Introduction
The methylotropic yeast Pichia pastoris is a model organism for the study of autophagy1 and peroxisome biogenesis2. Being able to look at the organism via transmission electron microscopy (TEM) can yield valuable data on the morphology of the secretory pathway and many other organelles of interest3. However, preparing the yeast for TEM work can be very arduous and costly. One of the reasons P. pastoris is so hard to prepare for visualization is because its cell wall is very thick and tough compared to the membrane of a mammalian cell. Thus, P. pastoris is notoriously difficult to infiltrate with fixatives, a step necessary to maintain its ultrastructure4. This article outlines an efficient and cost effective way to prepare P. pastoris for TEM without the need for certain specialized equipment. With this protocol, excellent pictures can be obtained by using the buffers, KMnO4, sorbitol, and PIPES, along with glutaraldehyde. These components preserve the ultrastructure of the yeast without any apparent artifactual change in morphology4.
Materials and Methods
Culturing procedure for P. pastoris grown in YPD
Inoculate 5 mL of YPD broth in a sterile 50 mL conical with P. pastoris colony grown from a plate, and grow overnight in a 30 degrees C shaker/incubator. Check the OD600 of the overnight culture, and then add the correct amount of the culture to 100 mL of the YPD (1% Yeast extract, 2% Peptone, 2% Dextrose) in a 500 mL sterile baffled Erlenmeyer flask so that the concentration is 0.01 OD600/ mL. Place the diluted culture in the shaker for approximately 12 hours. This will ensure that the culture will have an end OD600 reading between 0.5-1.0. Place yeast on ice before starting the fixation procedure.
Culturing procedure for P. pastoris grown in BMMY
Inoculate 5 mL of YPD broth in a sterile 50 mL conical with P. pastoris colony grown from a plate, and grow overnight in a 30 degrees C shaker/incubator. Check the OD600 of the overnight culture. Calculate the amount of the culture to add to 100 mL of the BMMY (Buffered Methanol-complex Medium with 1% Yeast extract, 0.5% methanol) so that the concentration is 0.012 OD600/ mL. Pipette the exact amount of the overnight culture into a 1.5 mL microfuge tube. Centrifuge for 1 minute at 5,000 × g. Remove the supernatant, resuspend in 1 mL BMMY, and add the cells to the 500 mL sterile baffled Erlenmeyer flask containing 100 mL of BMMY. Place the diluted culture in the shaker for approximately 24 hours. This will ensure that the culture will have an end OD600 reading between 0.5-1.0. Place yeast on ice before starting the fixation procedure.
Fixation
Using a procedure originally designed for S. cerevisiae, we have developed a fixation protocol to address the limitations of P. pastoris4. Measure 7.5 mL of culture into a 15 mL conical tube. Pour the culture into an equal volume of 2× fixative (7.50 mL 0.4M PIPES buffer + 1.25 mL 2.4M sorbitol + 0.03 mL 1.0M MgCl2 + 0.03 mL 1.0M CaCl2 + 7.50 mL 8% glutaraldehyde + ddH20 to 20 mL). Incubate at room temperature (∼25 degrees C) for 5 minutes. Spin down cells at 1060 × g for 10 minutes, and decant supernatant. Resuspend the cells in 9.5 mL of 1X (1:1 H2O) fixative. Incubate overnight at 4 degrees C. Centrifuge cells at 1060 × g for 5 minutes, and then aspirate fixative. Resuspend the cells in 8 mL of H2O and incubate for 10 minutes. Centrifuge at 1060 × g for 5 minutes. Repeat water wash 3 times. For the last centrifugation, aspirate off the