Abstract
Primary carnitine deficiency is an autosomal recessive disorder of fatty acid oxidation caused by defective carnitine transport. This disease presents early in life with hypoketotic hypoglycemia or later in life with skeletal myopathy or cardiomyopathy. The gene for this condition maps to 5q31.2–32 and OCTN2, an organic cation/carnitine transporter, also maps to the same chromosomal region. Here we test the causative role of OCTN2 in primary carnitine deficiency by searching for mutations in this gene in affected patients. Fibroblasts from patients with primary carnitine deficiency lacked mediated carnitine transport. Transfection of patient’s fibroblasts with the OCTN2 cDNA partially restored carnitine transport. Sequencing of the OCTN2 gene revealed different mutations in two unrelated patients. The first patient was homozygous (and both parents heterozygous) for a single base pair substitution converting the codon for Arg-282 to a STOP codon (R282X). The second patient was a compound heterozygote for a paternal 1-bp insertion producing a STOP codon (Y401X) and a maternal 1-bp deletion that produced a frameshift creating a subsequent STOP codon (458X). These mutations decreased the levels of mature OCTN2 mRNA and resulted in nonfunctional transporters, confirming that defects in the organic cation/carnitine transporter OCTN2 are responsible for primary carnitine deficiency.
Primary carnitine deficiency [On-Line Mendelian Inheritance in Man no. 212140] is a rare autosomal recessive disorder due to defective carnitine transport (1–3). Carnitine is essential for the transfer of long-chain fatty acids from the cytosol to mitochondria for subsequent beta oxidation and the lack of carnitine impairs the ability to use fat as fuel during periods of fasting or stress (1–3). Affected children present in infancy with nonketotic hypoglycemia, Reye syndrome or sudden infant death (1, 2). This syndrome can also present later in life with skeletal myopathy or cardiomyopathy (1–3). In several families, undiagnosed siblings of affected patients have died of hypoglycemia, sudden infant death syndrome, or intractable cardiomyopathy (1–10). By contrast, diagnosed patients respond promptly to dietary carnitine supplementation, with correction of metabolic abnormalities and even reversal of skeletal and heart muscle abnormalities (1–10). Patients waste carnitine in urine and their fibroblasts share the defective carnitine transporter with the kidney (4, 5). Heterozygous parents of these children have partially decreased plasma carnitine levels due to increased urinary loss (7). The defective transporter has high-affinity (Km = 4–10 μM) for carnitine, as compared with transporters in the liver and the brain that have low affinity (Km = 2–10 mM) (1, 2). The presence of genetically distinct carnitine transporters explains the restoration of carnitine stores in the liver, but not in the muscle of patients with primary carnitine deficiency after carnitine supplementation (1, 2).
The gene for primary carnitine deficiency was recently mapped to chromosome 5q31.1–32 (11). Independently, a cDNA encoding an organic cation transporter, OCTN2, has been isolated by its homology to the previously identified organic cation transporter OCTN1 and the OCTN2 gene also maps to the same chromosomal region (12). OCTN2 is a member of the family of organic cation transporters, integral membrane proteins believed to be important in detoxification, drug excretion, and transplacental solute transfer. Their prototypic substrate is tetraethylammonium and they differ from monoamine transporters and ATP-dependent multidrug exporting proteins (13). A recent study has shown that OCTN2 is also capable of mediating sodium-dependent, high-affinity carnitine transport (14). The OCTN2 gene is composed by 10 exons and spans ≈30 kb (12). The mature mRNA is 3500 nucleotides with an ORF of 1674 nucleotides. The gene is widely expressed, with high mRNA levels in the kidney, muscle, and placenta. The predicted protein is composed of 12 transmembrane spanning domains with a glucose transporter signature and a nucleotide binding fold motif (12, 14).
The substrate recognition and kinetic properties of this transporter (14) resembled those of the carnitine transporter of human fibroblasts (Y.W. and N.L., unpublished results). Therefore, we tested OCTN2 as the candidate gene for primary carnitine deficiency in two unrelated families. Our results indicate that patients with primary carnitine deficiency carry mutations that abolish production and function of OCTN2, confirming its role in this disease.
