Abstract
The transcriptional activity of steroid hormones is intimately associated with their structure. Deacylcortivazol (DAC) contains several features that were predicted to make it an inactive glucocorticoid. Nevertheless, gene induction and repression by complexes of glucocorticoid receptor (GR) with DAC occurs with greater potency (lower EC50) than, and equal efficacy (maximal activity, or Amax) to, the very active and smaller synthetic glucocorticoid dexamethasone (Dex). Guided by a recent x-ray structure of DAC bound to the GR ligand binding domain (LBD), we now report that several point mutants in the LBD have little effect on the binding of either agonist steroid. However, these same mutations dramatically alter the Amax and/or EC50 of exogenous and endogenous genes in a manner that depends on steroid structure. In some cases, Dex is no longer a full agonist. These properties appear to result from a preferential inactivation of the AF2 activation domain in the GR LBD of Dex-, but not DAC-, bound receptors. The Dex-bound receptors display normal binding to, but greatly reduced response to, the coactivator TIF2, thus indicating a defect in the transmission efficiency of GR-steroid complex information to the coactivator TIF2. In addition, all GR mutants that are active in gene induction with either Dex or DAC have greatly reduced activity in gene repression. This contrasts with the reports of GR mutations preferentially suppressing GR-mediated induction. The properties of these GR mutants in gene induction support the hypothesis that the Amax and EC50 of GR-controlled gene expression can be independently modified, indicate that the receptor can be modified to favor activity with a specific agonist steroid, and suggest that new ligands with suitable substituents may be able to affect the same LBD conformational changes and thereby broaden the therapeutic applications of glucocorticoid steroids
Keywords: mutant glucocorticoid receptors, potency, efficacy, partial agonist activity, transactivation, transrepression
Steroid binding to its cognate receptor protein is the obligate first step in steroid-regulated gene transcription. Specificity in steroid action is obtained by virtue of dissociation constants for steroid binding to receptors being in the nanomolar range or below. Not surprisingly, x-ray structures of steroids bound in the ligand binding domain (LBD) of receptors reveal a good fit with several amino acid side chains of the LBD cavity contacting the ligand (1; 2). For steroids with different structures, additional x-ray studies have documented ligand-induced conformational changes that were difficult to predict (3–5).
Early structure-activity relationships for glucocorticoid receptors (GRs) state that high affinity binding of steroid to GRs requires a C-3 ketone and no steric bulk on the Aring (6; 7). Shortly thereafter, it was reported that deacylcortivazol (DAC), which lacks a C-3 ketone and has a bulky phenylpyrazole substituent on the A-ring (Fig. 1), was about the highest affinity binding glucocorticoid steroid known and with the greatest potency for gene transcription (8). Because “efficacy” and “potency” are not uniquely described, we define greater potency in this report to mean a lower concentration of steroid required for half-maximal induction, or EC50. We use greater efficacy to mean a higher total amount of the maximal activity, or Amax, of the gene being expressed1. Thus, the EC50 for DAC is 10–40 fold lower than that for the very active synthetic glucocorticoid, dexamethasone (Dex), in both gene induction (8) and gene repression (9). The affinity of DAC for GRs is at least 10-fold higher than that of Dex (8). Thus, the higher affinity of DAC for GR can account for most of its lower EC50.
Fig. 1.
Structures of agonist glucocorticoids Dex and DAC.
GRs can effect both gene induction and gene repression but their mechanisms are often different (10; 11). Furthermore, most reported mutations and/or deletions in the amino terminal half of GR (12), the DNA binding domain (13–15), and the LBD (16–18) reduce GR-mediated induction more than repression. Nevertheless, DAC is more potent than Dex in both gene induction and repression. This suggests that DAC binding to GR does not dramatically alter the ability of GR to regulate gene transcription in either of two mechanistically different modes.
So how does DAC bind in the GR LBD cavity that is too small (2) without causing ligand-induced rearrangements of the LBD that would disrupt normal GR actions? The recent x-ray structure of a human GR-DAC complex indicates an almost identical binding as Dex except for the opening of an additional cavity in the LBD to accommodate the bulky A-ring substituents of DAC (19). Thus, most of the contacts of GR protein with Dex and DAC are the same. Human and rat GRs have been examined interchangeably with no apparent differences, as might be expected from the 96.4% identity in their LBDs and 90.8% overall identity (20; 21). Therefore, we used rat GRs in this study when making selected point mutations of the GR LBD to uncover possible differences between Dex- and DAC-bound GRs and thereby advance our understanding of the molecular determinants of steroid activity in two different GR conformations. We find that some mutations eliminate all detectable steroid binding. However, other novel mutations selectively alter not only the absolute Amax vs. EC50 of GR-mediated gene induction, but also the relative Amax of induction vs. repression, in an agonist steroid-specific manner. These results demonstrate that it is possible to separate the control of Amax and EC50, and to preferentially inhibit repression vs. induction. This ability to dissociate these parameters opens new avenues for the selective control of gene expression and possible therapeutic applications of glucocorticoids.
Materials and Methods
Unless otherwise indicated, all operations were performed at 0 °C.
Chemicals
Dexamethasone (Dex) and phorbol 12-myristate 13-acetate (PMA) were purchased from Sigma. Dex-21-mesylate (Dex-Mes) was synthesized as previously described (22). Cortivazol (gift from Roussel UCLAF) was converted to Deacylcortivazol (DAC) by Dr. Craig Thomas (NIDDK/NIH, Bethesda, MD). RU486 was a gift from Etienne Baulieu (Paris, France). [1,2,4,6,7-3H] Dexamethasone (Dex, 90.0 Ci/mmol) was purchased from Amersham Pharmacia Biotech, restriction enzymes and T4 DNA ligase from New England Biolabs, Fermentas, or Promega, and the dual-luciferase reporter assay from Promega (Madison, WI).
Plasmids
Renilla TS was a gift from Nasreldin M. Ibrahim, Otto Frohlich, and S. Russ Price (Emory University School of Medicine). pM vector and GAL/VP16 were purchased from CLONTECH (Palo Alto, CA), and pFR-LUC reporter from Stratagene (La Jolla, CA). Rat GR (pSG5-GR), GREtkLUC, TIF2/GRIP1, and GAL/GRIP1 (23), GAL/GR and VP16/GR (24), and GAL/GR525C (25) have been previously described. GAL/NCoR-RID [amino acids 1944-2453] was a gift from Mitch Lazar (University of Pennsylvania School of Medicine). MMTVLuc (pLTRLUC) was donated by Gordon Hager (NCI/NIH).
The site-directed mutagenesis kit (Stratagene) was used with the following primers (modified nucleotides of codon [bold} are underlined) to make mutant GRs. E558A, 5′ CTG GAG GTG ATT GCA CCC GAG GTG TTG 3′; L584D, 5′ C ACA CTC AAC ATG GAC GGT GGG CGT CAA GTG 3′; Q588K, 5′ TG TTA GGT GGG CGT AAA GTG ATT GCA GCA GTG AAA TGG 3′; A625I, 5′ GG ATG TTT CTC ATG GCA TTT ATC CTG GGT TGG AGA TC 3′; L626I, 5′ CTC ATG GCA TTT GCC ATC GGT TGG AGA TCA TAC 3′; R629Y, 5′ G GCA TTT GCC CTG GGT TGG TAC TCA TAC AGA CAA TC 3′. After verifying the sequences of all mutant DNAs, Smal I/Sap I fragments replaced the corresponding segment of wild type GR plasmid, pSG5-GR. For mutant VP/GR plasmids, Sal I/Xba I fragments replaced the wild type region. Pst I/Bsp119 I (Asu II) fragments replaced the wild type sequence of GAL/GR525C.
Cell culture, transient transfection, and reporter analysis
Triplicate samples of CV-1 or U2OS cells were transiently transfected with FuGENE 6 reagent (Roche Molecular Biochemicals, Indianapolis, IN) in 24-well plates with either GRE-regulated luciferase reporter plasmid or the GAL-regulated reporter plasmid, pFRLuc (Stratagene; for two-hybrid assays), plus other plasmids (total DNA = 300 ng/well), induced with steroid for 20 hr, and assayed for Luciferase activity as described (26; 27). For gene repression, cells were treated with steroid for 16 hr ± PMA for the last 2 hr in serum-free media as this treatment gives greater fold repression than with 2 hr of steroid ± PMA (27). The maximal induced activity (Amax) was obtained with saturating concentrations of agonist steroid, which was either ≥ 100-fold higher than the EC50, or 10 μM, whichever was lower. In all cases, the data were normalized for Renilla TS activity. In two-hybrid assays, the Amax values are normalized to that of one condition. For dose-response curves, the data are expressed as a percentage of the maximal response and then plotted to determine the EC50 and percent partial agonist activity. In gene repression, the maximal response is set as the activity without agonist steroid and the basal activity is the lower plateau activity seen with saturating concentrations of agonist steroid. For gene induction, the basal activity is that without steroid and maximal activity is that produced by saturating agonist steroid concentrations. The fold induction is (induced value)/(basal activity). Fold repression is similarly calculated as (basal activity)/(repressed value). The percent partial agonist activity of each antisteroid was calculated by expressing the activity of a saturating concentration of antagonist (1 or 10 μM) as percent of maximal activity of a saturating concentration of agonist (see above) under the same conditions. For dose-response curves, each point is the average of triplicate samples ± S.D. One curve of average points yields one value of EC50 (the concentration of agonist required for 50% of maximal response) via curve-fitting programs (see “Statistical analysis” below). For bar graphs giving average values of Amax, EC50, and percent partial agonist activity, the average of n replicates (each in triplicate but considered, statistically, as one observation) was plotted ± S.E.M. (n observations) unless otherwise noted.
Steroid binding and Scatchard assays
Cytosols from Cos-7 cells that had been transiently transfected with 5 μg/100-mm dish of GR plasmids were processed and analyzed for the binding of 50 nM [3H]Dex ± 100-fold excess of non-radioactive Dex in the presence of 20 mM sodium molybdate as described previously (28). For the competition assays, the only changes were to use increasing concentrations of non-radioactive Dex or DAC at 22 °C for 16 hr. For Scatchard assays, 5–6 concentrations of [3H]Dex (≤50 nM) ± 100-fold excess of non-radioactive Dex in the presence of 20 mM sodium molybdate was employed.
