program was specifically developed for application to admixed human populations, such as African-American and Hispanic-American populations [44].
Founding allele frequencies were estimated based on allele counts for each founding population. Estimated allele counts were then used as the prior allele frequencies for each plasmode analysis. Linkage was assumed and analyses were conducted using prior allele frequencies of the founding populations or with a set number of populations (K=2 for F2 and BC, K=3 for combined). AdmixMap allows for the specification of random or assortative mating populations, and all models were run with assortative mating assumed (default setting). Because AdmixMap utilizes pre-supplied allele frequencies, less iteration are required. A burn-in of 350 and 3500 iterations were used. Unlike Structure, AdmixMap does not automatically produce results for multiple replications with unique starting random number seeds, therefore three replications with unique random seeds, or starting values for the MCMC, were manually conducted.
4.4 MLE/FRAPPE
The frequentist method used in this study was developed by Tang et al. [20] as an extension of the approach first proposed by Hanis et al. [54,55] to allow for estimation of founding allele frequencies and individual admixture using maximum likelihood estimates. Code for the program is freely available from the website http://www.fhcrc.org/science/labs/tang/ under the section FRAPPE: Frequentist Estimation of Individual Ancestry Proportion. Details on the method are provided in Tang et al. [20]. Briefly, individual admixture proportions are represented by a vector Qi = (qi1, qi2, ….qiK) where qij represents the probability that an allele sampled at random for ith individual originates from the jth founding population K, where the number of founding populations is assumed to be known. Genotype data, from the same set of independent markers, for both admixed and representatives of the founding populations are required. Hardy-Weinberg equilibrium conditioned on the admixture proportion is assumed, as well as no genetic drift and well separated founding populations [20].
4.5 Statistical analysis
RMSE was chosen as the metric of the accuracy of the individual admixture estimates [20]. It can be written as: where q̄i and qi represent respectively the estimated and true individual admixture. For each program the RMSE for each individual was calculated for 3 different iterations and the average RMSE for the individual and combined plasmodes (BC and F2 separately for the combined) was then calculated for each algorithm.
Acknowledgments
This work was supported in part by: the National Institutes of Health grants, R01GM077490, R01ES09912, T32HL072757, 5R01AI52658, P30DK0563365, K25DK62817, and 5P60AR48095.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.McKeigue PM. Mapping genes that underlie ethnic differences in disease risk: Methods for detecting linkage in admixed populations, by conditioning on parental admixture. American Journal of Human Genetics. 1998;63:241–251. doi: 10.1086/301908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halder I, Shriver M. Measuring and using admixture to study the genetics of complex diseases. Human Genetics. 2003;1:52–62. doi: 10.1186/1479-7364-1-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McKeigue PM. Prospects for admixture mapping of complex traits. American Journal of Human Genetics. 2005;76:1–7. doi: 10.1086/426949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Darvasi A, Shifman S. The beauty of admixture. Nature Genetics. 2005;37:118–119. doi: 10.1038/ng0205-118. [DOI] [PubMed] [Google Scholar]
