Abstract
The generation of microRNAs is dependent on the RNase III enzyme Dicer, the levels of which vary in different normal cells and in disease states. We demonstrate that Dicer protein expression in JAR trophoblast cells, and several other cell types, was inhibited by multiple stresses including reactive oxygen species, phorbol esters and the Ras oncogene. Additionally, double-stranded RNA and Type I interferons repress Dicer protein in contrast to IFN-γ which induces Dicer. The effects of stresses and interferons are primarily post-transcriptional. The findings suggest that Dicer is a stress response component and identifies interferons as potentially important regulators of Dicer expression.
Keywords: RNAi, microRNA, Dicer, stress, interferon
Introduction
The cytoplasmic RNase III enzyme, Dicer, cleaves the pre-miRNAs transported from the nucleus to a ~22nt duplex miRNA (miR) (3). A single strand of the duplex is selected and assembled with Argonaute proteins into the RISC (RNA induced silencing complex), which guides miR to its 3′UTR target. In humans, miRs usually inhibit translational initiation (25) but, depending on the extent of complementarity with its target, miRs may also degrade mRNA (19). In proliferating cells miRs normally repress but miRs can become translational activators if the cells are growth arrested (40). Enhanced gene target expression has been reported by binding to a 5′UTR miR target site (27). MiRs are well-established regulators of cell growth, differentiation and development, which can not only destroy cancer cells but prevent further development of cancer over long time periods (12). However, miRs have also been recently shown to be involved in acute processes and responses to environmental insults, such as nutrient depletion, osmotic and cardiac load stress responses (4, 9, 22, 39). Apoptotic cells may have lower Dicer resulting from caspase cleavage (23) although downregulation of Dicer may occur in the absence of apoptosis as shown here.
Mammals have a single Dicer gene with several variants having different first exons, which, at least in part, may be responsible for the observed differences of Dicer expression in normal human tissues (15, 33). Dicer levels also vary between different tumor cells and these variations have been correlated with cancer progression (8, 15, 16). Previous studies have suggested a general downregulation of miRs in cancer cells (20, 36) and, that knockdown of the machinery components for miRs (e.g. Dicer and others) enhances tumorigenesis (17). The above mentioned tissue expression differences suggest that Dicer expression may be regulated although little is currently known of the mechanism(s). In this report, we examine the mechanistic basis for the regulation of Dicer in several human cells. We present evidence that the deacetylase inhibitor trichostatin A (TSA) and certain cellular stresses (reactive oxidative species [ROS], the phorbol ester PMA, Ras oncogene activation) inhibit Dicer protein expression in several cell types. Importantly, double-stranded RNA (Poly IC) and Type I interferons repress Dicer while IFN-γ enhances Dicer expression at the protein level. The data suggest that Dicer regulation is a component of the response to multiple cellular stresses and that post-transcriptional mechanisms are primarily involved.