MATERIALS AND METHODS
Cell Strains and Carnitine Transport.
Fibroblasts from a patient with primary carnitine deficiency (GM 10665) and DNA from both parents were obtained from the National Institute of General Medical Sciences Human Genetic Mutant Cell Repository, Coriell Cell Repositories (Camden, NJ). Fibroblasts from patient 2996 were obtained as previously described (7). Fibroblasts were grown in DMEM supplemented with 15% fetal bovine serum. Both patients presented early in life with Reye-like episodes (ref. 7; see description of patient GM 10665 in National Institute of General Medical Sciences catalog). Both patients responded well to carnitine supplementation with improvement of their symptoms and prevention of new attacks.
Carnitine transport was measured at 37°C with the cluster-tray method as described (7). Cells were grown to confluence in 24-well plates (Costar) and depleted of intracellular amino acids by incubation for 90 min in Earle’s balanced salt solution containing 5.5 mM d-glucose and supplemented with 0.5% BSA. Carnitine (0.5 μM, 0.5 μCi/ml; 1 Ci = 37 GBq) was then added to the cells for 2 h. Nonsaturable carnitine transport was measured in the presence of 2 mM cold carnitine. The transport reaction was stopped by rapidly washing the cells four times with ice-cold 0.1 M MgCl2. Intracellular carnitine was then corrected for intracellular water content and expressed as nmol/ml cell water (7). Saturable carnitine transport was calculated by subtracting nonsaturable carnitine transport from total transport and values are reported as means ± SE of three to six independent determinations.
Construction of the OCTN2 Expression Vector.
The OCTN2 expression vector was generated by digestion of the OCTN2 cDNA in pSPORT (12) with StuI. This procedure removed 911 bp of cDNA after the normal STOP codon (12). The resulting 2341 bp of OCTN2 cDNA was religated, excised from pSPORT with EcoRI and NotI, and inserted in the corresponding sites of pcDNA3. The final vector was sequenced, confirming the published sequence with the exception of a 1-bp change not affecting the amino acid sequence. This plasmid was transfected into carnitine transport deficient (2996) human fibroblasts by lipofectamine according to the manufacturer’s instructions (GIBCO/BRL). Cells were assayed for carnitine transport 48 h after transfection. Mutant full-length OCTN2 cDNAs were synthesized from patients’ mRNA by reverse transcriptase and amplification by using the Expand Long Template PCR system (Boehringer Mannheim). The following primers were used: 5′-GCGAATTCCCAGACCCCAGGCCGCGCT-3′ and 5′-GCGAATTCAGTTTCTCCCTTACTGGA-3′ corresponding to the 5′ and 3′ ends of the cDNA with attached EcoRI sites. The amplified cDNA was cloned in pcDNA3, sequenced, and clones containing the desired mutations were subsequently transiently transfected in Chinese hamster ovary (CHO) cells as described above for human fibroblasts.
Analysis of DNA and cDNA.
Genomic DNA and total RNA were extracted from fibroblasts by standard methods (15) and cDNA was synthesized by reverse transcriptase. The cDNA was amplified by PCR and subjected to direct sequencing in both directions, identifying both the maternal and paternal allele on the same reading (15, 16). Identified mutations were confirmed by sequencing genomic DNA. PCR primers for genomic DNA were designed based on the genomic sequence of OCTN2 reported in GenBank (loci AC004628 and AC L81760 on chromosome 5q). Primers were ≈60 bp upstream or downstream of each exon, allowing sequencing of splice donor and splice acceptor sequences and of the lariat branch site. Primers for cDNA amplification were designed 400-bp apart along the published cDNA sequence (12, 14). PCR amplification of cDNA and genomic DNA was performed according to standard conditions except that 10% dimethyl sulfoxide was included in the reaction when exon 1 or the 5′ of the cDNA were amplified. PCR products were visualized by agarose gel electrophoresis, purified by column, and sequenced directly without subcloning (15, 16) using an Applied Biosystems automated DNA sequencer. Alternatively, PCR products were cloned in the TA cloning vector and sequenced according to standard protocols.