- 5.Reich D, Patterson N. Will admixture mapping work to find disease genes? Philos Trans R Soc Lond B Biol Sci. 2005;360:1605–1607. doi: 10.1098/rstb.2005.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith MW, O’Brien SJ. Mapping by admixture linkage disequilibrium: Advances, limitations and guidelines. Nature Reviews Genetics. 2005;6:623–632. doi: 10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 7.Marchini J, Cardon LR, Phillips MS, Donnelly P. The effects of human population structure on large genetic association studies. Nature Genetics. 2004;36:512–517. doi: 10.1038/ng1337. [DOI] [PubMed] [Google Scholar]
- 8.Tsai HJ, Choudhry S, Naqvi M, Rodriguez-Cintron W, Burchard EG, Ziv E. Comparison of three methods to estimate genetic ancestry and control for stratification in genetic association studies among admixed populations. Human Genetics. 2005;118:424–433. doi: 10.1007/s00439-005-0067-z. [DOI] [PubMed] [Google Scholar]
- 9.Devlin B, Roeder K, Wasserman L. Genomic control, a new approach to genetic-based association studies. Theoretical Population Biology. 2001;60:155–166. doi: 10.1006/tpbi.2001.1542. [DOI] [PubMed] [Google Scholar]
- 10.Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. American Journal of Human Genetics. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoggart CJ, Shriver MD, Kittles RA, Clayton DG, McKeigue PM. Design and analysis of admixture mapping studies. American Journal of Human Genetics. 2004;74:965–978. doi: 10.1086/420855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patterson N, Hattangadi N, Lane B, Lohmueller KE, Hafler DA, Oksenberg JR, Hauser SL, Smith MW, O’Brien SJ, Altshuler D, Daly MJ, Reich D. Methods for high-density admixture mapping of disease genes. American Journal of Human Genetics. 2004;74:979–1000. doi: 10.1086/420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoggart CJ, Parra EJ, Shriver MD, Bonilla C, Kittles RA, Clayton DG, McKeigue PM. Control of confounding of genetic associations in stratified populations. American Journal of Human Genetics. 2003;72:1492–1504. doi: 10.1086/375613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu XF, Zhang SL, Zhao HY, Cooper RS. Association mapping, using a mixture model for complex traits. Genetic Epidemiology. 2002;23:181–196. doi: 10.1002/gepi.210. [DOI] [PubMed] [Google Scholar]
- 17.Zhang SL, Zhu XF, Zhao HY. On a semiparametric test to detect associations between quantitative traits and candidate genes using unrelated individuals. Genetic Epidemiology. 2003;24:44–56. doi: 10.1002/gepi.10196. [DOI] [PubMed] [Google Scholar]
- 18.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 19.Montana G, Hoggart C. Statistical software for gene mapping by admixture linkage disequilibrium. Briefings in Bioinformatics. 2007;8(6):393–395. doi: 10.1093/bib/bbm035. [DOI] [PubMed] [Google Scholar]
- 20.Tang H, Peng J, Wang P, Risch NJ. Estimation of individual admixture: Analytical and study design considerations. Genetic Epidemiology. 2005;28:289–301. doi: 10.1002/gepi.20064. [DOI] [PubMed] [Google Scholar]
- 21.Bonilla C, Parra EJ, Pfaff CL, Dios S, Marshall JA, Hamman RF, Ferrell RE, Hoggart CL, McKeigue PM, Shriver MD. Admixture in the Hispanics of the San Luis Valley, Colorado, and its implications for complex trait gene mapping. Annals of Human Genetics. 2004;68:139–153. doi: 10.1046/j.1529-8817.2003.00084.x. [DOI] [PubMed] [Google Scholar]
- 22.Beaumont M, Barratt EM, Gottelli D, Kitchener AC, Daniels MJ, Pritchard JK, Bruford MW. Genetic diversity and introgression in the Scottish wildcat. Molecular Ecology. 2001;10:319–336. doi: 10.1046/j.1365-294x.2001.01196.x. [DOI] [PubMed] [Google Scholar]
- 23.Fitzpatrick BM, Shaffer HB. Environment-dependent admixture dynamics in a tiger salamander hybrid zone. Evolution. 2004;58:1282–1293. doi: 10.1111/j.0014-3820.2004.tb01707.x. [DOI] [PubMed] [Google Scholar]