Materials and Methods
Cells and Reagents
The human trophoblast cell lines JAR and JEG-3, human cervical carcinoma cell line HeLa, human B-cell lines Raji and Daudi, human fibroblast cell line IMR-90, and mouse melanoma cell line B16 were from American Type Culture Collection (ATCC) (Manassas, VA) and cultured according to ATCC’s instructions. Freshly dissected murine kidneys and spleens, as well as excess surgical tissue from kidney specimens identified by Pathology as normal and released under IRB approval, were enzymatically dissociated and cultured in complete RPMI 1640 (Invitrogen, Carlsbad CA). Ras transduced IMR-90 cells were provided by A.W. Lin (18). Human and mouse IFN-α and IFN-γ were obtained from R&D Systems (Minneapolis, MN). IFN-α was used at a concentration of 1000U/mL or 10000U/mL for three days and IFN-γ at a concentration of 100U/mL or 500U/mL for 24 hours. TSA was from Wako Biochemical (Richmond, VA). TSA was used at a concentration of 50nM in JAR cells, 250nM in JEG-3 cells, and 100nM in B16 cells for 24 hours. Valproic acid (VA), hydrogen peroxide (H2O2), phorbol-12-myristate-13-acetate (PMA), and Ionomycin (IM) were obtained from Sigma (St Louis, MO). VA was used at 250μM for 24 hours. JAR cells were treated with 250μM H2O2 for 15 minutes, washed and cultured for an additional 24 hours or 500μM H2O2 for 4 hours, washed and cultured for an additional 20 hours. PMA treatments were 500ng/mL and IM was used in combination with PMA at a concentration of 10μM for 24 hours. Poly I:C was obtained from GE Healthcare (Piscataway, NJ) and was transfected into JAR cells (2μg/mL for 24 hours) using Effectene transfection reagent from Qiagen (Valencia, CA). The CpG oligodeoxynucleotide (5′-TCCATGACGTTCCTGACGTT-3′) and a non-CpG control ODN (5′-TCCATGAGCTTCCTGAGCTT-3′) were synthesized by Invitrogen and cells were treated with 6μg/mL ODN for 72 hours. Apoptotic cells were assayed by Annexin V-FITC staining (Caltag, Burlingame, CA) according to manufacturer’s instructions. Stained cells were analyzed on a FACScan (Becton Dickinson, San Jose, CA) with CellQuest software and subsequent analyses were performed with FCSExpress software (De Novo Software, Los Angeles, CA).
Western blotting
Cells harvested for whole cell lysates were pelleted and lysed on ice for 30 min in RIPA lysis buffer (Sigma) supplemented with protease inhibitor cocktail (Sigma), 1mM DTT and 2mM sodium orthovanadate. The extracts were centrifuged at 10,000 × g for 10 min and the supernatants collected. For all proteins, with the exception of βactin, 40μg of lysates were re-suspended in SDS sample buffer plus 0.13M dithiothreitol (Sigma), separated on 7% [for Dicer] or 10% [for all other proteins] SDS-PAGE gels and transferred to Immobion-P membrane (BioRad, Hercules CA). For βactin, 10μg of protein was used. The antibodies employed were anti-Dicer, anti-βactin, anti-GAPDH, anti-Mad1, anti-Calpain S1 (Abcam, Cambridge MA), anti-Ago1, anti-Hsp90 (Upstate Biotechnology, Lake Placid, NY), anti-TTP, anti-Stat1 (Santa Cruz Biotechnology, Santa Cruz, CA), anti-Caspase 3 (BD Biosciences, San Jose CA), anti-Caspase 8 (Calbiochem, Gibbstown NJ), goat anti-rabbit IgG-horseradish peroxidase and goat anti-mouse IgG-horseradish peroxidase (Promega, Madison WI). Blots were developed with a West Pico Chemiluminescent Kit (Pierce, Rockford IL).
Real-time Quantitative RT-PCR
Total RNA was isolated using the mirVana™ kit from Ambion (Austin, TX) and 2μg was used for reverse transcription with Superscript™ II (Invitrogen). Real-time PCR was performed as previously described on an ABI7900HT (Applied Biosystems, Foster City, CA) (21). Amplification of cDNA samples was carried out with either Taqman PCR Master Mix or SYBR Green Master Mix (Eurogentec, San Diego, CA) according to the manufacturer’s protocol. Primers used included human GAPDH 5′-GAAGGTGAAGGTCGGAGTC-3′ [forward] and 5′-GAAGATGGTGATGGGATTTC-3′ [reverse], mouse GAPDH 5′-TGCACCACCAACTGCTTAG-3′ [forward] and 5′-GGATGCAGGGATGATGTTC-3′ [reverse], human Dicer (15) 5′-GTACGACTACCACAAGTACTTC-3′ [forward] and 5′-ATAGTACACCTGCCAGACTGT-3′ [reverse], and mouse Dicer 5′ –TACACACGCCTCCTACCACTACAA-3′ [forward] and 5′ –CCAAAATCGCATCTCCCAGGAATT-3′ [reverse].
MicroRNA assessment by microarray
The miRNA expression profiles of JAR, HeLa and Raji cells (10μg total RNA) were assessed on mirVana miRNA Bioarrays (Ambion) by Assuragen Inc (Austin, TX).