Total RNA extraction and quantitative real time-PCR (qRT-PCR)
U2OS cells (100,000 cells/well in 6-well plates) were treated as described for the analysis of induction of ladinin 1 (27) or the repression of collagenase 3 (29). The relative levels of target mRNAs were quantitated using SyberGreen and the ABI 7900HT real-time PCR system for ladinin 1. Collagenase 3 and glyceraldehyde-3-phosphate dehydrogenase (primer from ABI, 4310884E) were quantitated by Taqman.
Western blotting
Western blots were prepared, probed with rabbit anti-GR antibodies (PA1-511A and PA1-516, ABR), rabbit VP16 polyclonal antibody (Santa Cruz), or mouse GAL4 DBD monoclonal antibody (Santa Cruz), and visualized by ECL detection reagents as described by the manufacturer (Amersham Biosciences).
Statistical analysis
Unless otherwise noted, the values of n independent experiments, performed in triplicate, were analyzed for statistical significance by the two-tailed Student’s t test using InStat 2.03 (GraphPad Software, San Diego, CA). In every case, each average of triplicates was treated as one value of the n experiments. Paired t-test is often used when n=3. When the difference between the SDs of two populations was significantly different, the Mann-Whitney or Alternate Welch t test was used. A nonparametric test was used if the distribution of values was non-Gaussian. Best-fit curves (R2 almost always ≥ 0.95) following Michaelis-Menten kinetics were obtained for the dose-response experiments with KaleidaGraph (Synergy Software, Reading, PA).
Results
Effect of selected GR LBD mutations on steroid binding affinity
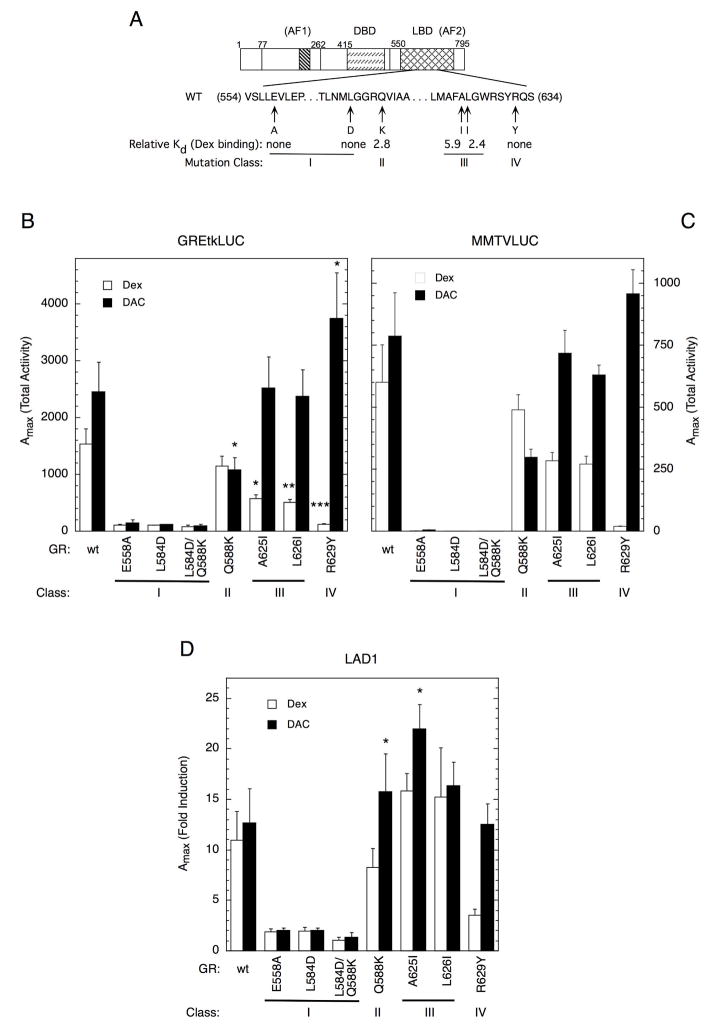
To probe the binding and activation properties of DAC and Dex, we performed detail structural comparisons between the DAC- and Dex-bound GR x-ray structures (2; 19). Superposition of these two structures reveals several major structural differences among the amino acids surrounding the different A-ring substitutes (Figure 1A, a phenylpyrazole group in DAC vs. a C3-ketone in Dex). Guided by these analyses, we prepared seven mutations in the GR pocket residues that are near the different substitutes of DAC and Dex (Fig. 2A). The rationale for these mutations is as follows. The first three mutations (E558A, L584D, and L584D/Q588K) are designed to change the charge properties of the ligand-binding pocket. The Q588K mutation is intended to mimic the wild type receptor as the mutated lysine can serve as a hydrogen bond donor, like the glutamine side chain. The A625I and L626I mutations are calculated to change the shape but not the charge properties of the pocket. The R629Y mutation is planned to disrupt the binding of Dex but not DAC to GR. In the GR/DAC structure, R629 forms packing interactions with DAC whereas in the GR/DEX structure R629 forms hydrogen bonds with the C-3 ketone of the steroid. The R629Y mutation is expected to disrupt the H-bond interaction with Dex but not the packing interactions with DAC.
Fig. 2.
Effects of GR mutations on level of gene induction. A. Schematic of position of point mutations in LBD with numbers above the drawing indicating the amino acids of the LBD with AF2 (cross hatched), AF1 (core is striped), and DBD (dashes) domains. The affinity of Dex binding (relative to wt GR) was determined by Scatchard assays (see Materials and Methods). B-D. The Amax, or total amounts of reporter gene induction (GREtkLUC in B, MMTVLUC in C), by EtOH ± saturating concentrations of Dex or DAC was determined in CV-1 cells as described in Materials and Methods. For the endogenous gene LAD1, the fold induction by 1 μM Dex or DAC in U2OS cells was determined by qRT-PCR as presented in Materials and Methods. The average values ± S.E.M. from 7 (GREtkLUC; 2 for Class I mutants), 2 (MMTVLUC) and 4 (LAD1) independent experiments are plotted. ND = not determined. Relative to wild type GR, * P < 0.050, ** P = 0.0070, *** P = 0.0006.
Ligand binding assays reveal that these seven mutations fall into four classes. The Class I (E558A, L584D, and L584D/Q588K) mutants no longer bind Dex, indicating that changing the hydrophobicity of the GR pocket has a deadly detrimental effect on ligand binding (Fig. 2A and Table 1). The Class II (Q588K, where Q588 moves dramatically upon DAC binding) and Class III (A625I and L626I, which involve small alterations) mutants bind Dex with only ~2.6- (Q588K and L626I) or 5.9-fold (A625I) reduced affinity. The Class IV mutant (R629Y) also displays negligible Dex binding, due to the disruption of the hydrogen bonds to the C-3 ketone of Dex, but it may retain DAC binding as R629Y is expected to form the same packing interactions with the phenylpyrazole ring of DAC. Interestingly, the binding affinity of DAC is largely unaffected in the mutants that still bind Dex, as determined by cell-free competition assays with [3H]Dex (Table 1). The different binding affinity in the GR mutants does not result from changes in protein abundance as Western blots show that each mutant GR is expressed at the same level (data not shown). To further assess changes in DAC- and Dex-mediated properties of the mutants, functional cell-based assays were employed below.
Table 1.
Affinity of mutant GRs for Dex and DAC
| DEX binding Kd | Relative Kd from Competition binding | ||||
|---|---|---|---|---|---|
| Mutation | Class | Scatchard (nM) | Relative to wt | Dex | DAC |
| Wt | 1.93 ± 0.56 | 1 | 1 | 0.32 ± 0.07 | |
| E558A | I | NSB | |||
| L584D | I | NSB | |||
| L584D/Q588K | I | NSB | |||
| Q588K | II | 5.95 ± 1.64 | 2.78 ± 1.05 | 1.67 ± 0.49 | 0.35 ± 0.06 |
| A625I | III | 13.64 ± 2.89 | 5.86 ± 1.07 | 3.24 ± 1.15 | 0.35 ± 0.03 |
| L626I | III | 6.18 ± 2.04 | 2.37 ± 0.30 | 2.14 ± 0.62 | 0.32 ± 0.05 |
| R629Y | IV | NSB | |||
Scatchard assay values were determined from binding assays with [3H]Dex, while competition binding assays involved displacement of [3H]Dex binding by non-labeled Dex or DAC, as described in Materials and Methods. Scatchard assay values are absolute. All other values are relative to Dex binding. Errors are S.E.M. of n = 5 (Scatchard plots for Dex binding Kd) and n = 4 (Competition binding) independent experiments. NSB, no significant binding detected above background. Blanks indicate no measurement.
Effect of selected GR LBD mutations on Amax of GR-regulated gene induction
Three important properties of GR-mediated gene transcription (Amax, EC50, and the percent partial agonist activity of antiglucocorticoids) (30; 31) were measured for induction of two exogenous genes (GREtkLUC with a simple tandem repeat of GREs from the tyrosine aminotransferase gene (32) and MMTVLuc with a more complex enhancer from the mouse mammary tumor virus (MMTV) (33)). Initial experiments with varying amounts of receptor plasmid revealed that the 0.5 ng of each mutant receptor plasmid provides about 70% of the activity seen with up to 100-fold higher levels of receptor plasmid (data not shown). Thus, we conclude that our below described assays with 0.5 ng of receptor plasmid involve approximately equal, and non-limiting, amounts of each mutant receptor. Figs. 2B&C show that maximal activity (Amax) of Class I mutants with saturating concentrations (1–10 μM) of agonist steroid (Dex or DAC) in CV-1 cells is barely above basal activity with both transfected reporters (= 5.6% for GREtkLUC), consistent with their lack of steroid binding activity (Table 1). The Amax of the Class II mutant (Q588K) appears to be reduced more for DAC induction (by ~55%) than for Dex induction (~15% reduction). The Class III mutants (A625I and L626I) retain ≤ 50% of the wild type activity when bound by Dex. In contrast, the same mutant receptors maintain >80% of the activity of wild type GR when bound by DAC. Thus, Dex shows much less activity than DAC and is no longer a full agonist with Class III mutants. This phenomenon is especially dramatic with the Class IV mutant, R629Y, where Dex has negligible activity (consistent with its undetectable binding of Dex) while DAC can be more active than with the wt GR. Therefore, the ability of the mutant receptors to induce gene expression is now very sensitive to the structure of “agonist” steroids.