RNAse Protection Assay.
Cellular RNA was extracted with guanidinium thiocyanate. Total RNA was hybridized with labeled antisense OCTN2 and actin RNA by using the RNAse protection assay from Ambion and following the manufacturer’s instructions. Briefly, the 704-1377 cDNA fragment of the OCTN2 cDNA was amplified by PCR by using Pfu high-fidelity polymerase and cloned in the TA cloning vector. The plasmid was then digested with DdeI and a 293-nucleotide antisense RNA was generated by using Sp6 polymerase and [32P]ATP. Labeled RNA was purified by acrylamide gel electrophoresis and hybridized overnight with 15–25 μg of cellular RNA. RNAse digestion was then performed and the protected bands were separated by gel electrophoresis. As internal control, a 127-nucleotide antisense RNA was also generated from the human actin cDNA by labeling to a 20-fold lower specific activity than the OCTN2 RNA. The resulting gel was dried and counts in each band were determined by using a microarray radioactivity detector (Instant Imager, Packard). Lane-specific background was subtracted from each lane. The gel was then exposed to film. Linearity of the assay was determined by using increasing amounts of control RNA.
RESULTS
Carnitine Transport in Fibroblasts from Patients with Primary Carnitine Deficiency.
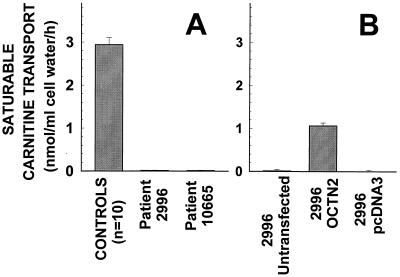
Fig. 1 shows carnitine transport by fibroblasts of patients with primary carnitine deficiency before and after transfection with the OCTN2 cDNA. Saturable carnitine transport was absent (<5% of controls and not significantly different from zero) in fibroblasts from two unrelated patients with primary carnitine deficiency (Fig. 1A). Transient transfection with the OCTN2 cDNA partially restored carnitine transport in 2996 fibroblasts to about one third of the levels of control cells (Fig. 1B). By contrast, transfection with the pcDNA3 vector alone failed to increase carnitine transport.
Figure 1.
Carnitine transport by human fibroblasts from controls and patients with primary carnitine deficiency. (A) Confluent cells were washed and incubated for 90 min in Earle’s balanced salt solution containing d-glucose (5.5 mM) and supplemented with 0.5% BSA. Cells were then incubated for 2 h with [3H]carnitine (0.5 μM, 0.5 μCi/ml) in the absence or presence of 2 mM cold carnitine, included to assess nonsaturable carnitine uptake. Mediated transport was obtained by subtracting the nonsaturable component from total transport. Carnitine accumulation was corrected for cellular protein and intracellular water content. Each point is the mean of at least three independent triplicates (10 for control cells) ± SE. (B) Fibroblasts from patients 2996 were transfected with lipofectamine and 5 μg of OCTN2-pcDNA3, pcDNA3 alone, or nothing (untransfected). One day after transfection, cells were trypsinized and plated in 24-well plates and carnitine transport was measured the following day as above in the presence and absence of 2 mM carnitine. Points are means ± SE of triplicates.
Identification of Mutations in the OCTN2 Gene.
Both cDNA, synthesized from fibroblast RNA, and genomic DNA were amplified by PCR using OCTN2 cDNA primers or primers in the flanking region of each of the 10 exons of the OCTN2 gene. Amplified DNA was sequenced directly without subcloning by using one of the original primers. cDNA mutations were confirmed by analysis of genomic DNA.