supernatant and resuspend the pellet in the tiny residual amount of water (∼100 μL) that remains. Add 5 mL of 2% KMnO4, and incubate 5 minutes at room temperature. Pellet cells at 1060 × g for 5 minutes. Aspirate fixative and overlay with fresh 2% KMnO4. Incubate for 45 minutes at room temperature. Centrifuge cells at 1060 × g for 5 minutes. Aspirate permanganate solution, making sure not to disturb the pellet. Fill the 15 mL conical with ddH2O, remove water, and repeat until no purple color is evident. Add 1% uranyl acetate and incubate at 4° C overnight. Remove the uranyl acetate with an aspirator. Add 5 mL H2O, mix, and centrifuge at 1060 × g until pellet is settled. Repeat this step 3 times. Dehydrate in a graded series of ethanol from 25%-100% EtOH. Resuspend pellet in 2:1 EtOH: Spurr’s resin (3 mL:1.5 mL) in a 20 mL glass scintillation vial. Rotate at slow speed (∼175 rpm) for 2 hours at room temperature. Remove the resin and replace with 1:1 EtOH: Spurr’s resin (1.5 mL:1.5 mL). Allow to rotate at slow speed uncapped overnight at room temperature. Remove the resin and add 2 mL of 100% resin. Rotate uncapped for 1 hour, and then remove resin after centrifugation by pipetting. Add 100% resin and rotate for 2 hours capped. Transfer the resin containing the cells into 1.5 mL microfuge tubes and centrifuge at 2000 × g for 10 minutes. Remove resin. Add 2 drops of pellet to each micromold with a Pasteur pipette and fill with additional 100% resin in micromolds. Place the micromolds in a 60 degrees C oven for 24 hours.
Post-fixation
Trim the blocks to a 0.5 mm trapezoid face, and section. Use an Emcorp Diamond knife for sectioning at a speed of 1.6 mm/s and at a thickness of 55-60 nm on a Leica Ultracut R microtome. Retrieve sections on 200 mesh copper grids, and capture the images on a computer. In our lab images were captured using a Zeiss 109 TEM with a Micromax ccd digital camera (Princeton Instruments, Roper Scientific, Trenton, NJ), with the software program Winview (Princeton Instruments, Acton, Trenton, NJ).
Results
This method allows researchers to look at various aspects of the morphology of P. pastoris. Structures commonly observed are: mitochondria, nuclei, vacuoles, peroxisomes (under methanol-growth conditions), endoplasmic reticuli, and Golgi (Figures 1 and 2). One of the most crucial aspects of working with P. pastoris for TEM work is the preparation of the cultures. The cell density should be between 0.5-1.0 OD600. When there was an optical density greater than 1.0, the following problems occurred: a) budding scars were present on the cells; b) the cells were poorly infiltrated; c) when sectioning, the cells were ripped out of the resin because they cut at a different rate than the resin in which they were embedded; and d) there were many holes in the specimen so that when the electron beam came in contact with these holes, the sample was destroyed. In addition, the correct thickness of the sections is also important for capturing clear pictures. When the sections were too thick, sharp focus was extremely difficult, and when they were too thin, the sections disintegrated under the beam. When the samples had the optimal concentration of cells and the sections were the correct thickness, clear images were obtained.
Figure 1.
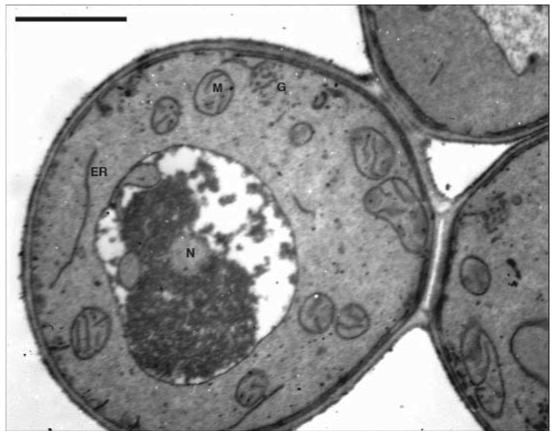
Electron micrograph of P. pastoris grown in YPD medium. G = Golgi vesicles; N = nucleus; ER = endoplasmic reticulum; M = mitochondria. Scale bar = 1 μm
Figure 2.