The Amax for DAC induction of the endogenous tyrosine aminotransferase gene in rat hepatoma tissue culture (HTC) cells is about 10% greater than for Dex induction (8). To examine the effect of the above mutations on the induction of an endogenous gene in greater detail, we selected the GR-responsive ladinin 1 (LAD1) gene in human U2OS cells (27; 34). The Amax for DAC induction of LAD1, as measured by quantitative RT-PCR, is again slightly higher than that for Dex (Fig. 2D). Likewise, the changes in Amax for induction of LAD1 by each mutant receptor relative to wild type GR show similarities to those seen for the exogenous reporters: negligible activity with the Class I mutations and much greater activity of DAC than Dex with R629Y. The higher activity of Class II and III mutants with Dex and DAC for LAD1 than the transfected reporters (Figs. 2B&C) is notable.
Effect of selected GR LBD mutations on EC50 of GR-regulated gene induction
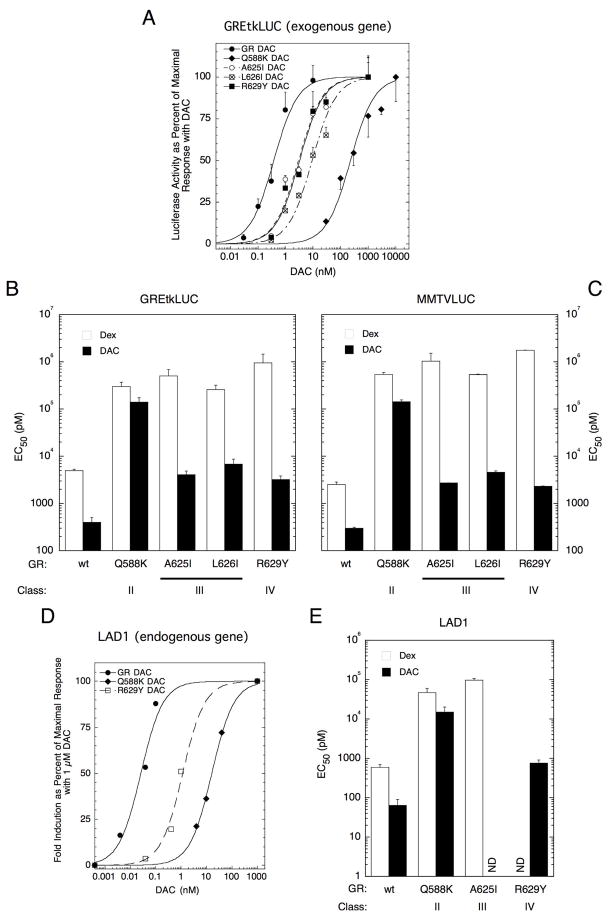
Dose-response experiments were performed with the above exogenous genes to confirm the affinity of each steroid for those receptors that bound steroid (Table 1) and afforded appreciable induced gene expression (Figs. 2B&C). Representative results are shown in Fig. 3A. In each experiment, the EC50 for gene induction was determined as described in Material and Methods and the average values plotted in Figs. 3B&C. Surprisingly, the EC50 for Dex induction of both genes induced by each mutant GR is increased 50–700 times that of the wild type GR (note logarithmic scale of y-axis), or 16-to 91-fold more than predicted from the affinity of Dex for each GR. The discrepancies were largest (66- to 91-fold) for induction of MMTVLuc. Similar, unexpectedly large right-shifts in the dose-response curves are observed for induction by DAC, especially the ≥ 300-fold difference with the Q588K mutant, despite marginal changes in steroid affinity. Selected mutants with significant levels of induction were chosen for studies with the endogenous LAD1 gene and displayed similar results (Figs. 3D&E). As noted above, the amount of biologically active receptor present is about the same for each mutant receptor. Thus, a change other than steroid binding affinity or amount of receptor protein must be responsible for these large increases in EC50.
Fig. 3.
Dose-response curves for mutant GR induction of reporter genes. A. Dose-response curves for DAC induction of exogenous GREtkLUC in CV-1 cells. For each receptor, the data of one experiment are plotted as percent of the maximal level of induction by DAC, as described in Materials and Methods. B-C. Change in EC50 with receptor mutation for induction of exogenous reporter genes by Dex or DAC. D. Dose-response curves for DAC induction of endogenous LAD1 gene in U2OS cells. A representative experiment is shown. E. Change in EC50 with receptor mutation for induction of LAD1 gene by Dex or DAC. Note that the average EC50 values (bar graphs) ± S.E.M. are plotted on a logarithmic y-axis for n = 5–8 (GREtkLUC; relative to wt GR, P = 0.0016 for all Dex-bound receptors and P = 0.0025 for all DAC-bound receptors), 2 (MMTVLUC) and 4 (LAD1; relative to wt GR, P = 0.029 for all Dex- and DAC-bound receptors) independent experiments. ND = not determined.
There is no established relationship between Amax and EC50. It is often anticipated that the Amax will decrease as the EC50 increases and this is seen when comparing the properties of Dex induction via the wild type vs. Class II and III mutants (Figs. 2B–D vs. 3B,C&E). However, the present data provide several exceptions to this expected behavior. The Amax values for Dex induction of the LAD1 gene by these mutants are not significantly different despite 75- to 190-fold increases in EC50 (Figs. 2D vs. 3E; note log scale of Y-axis in Fig. 3E). Similarly, the Amax for DAC induction of GREtkLUC or MMTVLUC reporters is unchanged (Class III) or increases (Class IV) while the EC50 values for gene induction increase by 8- to 19-fold (Figs. 2B&C vs. 3B&C). With DAC and the endogenous LAD1 gene, the differences are even more pronounced: less than 2-fold rises in Amax values are associated with 25- and 300-fold increases in EC50 (Fig. 3E; note log scale of Y-axis in Fig. 3E). These data clearly show that the Amax and EC50 can be independently modified and suggest that separate pathways or factors are involved.
Effect of selected GR LBD mutations on percent partial agonist activity of antiglucocorticoids in GR-regulated gene induction
We have reported that an increase in EC50 is associated with a decrease in percent partial agonist activity of antiglucocorticoids and vice versa (26; 27; 30; 35–38). This is an empirical observation with no theoretical explanation so far. With the present mutant GRs, an increase in EC50 for Dex induction is always coupled with a decrease in the percent partial agonist activity of the antiglucocorticoid Dex-Mes (Table 2). Thus, the data from our mutants further support a linkage of these two parameters.
Table 2.
Correlation of decreased percent partial agonist activity of Dex-Mes and increased EC50 for mutant GRs with different reporter genes
| Mutation | Class | GREtkLUC | MMTVLuc | LAD1 | LAD1 (re DAC) | ||||
|---|---|---|---|---|---|---|---|---|---|
| % Activ | Rel EC50 | % Activ | Rel EC50 | % Activ | Rel EC50 | % Activ Rel EC50 | |||
| Wt | 32.8 ± 3.4 | 1 | 36.1 ± 0.1 | 1 | 29 ± 10 | 1 | 25 ± 6 | 1 | |
| E558A | I | NMF | NMF | ||||||
| L584D | I | NMF | NMF | ||||||
| L584D/Q588K | I | NMF | NMF | ||||||
| Q588K | II | 2.5 ± 0.7 | 57 | 1.5 ± 0.1 | 215 | 17.2 ± 10.9 | 75 | 6.1 ± 1.5 | 319 |
| A625I | III | 5.6 ± 2.7 | 95 | 1.6 ± 0.0 | 391 | 10.2 ± 6.2 | 191 | ||
| L626I | III | 5.2 ± 1.9 | 50 | 1.9 ± 0.1 | 215 | ||||
| R629Y | IV | NMF | −0.6 ± 0.1 | 708 | 6.9 ± 4.2 | 26 | |||
The percent partial agonist activity of Dex-Mes, expressed as the percent of maximal activity with Dex for each gene, was determined as described in Materials and Methods. To obtain the percent agonist activity of Dex-Mes with R629Y (which has minimal binding of Dex) and the LAD1 gene, the activity of Dex-Mes is expressed as the percent of maximal activity with DAC and listed in the column labeled “LAD1 (re DAC)”. NMF, no meaningful figure. Blanks indicate no measurements. Averages are ± S.E.M. of n = 4–8 (GREtkLUC), n = 2 (MMTVLuc), and n = 4 (LAD1) independent experiments. For ease of comparison, the average EC50 values from Fig. 2 (without error bars) are listed.