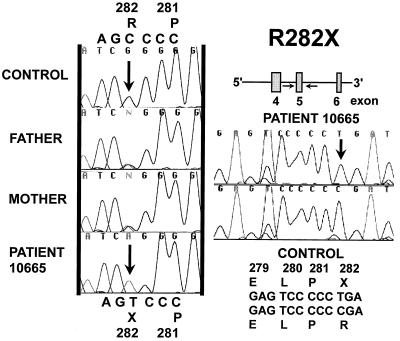
Patient 10665 was homozygous for a C to T transition in exon 5 converting the codon for Arg-282 to a STOP codon (R282X) (Fig. 2). Both parents were heterozygous for this mutation and presented both the normal CGA (Arg) and the mutant TGA (STOP) codon. All the other exons of the OCTN2 gene were sequenced and patient 10665 did not show any other variation affecting the amino acid sequence.
Figure 2.
Direct sequencing of exon 5 of the OCTN2 gene in patient 10665 and both parents (10666 and 10667). Exon 5 of the OCTN2 gene was amplified by PCR and sequenced directly without subcloning. Only the reverse direction is shown for the parents. The proband was homozygous and both parents heterozygous for a 1-bp substitution inserting a premature STOP codon.
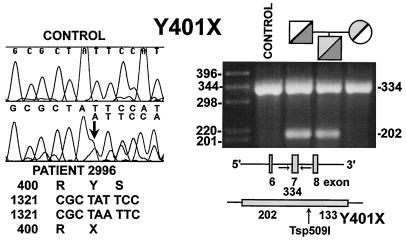
Patient 2996 was a compound heterozygote for two different mutations. The paternal allele carried a 1-bp (A) insertion in exon 7 converting the codon for Tyr-401 (TAT) to a STOP codon (TAA) (Y401X, Fig. 3). The frameshift produced a double nucleotide sequence initiating at the codon for Tyr-401. Presence of this mutation was confirmed by sequencing in the opposite direction and by cloning and sequencing the PCR-amplified exon 7. This 1-bp insertion created a Tsp509I restriction site in the mutant allele, which was cut into two fragments of 133 and 202 bp by the enzyme, while normal DNA was not. The proband was heterozygous for the Tsp509I site, which was also present in the paternal DNA (Fig. 3).
Figure 3.
Sequence of exon 7 of the OCTN2 gene in patient 2996 (Left). Genomic DNA was amplified by PCR and sequenced. Note the presence of double sequence at and after the codon for Y401. Tsp509I digestion of PCR amplified exon 7 of the patient’s family (Right). Both the proband and the father were heterozygous for a normal 334-bp band and an abnormal 202-bp band.
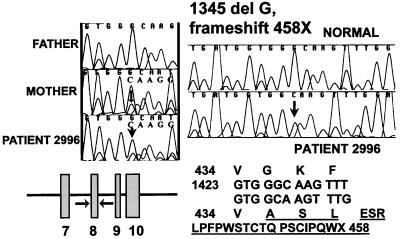
The maternal mutation was a 1-bp (G) deletion in exon 8, which caused a frameshift starting at the codon for Gly-435 (Fig. 4). This was evident as a double sequence that initiated at the second G of the Gly codon (GGC). Twenty-three new amino acids were inserted in the mutant protein, disrupting the predicted hydrophobic transmembrane domain 10. As a result of the frameshift, a premature termination codon was present in position 458 (458X). Presence of this mutation was confirmed by cloning and sequencing the PCR-amplified exon 8, which had the 1-bp deletion. This mutation did not create or remove any restriction site. Direct sequencing confirmed that the mother, but not the father carried the same mutation. All the other exons of the OCTN2 gene in patient 2996 did not have any other variation affecting the amino acid sequence.
Figure 4.
Sequence of exon 8 of the OCTN2 gene in patient 2996. Genomic DNA was amplified by PCR and sequenced. Note the presence of double sequence at and after the codon for G435 in the patient and the mother. The right side of the figure shows the normal sequence and that of a clone of exon 8 from the proband containing the one bp deletion. The predicted neo-amino acid sequence is shown below the sequencing chromatograms.
None of the mutations identified in patients 10665 and 2996 was present in the DNA of 20 controls (40 alleles) and one other unrelated patient with primary carnitine deficiency that were screened.
Effect of OCTN2 Mutations on mRNA Levels.