- 24.Wen B, Xie XH, Gao S, Li H, Shi H, Song XF, Qian TZ, Xiao CJ, Jin JZ, Su B, Lu D, Chakraborty R, Jin L. Analyses of genetic structure of Tibeto-Burman populations reveals sex-biased admixture in southern Tibeto-Burmans. American Journal of Human Genetics. 2004;74:856–865. doi: 10.1086/386292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Semon M, Nielsen R, Jones MP, McCouch SR. The population structure of African cultivated rice Oryza glaberrima (Steud.): Evidence for elevated levels of linkage disequilibrium caused by admixture with O. sativa and ecological adaptation. Genetics. 2005;169:1639–1647. doi: 10.1534/genetics.104.033175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Musani SK, Halbert ND, Redden DT, Allison DB, Derr JN. Marker genotypes, population admixture, and their association with body weight, height, and relative body mass in U.S. federal bison herds. Genetics. 2006;174:775–783. doi: 10.1534/genetics.106.057547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mehta T, Tanik M, Allison DB. Towards sound epistemological foundations of statistical methods for high-dimensional biology. Nature Genetics. 2004;36:943–947. doi: 10.1038/ng1422. [DOI] [PubMed] [Google Scholar]
- 28.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Molecular Ecology. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- 29.Eller E, Hawks J, Relethford JH. Local extinction and recolonization, species effective population size, and modern human origins. Human Biology. 2004;76:689–709. doi: 10.1353/hub.2005.0006. [DOI] [PubMed] [Google Scholar]
- 30.Waples RS, Gaggiotti O. What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Molecular Ecology. 2006;15:1419–1439. doi: 10.1111/j.1365-294X.2006.02890.x. [DOI] [PubMed] [Google Scholar]
- 31.Cattell R, Jaspers J. A general plasmode for factor analytic exercises and research. Multivar Behav Res Mono. 1967;3:1–12. [Google Scholar]
- 32.Mehta TS, Zakharkin SO, Gadbury GL, Allison DB. Epistemological issues in omics and high-dimensional biology: give the people what they want. Physiological Genomics. 2006;28:24–32. doi: 10.1152/physiolgenomics.00095.2006. [DOI] [PubMed] [Google Scholar]
- 33.Rocha JL, Eisen EJ, Van Vleck LD, Pomp D. A large-sample QTL study in mice: I. Growth. Mammalian Genome. 2004;15:83–99. doi: 10.1007/s00335-003-2312-x. [DOI] [PubMed] [Google Scholar]
- 34.Rocha JL, Eisen EJ, Van Vleck LD, Pomp D. A large-sample QTL study in mice: II. Body composition. Mammalian Genome. 2004;15:100–113. doi: 10.1007/s00335-003-2308-6. [DOI] [PubMed] [Google Scholar]
- 35.Wolf JB, Pomp D, Eisen EJ, Cheverud JM, Leamy LJ. The contribution of epistatic pleiotropy to the genetic architecture of covariation among polygenic traits in mice. Evolution & Development. 2006;8:468–476. doi: 10.1111/j.1525-142X.2006.00120.x. [DOI] [PubMed] [Google Scholar]
- 36.Leamy LJ, Pomp D, Eisen EJ, Cheverud JM. Pleiotropy of quantitative trait loci for organ weights and limb bone lengths in mice. Physiological Genomics. 2002;10:21–29. doi: 10.1152/physiolgenomics.00018.2002. [DOI] [PubMed] [Google Scholar]
- 37.Redden D, Divers J, Vaughan L, Tiwari H, Beasley T, Fernandez J, Kimberly R, Feng R, Padilla M, Lui N, Miller M, Allison D. Regional admixture mapping and structured association testing: conceptual unification and an extensible general linear model. PLoS Genetics. 2006;2:1254–1264. doi: 10.1371/journal.pgen.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Divers J, Vaughan L, Padilla MA, Fernandez JR, Allison DB, Redden DT. Correcting for measurement error in individual ancestry estimates in structured association tests. Genetics. 2007;176:1823–1833. doi: 10.1534/genetics.107.075408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu B, Liu N, Zhao H. PSMIX: an R package for population structure inference via maximum likelihood method. BMC Bioinformatics. 2006;7:317. doi: 10.1186/1471-2105-7-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reich D, Patterson N, Ramesh V, De Jager PL, McDonald GJ, Tandon A, Choy E, Hu DL, Tamraz B, Pawlikowska L, Wassel-Fyr C, Huntsman S, Waliszewska A, Rossin E, Li RL, Garcia M, Reiner A, Ferrell R, Cummings S, Kwok PY, Harris T, Zmuda JM, Ziv E. Admixture mapping of an allele affecting interleukin 6 soluble receptor and interleukin 6 levels. American Journal of Human Genetics. 