Dicer activity assay
Cytoplasmic extracts were prepared as described (6). S100 cytoplasmic extract preparation included an additional centrifugation at 100,000 × g for 2 hours. Protein concentrations were determined with the Micro BCA Assay Kit (Pierce). Assays contained 10pmol of a synthetic pre-miR-122a (Ambion) and 100μg S100 cytoplasmic extract or 1 unit Turbo Dicer enzyme (Genlantis, San Diego CA) in a total volume of 10μL (10mM Tris HCl pH 7.5, 75mM NaCl, 1mM MgCl2, 2.5mM DTT) (6) and were incubated at 37°C for 2 hours. Dicer activity was assessed by quantitative RT-PCR determination of mature miR-122a levels (target sequence UGGAGUGUGACAAUGGUGUUUGU) (Applied Biosystems) relative to recombinant Dicer. The validity of this Dicer activity assay was assessed by serial dilution and demonstrated that Dicer activity could be titrated. This titration series provided a standard curve which showed a linear change in Ct values (from real time RT-PCR) with the units of recombinant Dicer added. The linear range of this titration series covered a 20-fold dilution of recombinant Dicer.
Results and Discussion
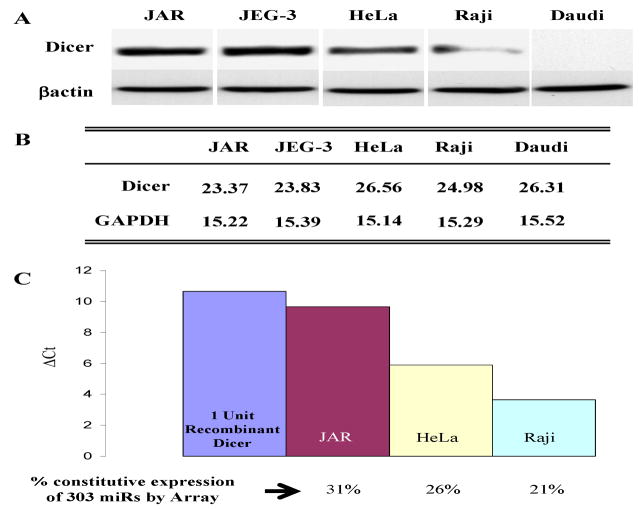
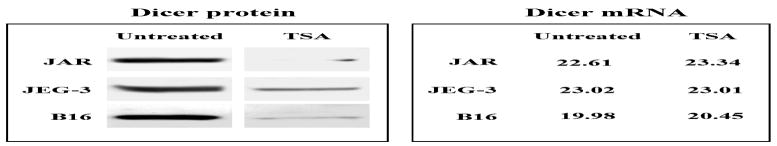
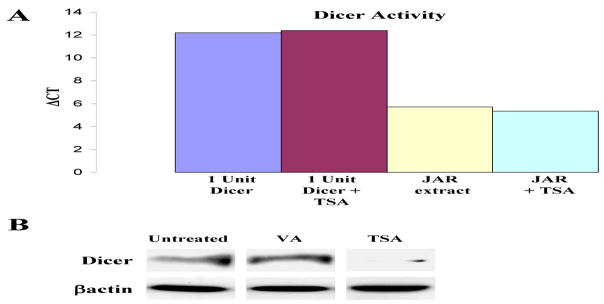
We initially studied three types of human cells varying in their immune gene expression patterns – JAR and JEG-3 trophoblast cells, in which MHC class II and costimulatory genes are silenced; HeLa cervical carcinoma cells, in which the immune genes are constitutively repressed but IFN-γ inducible; and Raji and Daudi B cells which, like normal B cells, have a high constitutive expression of MHC class II and multiple other immune genes (10). Figure 1A illustrates the high levels of Dicer protein in JAR and JEG-3 cells, intermediate levels in HeLa and low to absent in Raji and Daudi cells. As shown in Figure 1B, the cellular Dicer mRNA levels measured by PCR are not well correlated with protein expression. To determine the functional levels of Dicer in JAR cells we used an activity assay (see Materials and Methods) employing a pre-miRNA which, on treatment with S100 cell extracts containing Dicer, was spliced to the mature 22nt species. RT-PCR using primers specific for the 22nt product determined the quantitative levels of Dicer in cell extracts. The Dicer activity of cell extracts was assayed by comparison of the level of the 22nt spliced product with the level generated by recombinant Dicer as shown in Figure 1C. This Figure also illustrates the number of constitutively expressed miRs in each of the cell types determined by miR arrays. Notable are the cell type variations in Dicer levels and the correlation of its expression with the numbers of constitutively expressed miRs in each cell type. These data, although limited, suggest that global differences in Dicer protein expression levels may be reflected in the constitutive levels of cellular miRs. The above studies led us to further examine how Dicer might be regulated and differentially expressed under various cellular and environmental conditions. One possibility is that Dicer is regulated epigenetically. As a first step we tested