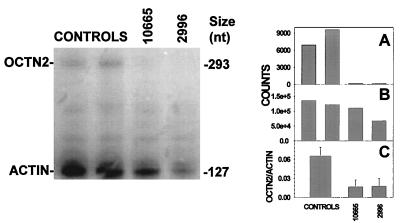
All the mutations identified caused the premature insertion of a STOP codon. The premature insertion of a STOP codon can produce a truncated protein, result in unstable RNA, or cause exon skipping. cDNA could be synthesized, indicating that at least minimal amounts of RNA had to be present. The OCTN2 gene has low levels of expression and cannot be easily detected by Northern blot analysis of total RNA (12, 14). Therefore, we used RNAse protection assay to determine whether these mutations reduced the levels of mature mRNA. Antisense RNA was generated by using the OCTN2 cDNA and hybridized to total RNA before RNAse protection. Antisense actin RNA was used to normalize for the amount of RNA. A representative experiment is shown in Fig. 5. The ratio of OCTN2/actin counts was reduced to ≈25% of control cells (average of three independent experiments) in fibroblasts from patients 10665 and 2996. This result indicates that the major effect of the premature insertion of these STOP codons was to decrease the levels of mature OCTN2 mRNA.
Figure 5.
OCTN2 mRNA levels in fibroblasts from patients with primary carnitine deficiency. (Left) Total RNA (15–25 μg) from controls and patients with primary carnitine deficiency was analyzed by RNAse protection assay. Right. The gel was counted for 48 h with an Instant Imager and the counts obtained for OCTN2 (A) and actin (B) are reported at the right of the autoradiogram for each band. Initial experiments with normal RNA indicated a linear increase in signal intensity of both bands by increasing the amount of total RNA. The experiment was repeated three times using twice the 293-nucleotide probe presented in this figure and once a probe in the 3′ region of the OCTN2 cDNA. C reports the average OCTN2/actin ratio from these three experiments in the two patients as compared with three different control strains.
Expression of Mutant cDNAs.
Because saturable carnitine transport was completely absent in the patients cells (Fig. 1), whereas the mutations identified reduced OCTN2 mRNA to 25% of normal, we expressed the cDNAs containing the mutations identified in patients 10665 and 2996 in CHO cells to confirm that they resulted in nonfunctional transporters. Transient transfection of the expression vector containing the normal OCTN2 cDNA in CHO cells increased carnitine (0.5 μM) transport 50-fold (from 0.41 ± 0.04 to 21.07 ± 0.38 nmol·ml cell water−1·h−1), while transfection with the vector alone (pcDNA3), pcDNA3 containing the cDNA obtained from 10665 with the R282X mutation, and pcDNA3 with the two mutations from cells of patient 2996 (Y401X and 458X) failed to increase carnitine transport, which remained between 0.45 and 0.50 nmol·ml cell water−1·h−1, values not significantly above the transport rate of parental CHO cells.
DISCUSSION
Primary carnitine deficiency, first reported in 1973 (3), is an autosomal recessive disorder caused by impaired carnitine transport. The defect affects renal tubular epithelial cells and is also expressed in fibroblasts (4, 5). The molecular defect does not involve the liver whose carnitine stores are nearly normalized in patients with carnitine deficiency after carnitine supplementation, in contrast with the muscle that remains carnitine depleted (1, 2). Early diagnosis of this condition with subsequent carnitine supplementation in the diet can prevent debilitating or fatal complications. The gene for this condition maps to 5q31.1–32 and a putative carnitine transporter, OCTN2, has been isolated from human placental and kidney cDNA libraries (12, 14). Here we confirm that defects in OCTN2 are causally involved in primary carnitine deficiency.