2007;80:716–726. doi: 10.1086/513206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reich D, Patterson N, De Jager PL, McDonald GJ, Waliszewska A, Tandon A, Lincoln RR, Deloa C, Fruhan SA, Cabre P, Bera O, Semana G, Kelly MA, Francis DA, Ardlie K, Khan O, Cree BAC, Hauser SL, Oksenberg JR, Hafler DA. A whole-genome admixture scan finds a candidate locus for multiple sclerosis susceptibility. Nature Genetics. 2005;37:1113–1118. doi: 10.1038/ng1646. [DOI] [PubMed] [Google Scholar]
- 42.Freedman ML, Haiman CA, Patterson N, McDonald GJ, Tandon A, et al. Admixture mapping identifies 8q24 as a prostate cancer risk locus in African-American men. PNAS. 2006;103:14068–14073. doi: 10.1073/pnas.0605832103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pritchard JK, Wen W. Documentation for Structure Software. 2004 Version 2 http://pritch.bsd.uchicago.edu/software/readme_structure2_1.pdf.
- 44.McKeigue P, O’Donnell D. ADMIXMAP- a Program to Model Admixture Using Marker Genotype Data v. 33- Manual . 2006 http://www.ucdie/genepi/admix%20manual.html.
- 45.Rosenberg NA, Mahajan S, Ramachandran S, Zhao CF, Pritchard JK, Feldman MW. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genetics. 2005;1:660–671. doi: 10.1371/journal.pgen.0010070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pritchard JK, Rosenberg NA. Use of unlinked genetic markers to detect population stratification in association studies. American Journal of Human Genetics. 1999;65:220–228. doi: 10.1086/302449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang BZ, Zhao HY, Kranzler HR, Gelernter J. Practical population group assignment with selected informative markers: characteristics and properties of Bayesian clustering via STRUCTURE. Genetic Epidemiology. 2005;28:302–312. doi: 10.1002/gepi.20070. [DOI] [PubMed] [Google Scholar]
- 48.Yang BZ, Zhao HY, Kranzler HR, Gelernter J. Characterization of a likelihood based method and effects of markers informativeness in evaluation of admixture and population group assignment. BMC Genetics. 2005;6:50. doi: 10.1186/1471-2156-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hellenthal G, Stephens M. Insights into recombination from population genetic variation. Current Opinion in Genetics & Development. 2006;16:565–572. doi: 10.1016/j.gde.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 50.Calabrese P. A population genetics model with recombination hotspots that are heterogeneous across the population. PNAS. 2007;104:4748–4752. doi: 10.1073/pnas.0610195104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Coop G, Przeworski M. An evolutionary view of human recombination. Nature Reviews Genetics. 2007;8:23–34. doi: 10.1038/nrg1947. [DOI] [PubMed] [Google Scholar]
- 52.McKeigue PM. Multipoint admixture mapping. Genetic Epidemiology. 2000;19:464–465. doi: 10.1002/1098-2272(200012)19:4<464::AID-GEPI17>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 53.Yi NJ, Zinniel DK, Kim K, Eisen EJ, Bartolucci A, Allison DB, Pomp D. Bayesian analyses of multiple epistatic QTL models for body weight and body composition in mice. Genetical Research. 2006;87:45–60. doi: 10.1017/S0016672306007944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hanis CL, Chakraborty R, Ferrell RE, Schull WJ. Individual admixture estimates - Disease associations and individual risk of Diabetes and gallbladder-disease among Mexican-Americans in Starr County, Texas. American Journal of Physical Anthropology. 1986;70:433–441. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]
- 55.Hanis CL, Chakraborty R, Schull WJ. Individual admixture estimates and genetic-marker - Disease associations. American Journal of Physical Anthropology. 1985;66:178–178. doi: 10.1002/ajpa.1330700404. [DOI] [PubMed] [Google Scholar]