Effect of selected GR LBD mutations on GR-regulated gene repression
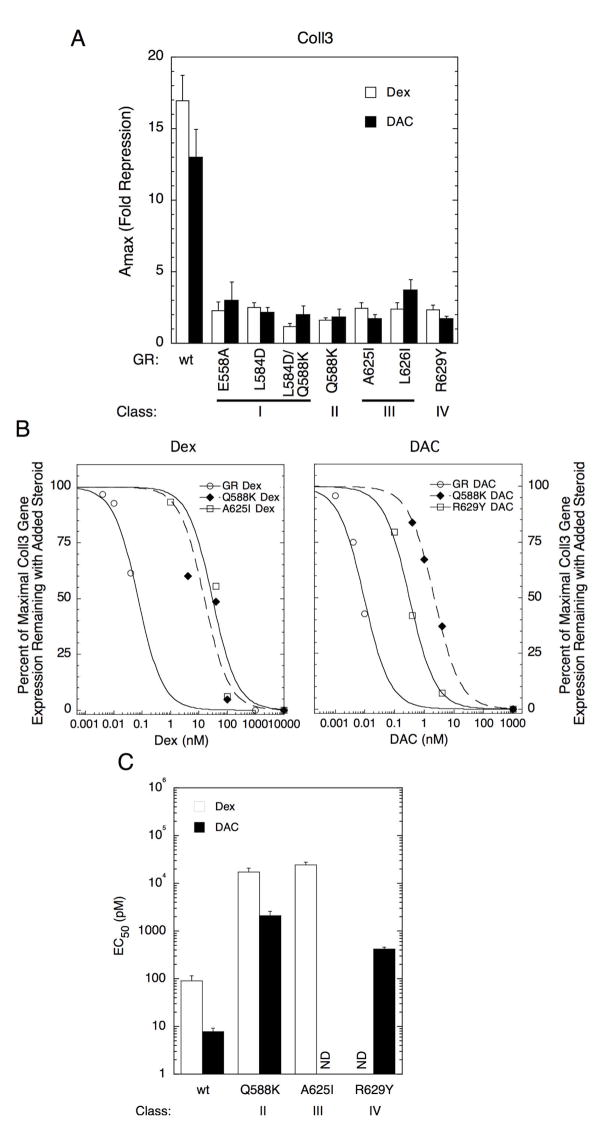
We next examined the ability of our mutants to repress the induction of an endogenous gene, collagenase 3 (Coll3), again in U2OS cells (27; 39). Most reported GR mutations reduce the Amax of induction much more than that of repression (12–16; 40; 41). Therefore, the ability of each mutant that bound steroid (i.e., Classes II-IV) to selectively suppress Amax for repression of the endogenous Coll3 gene (Fig. 4A) with little effect on Amax for induction of the endogenous LAD1 gene (Fig. 2D) in the same cells was unexpected. The Amax values of the various mutants (with significant steroid binding activity) for LAD1 induction by Dex or DAC are between 73 and 180% of that with wild type GR but only 1–23% of wild type GR for Coll3 repression. This behavior is opposite to the commonly observed response and suggests a new approach for preferential inactivation of GR-mediated gene repression. The dose-response curves for GR-mediated repression of the Coll3 gene by Dex and DAC with the same mutants as used with LAD1 in Fig. 3E were then determined by qRT-PCR. Representative experiments are shown in Fig. 4B. Interestingly, each mutation shifts the dose-response curve for Coll3 gene repression to higher steroid concentrations (Fig. 4C) by about the same amount as for induction of the LAD1 gene (Fig. 3E). Therefore, the effects of these mutations on endogenous gene induction and repression are the same for the determinants of the EC50 and different for the Amax. This is additional evidence that the pathways controlling the EC50 and Amax can be separated.
Fig. 4.
Repression of the endogenous Coll3 gene by mutant GRs in U2OS cells. A. Effect of mutations on fold repression of Coll3 mRNA by Dex and DAC. Data represent average of 4 independent experiments ± S.E.M. (relative to wt GR, P <0.05 for all except DAC with E558A and L626I). B. Dose-response curves for Dex or DAC repression of endogenous Coll3 by indicated mutant GRs. The Coll3 mRNA obtained with increasing steroid, expressed as percent of Coll3 mRNA remaining with a saturating concentration of steroid, is plotted for each mutant receptor in one representative experiment. C. Change in EC50 for repression of Coll3 mRNA expression with receptor mutation in presence of Dex or DAC. See Materials and Methods for experimental details. The average values ± S.E.M. of 4 independent experiments are plotted on a logarithmic y-axis (relative to wt GR, P <0.05 for all). ND = not determined.
Interaction of coactivators and corepressors with mutant GRs
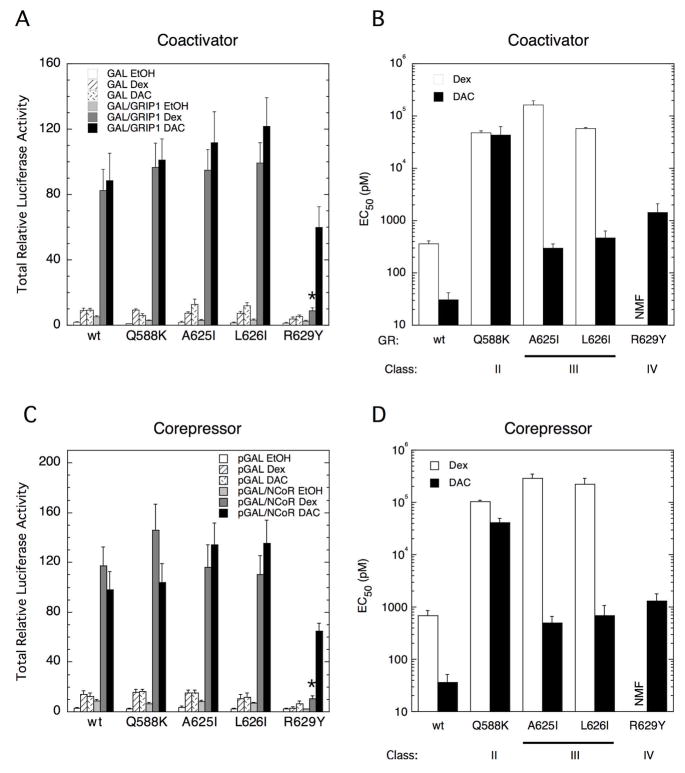
Changes in the ratio of receptor-associated coactivators vs. corepressors is one mechanism by which the EC50 for gene induction can be raised or lowered (26; 30; 31; 35–37; 42; 43). To determine whether the EC50 differences observed in Figs. 2–4 are due to alterations in coactivator and/or corepressor binding affinity to the mutant GRs, we used mammalian two-hybrid assays of interactions between full-length GR fused to the VP16 transactivation domain and chimeras of the GAL4 DBD with either full-length p160 coactivator TIF2 or the receptor interaction domain of the corepressor NCoR. In this assay, the total amount of product (Amax) reflects the strength or affinity of cofactor binding to GR. TIF2 is the human homolog of, is 94% identical to, and is biologically indistinguishable from, the mouse protein GRIP1. Therefore, although we use different constructs of each (designated in the figure legends), we will refer to both proteins as TIF2. Western blots indicate that each VP16/GR construct is expressed at similar levels (data not shown). The wt GR and most of the mutant GRs yield almost the same Amax with GAL/TIF2 chimera in the presence of excess Dex or DAC (Fig. 5A). The exception is R629Y, which displays no Dex-induced interaction with coactivator because this mutant has no appreciable Dex binding. Thus, these mutations do not significantly alter the amount or affinity of receptor-coactivator interactions at saturating concentrations of ligand. However, the EC50 of steroid-induced binding of receptor-agonist complexes with TIF2 is increased by at least a factor of 10 with each mutation (Fig. 5B), which is much more than expected from the changes in steroid binding affinity (Table 1). Also, the amount of GR-antagonist (RU486) binding to coactivator relative to that for DAC-bound complexes is reduced by ≥ 85% (Table 3). Therefore, the changes in EC50 in Fig. 5B and in percent partial agonist activity reflect different properties of the mutant GRs than their affinity for coactivators.
Fig. 5.
Two-hybrid assays for VP16/GR chimeras with GAL/coactivator or GAL/corepressor in CV-1 cells. A&C. Total Luciferase activity induced from FRLuc reporter by GAL ± GRIP1 (A) or NCoR-RID (C) with indicated mutant VP16/GRs plus EtOH, 1 μM Dex, or 0.1 μM DAC was determined, normalized to the value for GAL/EtOH with the Q588K mutant, and plotted as described in Materials and Methods. *P < 0.05 for mutant vs. wt GR with GAL/GRIP1 or GAL/NCoR-RID and steroid. B&D. EC50 values for Dex or DAC induction of FRLuc reporter by GAL/GRIP1 (B) or NCoR-RID (D) with indicated mutant VP16/GRs was determined and plotted as described in Materials and Methods. Error bars for all graphs are S.E.M of 5 independent experiments (4 experiments for pGAL controls for NCoR-RID; 3 experiments for VP16/GRR629Y). NMF = no meaningful figure because the fold induction approached the usual error bars in the data points, thereby precluding a meaningful dose-response curve. In all cases, P = 0.029 for mutant vs. wt GR with GAL/GRIP1 or GAL/NCoR-RID and steroid.
Table 3.
Interaction of RU486-bound GRs with cofactors in two-hybrid assays as percent of maximal activity with DAC (= 100)
| Mutation | Class | GAL/TIF2 | GAL/NCoR-RID |
|---|---|---|---|
| Wt | 66 ± 4 | 590 ± 16 | |
| Q588K | II | 1.9 ± 0.4 | 32 ± 2 |
| A625I | III | 6.9 ± 0.6 | 63 ± 7 |
| L626I | III | 9.2 ± 2.5 | 64 ± 3 |
| R629Y | IV | 0.6 ± 0.2 | 7.1 ± 1.8 |
The percent partial agonist activity of RU486, expressed as the percent of maximal activity with DAC, in the two-hybrid assays with VP16/GR chimeras and GAL/TIF2 (= full-length GRIP) or GAL/NCoR-RID was determined as described in Materials and Methods. The percent partial agonist activity of RU486 with GAL/NCoR-RID and wt GR is >100 because interaction of DAC-bound GR is weaker than with RU486-bound GR. Averages are ± S.E.M. of n = 3 independent experiments.
In two-hybrid interactions of GR with the corepressor NCoR, there is also very little change in the Amax with any mutant (Fig. 5C), except for Dex with the R629Y mutant that minimally binds Dex. This similarity in the total luciferase activity produced with Dex or DAC in the interaction of each mutant receptor, except R629Y, with either coactivator (Fig. 5A) or corepressor (Fig. 5C) suggests that the capacity of both receptor-Dex and receptor-DAC complexes to interact with these cofactors, which is dictated by GR-cofactor affinity, is independent of most of the mutations examined. At the same time, there are dramatic and steroid-specific increases in the EC50 for steroid-induced reporter gene induction by the corepressor/mutant receptor interactions (Fig. 5D) that are nearly identical to those observed with coactivator (Fig. 5B). Each receptor mutation also eliminates most of the ability of the antagonist RU486 to promote a productive interaction between GR and corepressor, just as is seen for GR-coactivator interactions (Table 3). Collectively, these results suggest that the mutation-induced changes in the EC50 of GR-mediated induction (Figs. 3B, C, E, and 4D) are independent of coactivator or corepressor affinity and, instead, that some other property of receptor-cofactor interaction is involved.