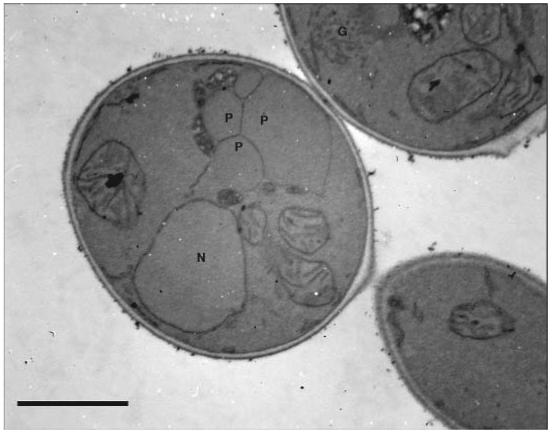
Electron micrograph of P. pastoris grown in BMMY medium. Methanol selectively induces the production of peroxisomes. G = Golgi vesicles; N = nucleus; ER = endoplasmic reticulum; M = mitochondria; P = peroxisome. Scale bar = 1 μm
Conclusion
There are many possible research projects that can be carried out with the use of this protocol. For instance, organelle and cell structure can be investigated as a function of nutrition or the mutant background of a strain6. Although several methodologies are available for TEM of yeast, this protocol has been optimized specifically for P. pastoris. Starting with a culture inoculated with P. pastoris to ending with electron micrographs of the organism will take approximately 9 days using this protocol. Strict adherence to the protocol is essential. Skipping or changing one step can potentially ruin one’s results. For instance, using formaldehyde in place of glutaraldehyde will cause poor fixation, leading to various problems described above. Using the traditional sodium cacodylate-osmium tetroxide fixation5 resulted in little to no fixation of P. pastoris. In addition, the use of counter stains, such as Reynold’s lead citrate and uranyl acetate, is not necessary to increase the contrast of the samples because there was no apparent change in the pictures with or without these counter stains. Another attribute of this method is its minimal investment in advanced instrumentation, such as a speed vac or a specialized microwave oven. Thus, this method is particularly suitable for a research scientist with limited resources and time.
Acknowledgements
This work was supported by NIH AREA (GM65882) grant to J.L-C. and G.P.L-C. This research is dedicated to the memory of the late Dr. Paul Richmond (University of the Pacific), who had a passion for microscopy and was the inspiration for this project. We are grateful to Dr. John Livesey (University of the Pacific) for his assistance with calculations
References
- 1.Yuan W, Stromhaug PE, Dunn WA., Jr. Glucose-induced Autophagy of Peroxisomes in Pichia pastoris Requires a Unique E1-like Protein. Molecular Biology of the Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sakai Y, Koller A, Rangell LK, Keller GA, Subramani S. Pex19p Interacts with Pex3p and Pex10p and Is Essential for Peroxisome Biogenesis in Pichia pastoris. Molecular Biology of the Cell. 1999;10:1745–1761. doi: 10.1091/mbc.10.6.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Soderholm J, Bhattacharyya D, Strongin D, Markovitz V, Connerly PL, Reinke CA, Glick BS. The Translational ER Localization Mechanism of Pichia pastoris Sec12. Developmental Cell. 2004;6:649–659. doi: 10.1016/s1534-5807(04)00129-7. [DOI] [PubMed] [Google Scholar]
- 4.Wright Robin. Transmission Electron Microscopy of Yeast. Microscopy Research and Technique. 2000;51:496–510. doi: 10.1002/1097-0029(20001215)51:6<496::AID-JEMT2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 5.Bozzola JJ, Russell LD. Electron Microscopy. 2nd edition Jones and Bartlett Publishers; Sudbury: 1992. p. 23. [Google Scholar]
- 6.Lin Cereghino J, Cregg JM. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Micro. Rev. 2000;24:45–66. doi: 10.1111/j.1574-6976.2000.tb00532.x. [DOI] [PubMed] [Google Scholar]