Fibroblasts from patients with primary carnitine deficiency lacked saturable carnitine transport (Fig. 1). Carnitine transport was partially restored by transfection with the normal OCTN2 cDNA in fibroblasts of patient 2996 (Fig. 1B), indicating that OCTN2 complemented the genetic defect of these fibroblasts. Sequencing of the OCTN2 cDNA and gene in our two patients with primary carnitine deficiency revealed homozygosity for a nonsense mutation (R282X) and compound heterozygosity for a 1-bp insertion creating a STOP codon (Y401X) and a 1-bp deletion causing a frameshift creating a premature termination codon (458X) (Figs. 2–4). Homozygosity was not expected in patient 10665 because the parents are not related, although both are of Indian descent (see description of GM 10665 in the National Institute of General Medical Sciences catalog and patient 10 of ref. 6). However, Southern blot analysis of the patient’s DNA digested with EcoRI and BamHI and hybridized to the full-length OCTN2 cDNA gave normal bands of normal intensity when normalized to the insulin receptor gene signal (data not shown). In addition, sequencing of genomic DNA from both parents indicated their heterozygosity for the R282X mutation (Fig. 2), excluding that patient 10665 was hemizygous for this mutation.
The mutations identified in our patients created premature STOP codons and reduced the levels of mature OCTN2 mRNA to ≈25% of controls (Fig. 5). This was not unexpected because premature STOP codons decrease mRNA stability in a number of diseases including inherited insulin-resistant syndromes (15). Translation of the residual OCTN2 mRNA would result in the production of a truncated membrane transporter with 6 (R282X), 8 (Y401X), or 9 (458X) predicted transmembrane domains instead of the normal 12. The lack of several transmembrane domains would likely result in a protein that is either rapidly degraded or not functional. In accord with this hypothesis, transient transfection of the normal OCTN2 cDNA in CHO cells greatly increased carnitine transport, while transfection with the mutant cDNAs failed to increase carnitine transport. Therefore, all these mutations are predicted to result in no functional carnitine transporters due to both reduced RNA levels and absent function of the translated protein. This agrees well with the markedly reduced saturable carnitine transport measured in cells from both patients (Fig. 1). The partial complementation of the transport defect in the patient’s cells by the OCTN2 cDNA and the presence of null alleles in both unrelated patients confirm that OCTN2 is the gene defective in primary carnitine deficiency.
Both of our patients presented relatively early in life (8 months and 2 years of age) with hypoglycemia and Reye-like episodes and it is unclear whether this early presentation correlates with the severe degree of impairment of carnitine transport caused by null alleles. The myopathic and hypoglycemic presentation of primary carnitine deficiency have been described within the same family (6, 17) and it is believed that environmental factors such as a diet poor in carnitine (4, 6, 18) or concurrent infections or stress (6) trigger early presentation of this disease. However, some families have only the late-onset presentation with cardiomyopathy or myopathy in more than one affected child (6, 10). It is possible that in some cases of late-onset disease less severe mutations may allow residual carnitine transport. Extension of mutational analysis to other families with milder, late-onset initial presentation of primary carnitine deficiency will allow to test this hypothesis.
While this manuscript was under review, abnormalities of the OCTN2 cDNA were reported in two patients with primary carnitine deficiency (19). cDNA analysis indicated that two unrelated patients were compound heterozygotes for a short OCTN2 cDNA lacking nucleotides 255-1649 and either a 19-nucleotide insertion or the deletion of nucleotide 875-1046 (19), corresponding to splicing of exon 4. It is unclear whether a single mutation or compound heterozygosity for two noncomplementing mutations was responsible for the abnormal findings in the cDNA of these patients, because studies of parental cells were not reported (19). Mutations of splicing sites, insertions, or deletions in genomic DNA could cause these abnormalities in the cDNA. However, no studies of genomic DNA were reported in this study (19) and the mutations reported in this paper represent the first identified in patients with primary carnitine deficiency.
Acknowledgments
We thank Dr. Fernando Scaglia for initiating the physiological study of these patients. This work was supported by National Institutes of Health Grants DK 48742 (N.L.) and DA 10045 (V.G.).