Steroid-specific inactivation of AF2 domain by mutations in truncated GRs
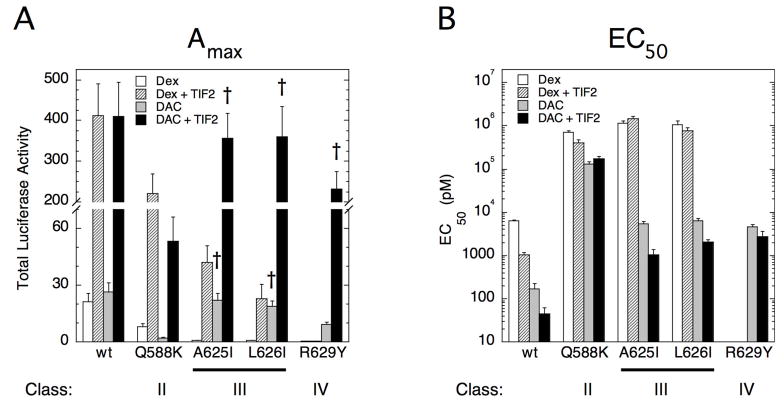
The above observations that mutations in GR affect GR-steroid complexes to separately alter their Amax and EC50 for gene induction is reminiscent of the ability of coactivators and corepressors to independently influence the Amax and EC50 of gene expression (23; 25; 30; 31; 36; 38). Coactivators bind more extensively to agonist-bound GRs than do corepressors in a competitive equilibrium manner (26; 42), suggesting that coactivator functions may also have a more prominent role in the current responses. This possibility was examined by looking at the induction properties of the GR LBD (amino acids 525 to the C-terminus). Because the GR LBD does not associate with DNA, it was fused to the GAL4 DNA binding domain (DBD) to give GAL/GR525C, which binds to and induces gene transactivation from a GAL4 regulated gene such as FRLuc. The GAL/GR525C construct has two especially useful properties for the present study. First, it contains only one activation function, AF2, so that significant effects of added TIF2 can be unambiguously assigned. Second, it is known that TIF2 causes a larger increase in Amax for GAL/GR525C than for full-length GR with activation functions AF1 and AF2 (37). Therefore, if our mutations are affecting coactivator function as opposed to binding, we predict that the changes in Amax with and without exogenous coactivator will be magnified when these mutations are in the context of the GAL/GR525C.
All of the mutant GAL/GR525C plasmids expressed equal levels of protein (data not shown). With Dex, most mutants display marginal activity (Amax) while that for the Class II mutant Q588K is reduced by ~65% (Fig. 6A; note split Y-axis and that † = no significant difference). For DAC-bound receptors, only the Q588K mutation causes a >65% decrease in Amax (Fig. 6A). Thus the AF2 domain, the only activation domain in the GAL/GR525C constructs and a domain known to interact with coactivators, has been largely inactivated by these mutations in Dex-bound receptors. In contrast, much more AF2 transactivation activity remains with all of the DAC-bound mutants except for Q588K. When TIF2 is cotransfected, the Amax increases for every receptor-steroid complex other than Dex-bound R629Y. However, those receptor-steroid combinations with low Amax in the absence of TIF2 (Dex with Class III [A625I and L626I] and Class IV [R629Y] mutants and DAC with the Class II mutant [Q588K]) still display a dramatically lower Amax compared to wild type controls with added TIF2 (Fig. 6A). This argues that the Class III mutations greatly reduce the ability of the AF2 domain of Dex-, but not DAC-, bound receptors to increase the Amax in response to both the endogenous coactivators in CV-1 cells and to the elevated levels of exogenous TIF2. The Class II and IV mutations also reduce this response of the AF2 domain to coactivators in DAC-and Dex-bound receptors respectively but are less inhibitory.
Fig. 6.
Transcriptional properties of GAL/GR525C mutants ± coactivator TIF2 in CV-1 cells. Total Luciferase activity (Amax) (A) and EC50 (B) for induction by GAL/GR525C alone from the FRLuc reporter with Dex or DAC ± TIF2 was determined and plotted as described in Materials and Methods. Error bars are S.E.M of 4 (A) or 5 (B) independent experiments. For all cases, † = not significant (P > 0.05) for mutant vs. wt GR ± GRIP1 and steroid. No symbol above bar indicates P ≤ 0.029.
As expected (37), cotransfected TIF2 also reduces the EC50 (increases the potency) both of Dex- and DAC-bound wild type GR525C. A reduction similar to that for the wild type chimera is seen for DAC-bound A625I and L626I mutant receptors; but, TIF2 is much less effective with the Dex-bound mutant receptors (Fig. 6B; note logarithmic Y-axis). The ability of TIF2 to increase the percent partial agonist activity of an antisteroid (37) is likewise lost with the mutant receptors of GAL/GR525C (Table 4). These results suggest that the various mutations in GR have not caused the loss of TIF2 binding, because TIF2 still produces increases in Amax. However, the effectiveness of TIF2 activity has been diminished for most of the Dex-bound mutants and for DAC-bound Class II and IV mutants. This is seen by the inability of added TIF2 to restore the Amax, EC50, and the percent partial agonist activity values of the affected mutants to wild type GR values. Importantly, this ineffectiveness of TIF2 is determined by the steroid bound to the mutant GR and not the mutation itself (e.g., compare responses of the A625I mutant receptor with Dex or Dex-Mes vs. DAC).
Table 4.
Percent partial agonist activity of Dex-Mes with mutant GAL/GR525C or full-length GR ± TIF2
| GAL/GR525C | GR | ||||
|---|---|---|---|---|---|
| Mutation | Class | w/o TIF2 | with TIF2 | w/o TIF2 | with TIF2 |
| Wt | 27 ± 1 | 57 ± 2 | 21 ± 3 | 45 ± 4 | |
| Q588K | II | 2.0 ± 0.9 | 1.9 ± 0.1 | 0.6 ± 0.7 | 9.9 ± 0.6 |
| A625I | III | NMF | 1.8 ± 0.3 | 0.3 ± 0.9 | 8.6 ± 1.0 |
| L626I | III | NMF | 2.9 ± 0.9 | −0.2 ± 0.5 | 7.6 ± 2.2 |
| R629Y | IV | NMF | NMF | −9.7 ± 2.9 | −0.4 ± 0.1 |
The percent partial agonist activity of Dex-Mes, expressed as the percent of maximal activity with Dex, in assays with GAL/GR525C chimera and FRLuc, or full-length GR (GR) and GREtkLUC, ± the coactivator TIF2 was determined as described in Materials and Methods. NMF, no meaningful figure. Averages are ± S.E.M. of n = 4–5 independent experiments.
Steroid-specific inactivation of AF2 domain by mutations in full-length GRs
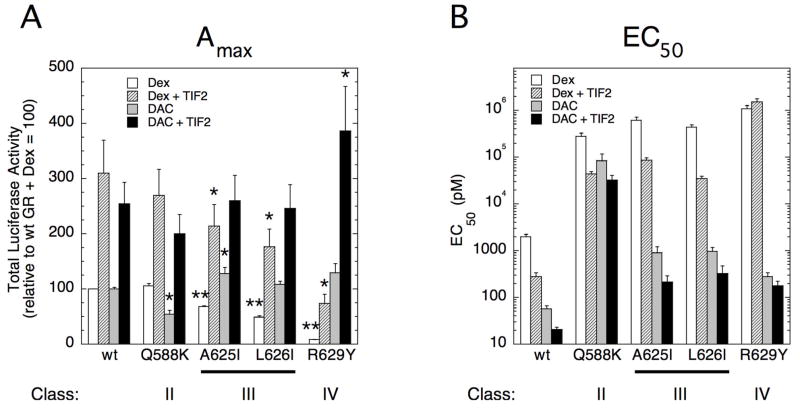
In order to determine whether the various mutations can also disable the AF2 domain in the context of full-length GRs, we next examined the effect of added coactivator on the properties of full-length receptors containing both activation functions: the C-terminal AF2 domain of GAL/GR525C and the N-terminal AF1 domain. We previously documented reduced responses of full-length GR vs. GAL/GR525C with added coactivator on the Amax, but approximately equal effects on the EC50, of induction by receptors containing the AF1 domain (37), which can interact directly with TIF2 (43). Therefore, we predicted that the differences ± TIF2 with full-length GR mutants would be less for the Amax than those with the GAL/GR525 constructs and more similar for the changes in EC50.
As shown in Fig. 7, the predicted behavior of full-length GRs is precisely what is observed. In the absence of exogenous TIF2, the differences between the Amaxs of Dex- and DAC-bound wild type and mutant GRs is much less than with the GAL/GR525 constructs (Fig. 6A), except for the non-Dex binding R629Y, due to the presence of the AF1 domain. Added TIF2 elevates the Amax in all cases; however, those receptor-steroid combinations with a defective AF2 domain in the context of GAL/GR525 (Classes III and IV with Dex and Class II with DAC) display lower Amax values (Figs. 7A vs. 6A). For the EC50 values of gene induction, TIF2 lowers the EC50 of almost all mutant GRs, but wild type GR values are more closely approximated by receptor-steroid combinations with a functional AF2 domain in this assay (e.g, DAC-bound Class III and IV mutants). It should be noted that the ability of added TIF2 to almost completely restore wild type properties to the mutant GRs for Amax but not for EC50 is yet another example of the ability of these mutations to differentially affect these two important properties of transactivation. Furthermore, the efficiency of TIF2 in the EC50 assay is sensitive to steroid structure (e.g., values for A625I are much closer to wild type GR when DAC is the ligand).
Fig. 7.
Transcriptional properties of full-length GR mutants ± coactivator TIF2 in CV-1 cells. Total Luciferase activity (Amax) (A) and EC50 (B) for induction of GREtkLUC reporter by Dex or DAC ± TIF2 was determined and plotted as described in Materials and Methods. Error bars are S.E.M of 5 independent experiments. In all cases either with or without TIF2, *P < 0.05, **P < 0.0005 for mutant vs. wt GR in (A). In (B), P = 0.0079 for all comparisons of mutant vs. wt GR.
Finally, because the AF2 domain is largely responsible for the percent partial agonist activity of Dex-Mes (37), Dex-Mes displays the predicted minimal percent partial agonist activity with the mutant full-length GRs containing a partially (Class II) or more completely (Classes III and IV) inactivated AF2, even in the presence of a functional AF1 domain (Table 4). Furthermore, added TIF2 only minimally increases the partial agonist activity of Dex-Mes with the mutant full-length GRs to a value that is dramatically lower than that for the wild type GR with TIF2. Collectively, these results show that selected mutations in the GR LBD can unequally affect several properties of GR induction (EC50 and Amax of agonist steroids and the percent partial agonist activity of antiglucocorticoids) in a manner that introduces previously unobserved differences among potent agonist glucocorticoids. In several cases, the mechanism of this new, steroid-selective behavior involves a steroid-specific inactivation of the AF2 domain and its ability to communicate with coactivators. Several of these mutations are also able to preferentially inhibit the repression vs. induction of responsive genes, although the mechanism of this response has not yet been elucidated.
Discussion
DAC is 25% larger than Dex (44) but displays a 10- to 40-fold lower EC50 (or potency) for GR-mediated gene induction and repression while producing the same Amax (= maximal activity or efficacy). The recent x-ray structure of DAC bound to the GR LBD (19) reveals significant reorganization of several residues to accommodate the bulky steroid but an otherwise similar binding in the LBD. Here we report that mutations of some amino acids near the A-ring of the bound steroid eliminate steroid binding. However, other mutations unexpectedly have little effect on the affinity of either Dex or DAC compared to the large changes in Amax and EC50 of agonists and in percent partial agonist activity of antiglucocorticoids. Interestingly, the effects of these latter mutations depend upon steroid structure and the type response (induction or repression) to give a spectrum of activities that has not been previously described.
Class I mutations (E558A, L584D, and L584D/A588K) eliminate Dex binding as predicted. Because DAC displays no biological activity with these mutants, we presume that DAC binding is also destroyed. In contrast, Class II and III mutations produce relatively small reductions in Dex binding affinity (≤ 6-fold) and activity (Amax) and almost negligible decreases in DAC binding or Amax. Surprisingly, the Class IV mutation (R629Y) affords barely detectable Dex binding in Scatchard assays and Dex activity (Amax) in bioassays but a large Amax with DAC in the same bioactivity assays. We conclude that this mutant displays significant binding affinity for DAC but not Dex. This suggests that both the binding affinity and transactivation activity can vary not only with receptor mutation but also with steroid structure. With GREtkLUC reporter, the Amax of DAC with the R629Y mutant is also significantly greater than that with the wild type GR (Fig. 2B). Even more unexpected is the much larger, steroid-dependent effects of Class II-IV mutations on the EC50 (Fig. 3) and percent partial agonist activity (Table 2) of gene induction than on steroid binding affinity. The selectivity is greatest with Q588K and R629Y, both for exogenous and endogenous genes, and thus is insensitive of promoter sequence and DNA architecture. There are few precedents for this behavior (45) and no mechanistic studies of which we are aware.
The mutations cause the EC50s for Dex and DAC to vary in unpredictable ways. The EC50 of DAC is 8–12 fold lower than Dex with the wt GR, 20–90 times lower with R629Y, and 2–6 times higher with Q588K. More remarkable is that Class II–IV mutations reduce the Amax of GR-mediated repression (Fig. 4A) more than that for induction (Fig. 2D), which is opposite to most reports and rarely observed (40) in a manner that can depend upon the cell line (41). This cell-specificity explanation does not appear to be a factor here, though, as the same U2OS cells display preferential inhibition of induction over repression with other GR mutants (12; 15). The variations in EC50 and Amax cannot be accounted for by effects on steroid binding affinity or receptor abundance. Therefore other phenomena must be involved.
The greatly reduced Amaxs for induction of FRLuc by Dex bound to chimeras of GAL4 DBD fused to mutant GR LBDs indicate that Class II, III, and IV mutations disrupt the activity of the AF2 domain, which is the only transactivation domain present (Figs. 2A and 6A). Unexpectedly, this nearly complete inactivation of the AF2 domain depends upon steroid structure. The AF2 activity of mutants binding the bulkier DAC is largely intact for Class III and IV mutations and is dramatically reduced only for the Class II mutant, Q588K. The biological activities of full-length GR (Fig. 7) are consistent with the predicted properties of Dex complexes being without AF2 activity while DAC complexes (except with Q588K) enjoy contributions of both AF1 and AF2 transactivation domains. A particularly clear demonstration is the minimal percent partial agonist activity of Dex-Mes ± TIF2 with both GAL/GR525C and full-length GR mutants (Table 4), which is predicted from our demonstration that the AF2 domain is necessary and sufficient for the expression of Dex-Mes percent partial agonist activity ± TIF2 (46). Thus, we have identified several amino acid mutations in the GR LBD that inactivate the AF2 domain in a manner that depends upon both the structure of the bound ligand and the nature of gene expression (induction vs. repression), thereby imparting major differences in biological activity to various steroid-receptor complexes.
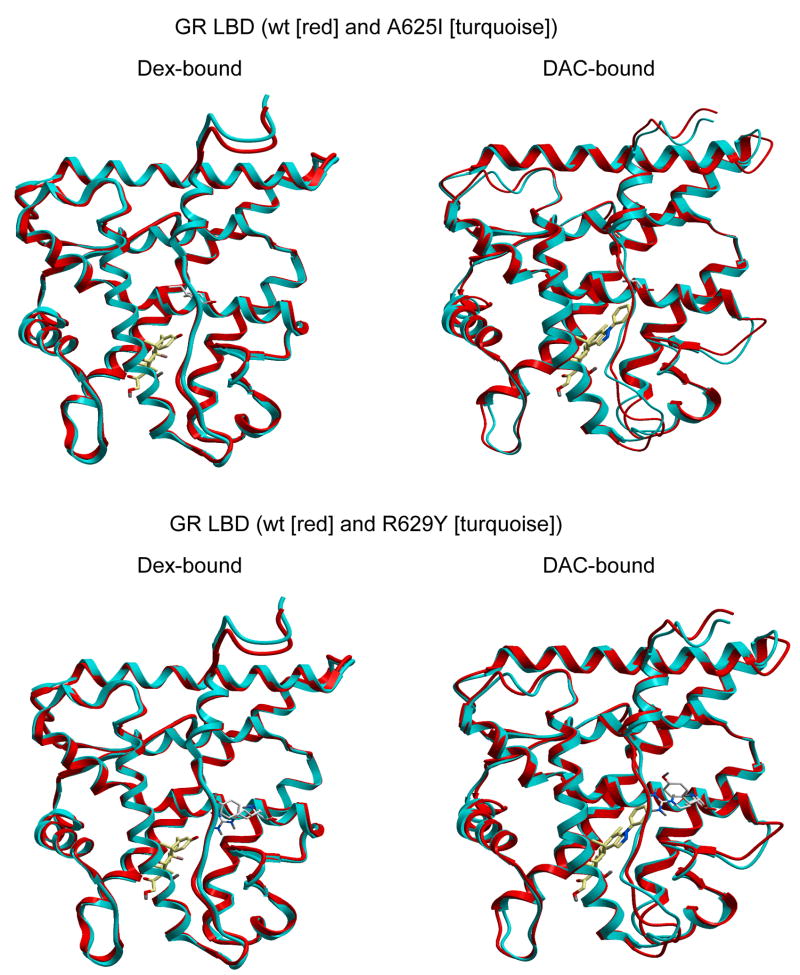
The underlying molecular explanations for the observed mutation-induced changes in AF2 and coactivator activity in both GR-mediated transactivation and repression are not yet clear. Our mutations of residues in the ligand-binding pocket do not appear to alter the binding affinity of coactivators and corepressors (Figs. 5A&B). Computer modeling predicts no dramatic mutation-induced changes in the peptide backbone of the coactivator binding pocket of GR with Dex or DAC (Fig. 8). However, the effects of mutations can be propagated throughout the protein even when the backbone conformation is preserved and there is no clear path of contacts (47; 48). Thus, it is likely that the small conformational changes of Fig. 8 resulting from Class II–IV mutations of Dex-bound receptors are transmitted to the amino acid side-chains at the surface of the LBD and may even be magnified, as seen for GR DBD mutations that are amplified at a distant location in the DBD (49). Similarly, small changes in ligand binding position can yield much larger effects on LBD structure (50). These surface topology changes could affect the nature of protein-induced conformational changes in bound coactivators and thus influence the efficacy of cofactor-protein interactions and eventually the EC50 (38; 51). This proposed mechanism is thus similar to the recent report that phosphorylation of the human estrogen receptor alpha at Ser305, which is just before helix 1, alters the orientation (and presumably activity) of bound coactivator SRC-1 without affecting the affinity of SRC-1 binding (52). This mechanism is in contrast to the ability of Y753 mutations in rat GR to reduce the binding of SRC-1 coactivator and preferentially reduce transactivation as opposed to transrepression (53), which is inhibited in our study (Figs. 4A vs. 2D).
Fig. 8.
Predicted structures of mutant GR LBDs. Computer modeling of Dex and DAC (both in yellow) binding to GR LBD of wt vs. A625I (top) and wt vs. R629Y (bottom) mutants. See Discussion for further details.
The reported ability of coactivators to selectively modify Amax vs. EC50 of agonists, and the percent partial agonist activity of antagonists, in gene induction by GRs (23; 25; 30; 31; 36; 38) suggests that these biological properties are uniquely sensitive to structural changes. The current results support this conclusion. For example, the Amax of full-length Q588K, either with or without TIF2, is similar to that of the wild type receptor with both Dex and DAC while the EC50 of Q558K ± TIF2 with either steroid is orders of magnitude higher than that of the wild type GR (Fig. 7). Furthermore, TIF2 increases the Amax of all GR mutants but has greatly reduced ability both to lower the EC50 of most mutant GAL/GR525C complexes and to increase the percent partial agonist activity of Dex-Mes (Fig. 6 and Table 4). The generally greater fold decrease in EC50 with added TIF2 for the full-length vs. truncated mutant GR-Dex complexes (Fig. 7B vs. Fig. 6B) is consistent with our recent finding that an amino terminal sequence of TIF2 interacts with the N-terminal region of full-length GRs (43). Nevertheless, even with full-length GRs, added TIF2 is less able to reverse the effects of each mutation on the EC50 of Dex than of DAC gene induction, presumably due to the presence of the inactivated AF2 domain in the Dex-bound receptors. We have not examined other coactivators under similar situations. However, the fact that the same increases in EC50 are seen in cells containing other coactivators (Fig. 3) suggests that the behavior with TIF2 is shared by the other coactivators. Thus, the mutational inactivation of the steroid-bound AF2 domain appears to unequally impact the ability of TIF2, and probably all p160 coactivators, to modulate the EC50 vs. Amax of GR-regulated gene expression. These differential effects on two aspects of regulated gene transcription are conceptually similar to the emerging hypothesis that steroid hormone induced changes in gene transcription and alternative splicing of transcripts are controlled by different processes (54; 55).