ABBREVIATION
- CHO
Chinese hamster ovary
References
- 1.Roe C R, Coates P M. In: The Metabolic and Molecular Basis of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 1501–1533. [Google Scholar]
- 2.Pons, R. & De Vivo, D. C. (1995) J. Child Neurol. 10, Suppl. 2, 2S8–2S24. [PubMed]
- 3.Engel A G, Angelini C. Science. 1973;179:899–902. doi: 10.1126/science.179.4076.899. [DOI] [PubMed] [Google Scholar]
- 4.Treem W R, Stanley C A, Finegold D N, Hale D E, Coates P M. N Engl J Med. 1988;319:1331–1336. doi: 10.1056/NEJM198811173192006. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson B O, Gustafson B, Lindstedt S, Nordin I. J Inherited Metab Dis. 1989;12:108–111. doi: 10.1007/BF01800711. [DOI] [PubMed] [Google Scholar]
- 6.Stanley C A, DeLeeuw S, Coates P M, Vianey-Liaud C, Divry P, Bonnefont J P, Saudubray J M, Haymond M, Trefz F K, Breningstall G N, et al. Ann Neurol. 1991;30:709–716. doi: 10.1002/ana.410300512. [DOI] [PubMed] [Google Scholar]
- 7.Scaglia F, Wang Y, Singh R H, Dembure P P, Pasquali M, Fernhoff P M, Longo N. Genet Med. 1998;1:34–39. doi: 10.1097/00125817-199811000-00008. [DOI] [PubMed] [Google Scholar]
- 8.Waber L J, Valle D, Neill C, DiMauro S, Shug A. J Pediatr. 1982;101:700–704. doi: 10.1016/s0022-3476(82)80294-1. [DOI] [PubMed] [Google Scholar]
- 9.Bennett M J, Hale D E, Pollitt R J, Stanley C A, Variend S. Clin Cardiol. 1996;19:243–246. doi: 10.1002/clc.4960190320. [DOI] [PubMed] [Google Scholar]
- 10. Tein I, DeVivo C D, Bierman F, Pulver P, De Meirleir L J, Cvitanovic-Sojat L, Pagon R A, Bertini E, Dionisi-Vici C, Servidei S, DiMauro S. Pediatr Res. 1990;28:247–255. doi: 10.1203/00006450-199009000-00020. [DOI] [PubMed] [Google Scholar]
- 11.Shoji Y, Koizumi A, Kayo T, Ohata T, Takahashi T, Harada K, Takada G. Am J Hum Genet. 1998;63:101–108. doi: 10.1086/301911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wu X, Prasad P D, Leibach F H, Ganapathy V. Biochem Biophys Res Commun. 1998;246:589–595. doi: 10.1006/bbrc.1998.8669. [DOI] [PubMed] [Google Scholar]
- 13.Grundemann D, Gorboulev V, Gambaryan S, Veyhi M, Koepsell H. Nature (London) 1994;372:549–552. doi: 10.1038/372549a0. [DOI] [PubMed] [Google Scholar]
- 14.Tamai I, Ohashi R, Nezu J, Yabuuchi H, Oku A, Shimane M, Sai Y, Tsuji A. J Biol Chem. 1998;273:20378–20382. doi: 10.1074/jbc.273.32.20378. [DOI] [PubMed] [Google Scholar]
- 15.Longo N, Langley S D, Griffin L D, Elsas L J. Am J Hum Genet. 1992;50:998–1007. [PMC free article] [PubMed] [Google Scholar]
- 16.Longo N, Langley S D, Griffin L D, Elsas L J. Proc Natl Acad Sci USA. 1993;90:60–64. doi: 10.1073/pnas.90.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garavaglia B, Uziel G, Dworzak F, Carrara F, Di Donato S. Neurology. 1991;41:1691–1693. doi: 10.1212/wnl.41.10.1691. [DOI] [PubMed] [Google Scholar]
- 18.Rinaldo P, Stanley C A, Hsu B Y L, Sanchez L A, Stern H J. J Pediatr. 1997;134:304–305. doi: 10.1016/s0022-3476(97)70171-9. [DOI] [PubMed] [Google Scholar]
- 19.Lamhonwah A M, Tein I. Biochem Biophys Res Commun. 1998;252:396–401. doi: 10.1006/bbrc.1998.9679. [DOI] [PubMed] [Google Scholar]