In summary, this study demonstrates that mutations within the GR steroid-binding cavity can selectively affect AF2 activity, and thus the EC50 and/or Amax of GR-regulated gene induction and repression, in a manner that is further regulated by steroid structure. Such discrimination is unique among glucocorticoid agonists. These results also suggest that, in a variation of reverse chemical genetics (56; 57), modifications of steroid structure could mimic the effects of the current receptor mutations to reproduce the separation of GR induction and repression properties, thereby generating new steroids with greater therapeutic selectivity. Some examples of such steroid derivatives appear to exist (58–62). However, our current x-ray structure based approach offers a more structurally directed avenue by which to design new derivatives. A similar method is being used to develop ligands that will induce the orphan receptor ERR to accept a molecule in the normally unoccupied LBD and induce desirable changes in receptor activity (63). The present data also suggest that the determinants of EC50 are more responsive than those of Amax to changes in the GR binding cavity, possibly due to more efficient transmission of critical structural changes in steroid-bound GRs through the receptor surface to the coupled target proteins. The fact that several mutations in helices 3 and 5 can preferentially reduce GR repression of at least one endogenous gene offers hope that this will prove to be more common. Together, these structural modifications suggest new avenues for the differential control of gene expression by glucocorticoids, and probably other steroid/nuclear receptor ligands, during development, differentiation, homeostasis, and endocrine therapies.
Acknowledgments
We thank Stefano Costanzi (NIDDK, NIH) for discussions during the planning of this study and Tomoshigo Kino (NICHD, NIH) for critical review.
Abbreviations
- Amax
maximal activity
- AF1 and AF2
activation function 1 and 2
- coll3
collagenase 3
- DAC
deacylcortivazol
- Dex
dexamethasone
- Dex-Mes
Dex-21-mesylate
- DBD
DNA binding domain
- GR
glucocorticoid receptor
- LAD1
ladinin 1
- LBD
ligand binding domain
- MMTV
mouse mammary tumor virus
- PMA
phorbol 12-myristate 13-acetate
Footnotes
This research was supported by the Intramural Research Program of the NIH, NIDDK.
In previous reports, we used the term Vmax for the maximal amount of activity. To avoid misinterpretation of our intent, and inappropriate association with the Vmax of enzyme kinetics, we here introduce the term Amax.
References
- 1.Williams SP, Sigler PB. Atomic structure of progesterone complexed with its receptor. Nature. 1998;393:392–396. doi: 10.1038/30775. [DOI] [PubMed] [Google Scholar]
- 2.Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM, Lambert MH, Moore JT, Pearce KH, Xu HE. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- 3.Brzozowski AM, Pike ACW, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson J-A, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- 4.Shiau AK, Barstad D, Loria PM, Cheng L, Kushner PJ, Agard DA, Greene GL. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 5.Kauppi B, Jakob C, Farnegardh M, Yang J, Ahola H, Alarcon M, Calles K, Engstrom O, Harlan J, Muchmore S, Ramqvist AK, Thorell S, Ohman L, Greer J, Gustafsson JA, Carlstedt-Duke J, Carlquist M. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J Biol Chem. 2003;278:22748–22754. doi: 10.1074/jbc.M212711200. [DOI] [PubMed] [Google Scholar]
- 6.Samuels HH, Tomkins GM. Relation of steroid structure to enzyme induction in hepatoma tissue culture cells. J Mol Biol. 1970;52:57–74. doi: 10.1016/0022-2836(70)90177-4. [DOI] [PubMed] [Google Scholar]
- 7.Rousseau GG, Schmit JP. Structure-activity relationships for glucocorticoids. Determination of receptor binding and biological activity. J Steroid Biochem. 1977;8:911–919. doi: 10.1016/0022-4731(77)90187-x. [DOI] [PubMed] [Google Scholar]
- 8.Simons SS, Jr, Thompson EB, Johnson DF. Anti-inflammatory pyrazolo-steroids: potent glucocorticoids containing bulky A-ring substituents and no C3-carbonyl. Biochem Biophys Res Comm. 1979;86:793–800. doi: 10.1016/0006-291x(79)91782-0. [DOI] [PubMed] [Google Scholar]
- 9.Harmon JM, Schmidt TJ, Thompson EB. Deacylcortivazol acts through glucocorticoid receptors. J Steroid Biochem. 1981;14:273–279. doi: 10.1016/0022-4731(81)90136-9. [DOI] [PubMed] [Google Scholar]
- 10.Webster JC, Cidlowski JA. Mechanisms of glucocorticoid-receptor-mediated repression of gene expression. Trends Endocrinol Metab. 1999;10:396–402. doi: 10.1016/s1043-2760(99)00186-1. [DOI] [PubMed] [Google Scholar]
- 11.Jakacka M, Ito M, Weiss J, Chien PY, Gehm BD, Jameson JL. Estrogen receptor binding to DNA is not required for its activity through the nonclassical AP1 pathway. J Biol Chem. 2001;276:13615–13621. doi: 10.1074/jbc.M008384200. [DOI] [PubMed] [Google Scholar]
- 12.Rogatsky I, Trowbridge JM, Garabedian MJ. Glucocorticoid receptor-mediated cell cycle arrest is achieved through distinct cell-specific transcriptional regulatory mechanisms. Mol Cell Biol. 1997;17:3181–3193. doi: 10.1128/mcb.17.6.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schena M, Freedman LP, Yamamoto KR. Mutations in the glucocorticoid receptor zinc finger region that distinguish interdigitated DNA binding and transcriptional enhancement activities. Genes and Develop. 1989;3:1590–1601. doi: 10.1101/gad.3.10.1590. [DOI] [PubMed] [Google Scholar]
- 14.Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 15.Rogatsky I, Hittelman AB, Pearce D, Garabedian MJ. Distinct glucocorticoid receptor transcriptional regulatory surfaces mediate the cytotoxic and cytostatic effects of glucocorticoids. Mol Cell Biol. 1999;19:5036–5049. doi: 10.1128/mcb.19.7.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu W, Hillmann A, Harmon J. Hormone-independent repression of AP-1-inducible collagenase promoter activity by glucocorticoid receptors. Mol Cell Biol. 1995;15:1005–1013. doi: 10.1128/mcb.15.2.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.deLange P, JW K, Huizenga NATM, Brinkmann AO, deJong FH, Karl M, Chrousos GP, Lamberts SWJ. Differential hormone-dependent transcriptional activation and repression by naturally occurring human glucocorticoid receptor variants. Mol Endo. 1997;11:1156–1164. doi: 10.1210/mend.11.8.9949. [DOI] [PubMed] [Google Scholar]
- 18.Ray DW, Suen CS, Brass A, Soden J, White A. Structure/function of the human glucocorticoid receptor: tyrosine 735 is important for transactivation. Mol Endo. 1999;13:1855–1863. doi: 10.1210/mend.13.11.0376. [DOI] [PubMed] [Google Scholar]
- 19.Suino-Powell K, Xu Y, Zhang C, Tao YG, Tolbert WD, Simons SSJ, Xu HE. Doubling the size of the glucocorticoid receptor ligand binding pocket by deacylcortivazol. Mol Cell Biol. 2008;28:1915–1923. doi: 10.1128/MCB.01541-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simons SS., Jr Function/activity of specific amino acids in glucocorticoid receptors. Vitamins and Hormones. 1994;48:49–130. doi: 10.1016/s0083-6729(08)61146-2. [DOI] [PubMed] [Google Scholar]
- 21.Simons SS., Jr . Structure and function of the steroid and nuclear receptor ligand-binding domain. In: Freedman LP, editor. The molecular biology of steroid and nuclear hormone receptors. 1998. pp. 35–104. [Google Scholar]
- 22.Simons SS, Jr, Pons M, Johnson DF. α-Keto mesylate: a reactive thiol-specific functional group. J Org Chem. 1980;45:3084–3088. [Google Scholar]
- 23.He Y, Szapary D, Simons SS., Jr Modulation of induction properties of glucocorticoid receptor-agonist and -antagonist complexes by coactivators involves binding to receptors but is independent of ability of coactivators to augment transactivation. J Biol Chem. 2002;277:49256–49266. doi: 10.1074/jbc.M205536200. [DOI] [PubMed] [Google Scholar]
- 24.Kaul S, Blackford JA, Jr, Chen J, Ogryzko VV, Simons SS., Jr Properties of the glucocorticoid modulatory element binding proteins GMEB-1 and -2: potential new modifiers of glucocorticoid receptor transactivation and members of the family of KDWK proteins. Mol Endocrinol. 2000;14:1010–1027. doi: 10.1210/mend.14.7.0494. [DOI] [PubMed] [Google Scholar]
- 25.Kaul S, Blackford JA, Jr, Cho S, Simons SS., Jr Ubc9 is a novel modulator of the induction properties of glucocorticoid receptors. J Biol Chem. 2002;277:12541–12549. doi: 10.1074/jbc.M112330200. [DOI] [PubMed] [Google Scholar]
- 26.Wang Q, Blackford JA, Jr, Song LN, Huang Y, Simons SS., Jr Equilibrium interactions of corepressors and coactivators modulate the properties of agonist and antagonist complexes of glucocorticoid receptors. Mol Endocrinol. 2004;18:1376–1395. doi: 10.1210/me.2003-0421. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Simons SS., Jr STAMP: a novel predicted factor assisting TIF2 actions in glucocorticoid receptor-mediated induction and repression. Mol Cell Biol. 2007;27:1467–1485. doi: 10.1128/MCB.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giannoukos G, Silverstein AM, Pratt WB, Simons SS., Jr The seven amino acids (547–553) of rat glucocorticoid receptor required for steroid and hsp90 binding contain a functionally independent LxxLL motif that is critical for steroid binding. J Biol Chem. 1999;274:36527–36536. doi: 10.1074/jbc.274.51.36527. [DOI] [PubMed] [Google Scholar]
- 29.Rogatsky I, Zarember KA, Yamamoto KR. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 2001;20:6071–6083. doi: 10.1093/emboj/20.21.6071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Simons SS., Jr The importance of being varied in steroid receptor transactivation. TIPS. 2003;24:253–259. doi: 10.1016/S0165-6147(03)00101-9. [DOI] [PubMed] [Google Scholar]
- 31.Simons SS., Jr How much is enough? Modulation of dose-response curve for steroid receptor-regulated gene expression by changing concentrations of transcription factor. Current Topics in Medicinal Chemistry. 2006;6:271–285. doi: 10.2174/156802606776173465. [DOI] [PubMed] [Google Scholar]
- 32.Sarlis NJ, Bayly SF, Szapary D, Simons SS., Jr Quantity of partial agonist activity for antiglucocorticoids complexed with mutant glucocorticoid receptors is constant in two different transactivation assays but not predictable from steroid structure. J Steroid Biochem Molec Biol. 1999;68:89–102. doi: 10.1016/s0960-0760(99)00021-7. [DOI] [PubMed] [Google Scholar]
- 33.Bresnick EH, John S, Berard DS, LeFebvre P, Hager GL. Glucocorticoid receptor-dependent disruption of a specific nucleosome on the mouse mammary tumor virus promoter is prevented by sodium butyrate. Proc Natl Acad Sci USA. 1990;87:3977–3981. doi: 10.1073/pnas.87.10.3977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rogatsky I, Wang JC, Derynck MK, Nonaka DF, Khodabakhsh DB, Haqq CM, Darimont BD, Garabedian MJ, Yamamoto KR. Target-specific utilization of transcriptional regulatory surfaces by the glucocorticoid receptor. Proc Natl Acad Sci U S A. 2003;100:13845–13850. doi: 10.1073/pnas.2336092100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Szapary D, Huang Y, Simons SS., Jr Opposing effects of corepressor and coactivators in determining the dose-response curve of agonists, and residual agonist activity of antagonists, for glucocorticoid receptor regulated gene expression. Mol Endocrinol. 1999;13:2108–2121. doi: 10.1210/mend.13.12.0384. [DOI] [PubMed] [Google Scholar]
- 36.Song LN, Huse B, Rusconi S, Simons SS., Jr Transactivation specificity of glucocorticoid vs. progesterone receptors: role of functionally different interactions of transcription factors with amino- and carboxyl-terminal receptor domains. J Biol Chem. 2001;276:24806–24816. doi: 10.1074/jbc.M102610200. [DOI] [PubMed] [Google Scholar]
- 37.Cho S, Kagan BL, Blackford JA, Jr, Szapary D, Simons SS., Jr Glucocorticoid receptor ligand binding domain is sufficient for the modulation of both the dose-response curve of receptor-agonist complexes and the partial agonist activity of receptor-antisteroid complexes by glucocorticoid receptors, coactivator TIF2, and Ubc9. Mol Endo. 2005;19:290–311. doi: 10.1210/me.2004-0134. [DOI] [PubMed] [Google Scholar]
- 38.Kim Y, Sun Y, Chow C, Pommier YG, Simons SS., Jr Effects of acetylation, polymerase phosphorylation, and DNA unwinding in glucocorticoid receptor transactivation. J Steroid Biochem Molec Biol. 2006;100:3–17. doi: 10.1016/j.jsbmb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 39.Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci U S A. 2002;99:16701–16706. doi: 10.1073/pnas.262671599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heck S, Kullmann M, Gast A, Ponta H, Rahmsdorf HJ, Herrlich P, Cato ACB. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 1994;13:4087–4095. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tao Y, Williams-Skipp C, Scheinman RI. Mapping of glucocorticoid receptor DNA binding domain surfaces contributing to transrepression of NF-{kappa}B and induction of apoptosis. J Biol Chem. 2001;276:2329–2332. doi: 10.1074/jbc.C000526200. [DOI] [PubMed] [Google Scholar]
- 42.Wang D, Simons SS., Jr Corepressor binding to progesterone and glucocorticoid receptors involves the AF-1 domain and is inhibited by molybdate. Mol Endo. 2005;19:1483–1500. doi: 10.1210/me.2005-0012. [DOI] [PubMed] [Google Scholar]
- 43.Wang D, Wang Q, Awasthi S, Simons SS., Jr Amino-terminal domain of TIF2 is involved in competing for corepressor binding to glucocorticoid and progesterone receptors. Biochemistry. 2007;48:8036–8049. doi: 10.1021/bi7004575. [DOI] [PubMed] [Google Scholar]
- 44.Huang Y, Simons SS., Jr Functional analysis of R651 mutations in the putative helix 6 of rat glucocorticoid receptors. Mol Cell Endo. 1999;158:117–130. doi: 10.1016/s0303-7207(99)00171-9. [DOI] [PubMed] [Google Scholar]
- 45.Vottero A, Kino T, Combe H, Lecomte P, Chrousos GP. A novel, C-terminal dominant negative mutation of the GR causes familial glucocorticoid resistance through abnormal interactions with p160 steroid receptor coactivators. J Clin Endocrinol Metab. 2002;87:2658–2667. doi: 10.1210/jcem.87.6.8520. [DOI] [PubMed] [Google Scholar]
- 46.Cho S, Blackford JA, Jr, Simons SS., Jr Role of activation function domain 1, DNA binding, and coactivator in the expression of partial agonist activity of glucocorticoid receptor complexes. Biochemistry. 2005;44:3547–3561. doi: 10.1021/bi048777i. [DOI] [PubMed] [Google Scholar]
- 47.Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hilser VJ, Thompson EB. Intrinsic disorder as a mechanism to optimize allosteric coupling in proteins. Proc Natl Acad Sci U S A. 2007;104:8311–8315. doi: 10.1073/pnas.0700329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Tilborg MAA, Lefstin JA, Kruiskamp M, Teuben J, Boelens R, Yamamoto KR, Kaptein R. Mutations in the glucocorticoid receptor DNA-binding domain mimic an allosteric effect of DNA. J Mol Biol. 2000;301:947–958. doi: 10.1006/jmbi.2000.4001. [DOI] [PubMed] [Google Scholar]
- 50.Shiau AK, Barstad D, Radek JT, Meyers MJ, Nettles KW, Katzenellenbogen BS, Katzenellenbogen JA, Agard DA, Greene GL. Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism. Nat Struct Biol. 2002;9:359–364. doi: 10.1038/nsb787. [DOI] [PubMed] [Google Scholar]
- 51.Shulman AI, Larson C, Mangelsdorf DJ, Ranganathan R. Structural determinants of allosteric ligand activation in RXR heterodimers. Cell. 2004;116:417–429. doi: 10.1016/s0092-8674(04)00119-9. [DOI] [PubMed] [Google Scholar]
- 52.Zwart W, Griekspoor A, Berno V, Lakeman K, Jalink K, Mancini M, Neefjes J, Michalides R. PKA-induced resistance to tamoxifen is associated with an altered orientation of ERalpha towards co-activator SRC-1. EMBO J. 2007;26:3534–3544. doi: 10.1038/sj.emboj.7601791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stevens A, Garside H, Berry A, Waters C, White A, Ray D. Dissociation of steroid receptor coactivator 1 and nuclear receptor corepressor recruitment to the human glucocorticoid receptor by modification of the ligand-receptor interface: the role of tyrosine 735. Mol Endocrinol. 2003;17:845–859. doi: 10.1210/me.2002-0320. [DOI] [PubMed] [Google Scholar]
- 54.Pan Q, Shai O, Misquitta C, Zhang W, Saltzman AL, Mohammad N, Babak T, Siu H, Hughes TR, Morris QD, Frey BJ, Blencowe BJ. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 55.Ip JY, Tong A, Pan Q, Topp JD, Blencowe BJ, Lynch KW. Global analysis of alternative splicing during T-cell activation. RNA. 2007;13:563–572. doi: 10.1261/rna.457207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bishop AC, Ubersax JA, Petsch DT, Matheos DP, Gray NS, Blethrow J, Shimizu E, Tsien JZ, Schultz PG, Rose MD, Wood JL, Morgan DO, Shokat KM. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- 57.Cohen MS, Zhang C, Shokat KM, Taunton J. Structural bioinformatics-based design of selective, irreversible kinase inhibitors. Science. 2005;308:1318–1321. doi: 10.1126/science1108367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Heck S, Bender K, Kullmann M, Gottlicher M, Herrlich P, Cato ACB. IkappaBalpha independent downregulation of NF-kappaB activity by glucocorticoid receptor. EMBO. 1997;16:4698–4707. doi: 10.1093/emboj/16.15.4698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vayssiere BM, Dupont S, Choquart A, Petit F, Garcia T, Marchandeau C, Gronemeyer H, Resche-Rigon M. Synthetic glucocorticoids that dissociate transactivation and AP-1 transrepression exhibit antiinflammatory activity in vivo. Mol Endocrinol. 1997;11:1245–1255. doi: 10.1210/mend.11.9.9979. [DOI] [PubMed] [Google Scholar]
- 60.Ali A, Thompson CF, Balkovec JM, Graham DW, Hammond ML, Quraishi N, Tata JR, Einstein M, Ge L, Harris G, Kelly TM, Mazur P, Pandit S, Santoro J, Sitlani A, Wang C, Williamson J, Miller DK, Thompson CM, Zaller DM, Forrest MJ, Carballo-Jane E, Luell S. Novel N-arylpyrazolo[3,2-c]-based ligands for the glucocorticoid receptor: receptor binding and in vivo activity. J Med Chem. 2004;47:2441–2452. doi: 10.1021/jm030585i. [DOI] [PubMed] [Google Scholar]
- 61.Schacke H, Schottelius A, Docke WD, Strehlke P, Jaroch S, Schmees N, Rehwinkel H, Hennekes H, Asadullah K. Dissociation of transactivation from transrepression by a selective glucocorticoid receptor agonist leads to separation of therapeutic effects from side effects. Proc Natl Acad Sci U S A. 2004;101:227–232. doi: 10.1073/pnas.0300372101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang JC, Shah N, Pantoja C, Meijsing SH, Ho JD, Scanlan TS, Yamamoto KR. Novel arylpyrazole compounds selectively modulate glucocorticoid receptor regulatory activity. Genes Dev. 2006;20:689–699. doi: 10.1101/gad.1400506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kallen J, Lattmann R, Beerli R, Blechschmidt A, Blommers MJ, Geiser M, Ottl J, Schlaeppi JM, Strauss A, Fournier B. Crystal structure of human estrogen-related receptor alpha in complex with a synthetic inverse agonist reveals its novel molecular mechanism. J Biol Chem. 2007;282:23231–23239. doi: 10.1074/jbc.M703337200. [DOI] [PubMed] [Google Scholar]