whether agents which alter chromatin, such as histone deacetylase inhibitors (HDACi), might enhance Dicer expression in cells. However, the HDACi (TSA) minimally altered Dicer message levels and, unexpectedly, substantially inhibited Dicer protein levels in JAR cells. A similar inhibition of Dicer protein with little or no change in mRNA levels was noted in JEG-3 trophoblasts, HeLa and the B16 melanoma (Fig. 2) or in low Dicer cells like Raji (data not shown). TSA could inhibit by directly acetylating Dicer protein and enhance its proteolysis as has been noted for several proteins, for example IRF7 (7). However, immunoprecipitation of Dicer from TSA-treated JAR cell extracts followed by western analysis for acetylated lysines did not detect acetylation of Dicer (data not shown). Moreover, although JAR extracts contained Dicer activity, adding TSA directly to cell extracts containing Dicer did not alter Dicer activity in a functional assay (Fig 3A). TSA also did not alter recombinant Dicer activity. However, cells treated with another HDACi, valproic acid, at concentrations known to activate several repressed immune genes, did not inhibit Dicer levels (Fig 3B). This suggested the possibility that properties of TSA, other than, or in addition to, its deacetylase activity, could be involved. For example in addition to its affect on chromatin TSA can activate cellular stress pathways, including NF-κB, MAPK and PI3K (24, 29, 32, 42) and therefore, TSA’s affect on Dicer expression could possibly be related to activation of stress pathways. Moreover, TSA, via one or more of the above kinase pathways, has been reported to induce ROS via the generation of H2O2 (31, 32). We therefore examined the effect of oxidative stress on Dicer using H2O2 treated JAR cells. Figure 4 illustrates the effect of short treatments (4hrs) with high concentrations (500 μM) of H2O2 and demonstrates selective inhibition of Dicer but not Argonaute1 or other control proteins in JAR cells which is discussed below and in Figure 6. Oncogenes, such as Ras, can also activate MAPK pathways and studies on Ras were seminal in the description of MAPK induced senescent pathways (5). As shown in Figure 4, activated Ras inhibits Dicer protein expression in the non-transformed human fibroblast cell line IMR-90. The phorbol ester PMA is a Ras agonist and, similar to Ras, inhibits Dicer protein expression in IMR-90 human fibroblasts (data not shown) and in JAR (Fig. 4). In each of the above stresses comparison of mRNA levels by qRT-PCR of controls versus treated samples demonstrates a ΔΔCt less than 2 indicating small if any changes in message transcript levels.
Figure 1. Dicer protein and activity are differentially regulated between cell types.
A. Untreated JAR, JEG-3, HeLa, Raji, and Daudi cells were analyzed by western blotting for Dicer and βactin levels. B. Dicer and GAPDH mRNA levels were assessed by quantitative real time RT-PCR for each of the above cell lines and are expressed as CT values. C. Dicer activity was assessed for JAR, HeLa, and Raji cells. Cytoplasmic extract from each cell line or recombinant Dicer enzyme was assayed with a synthetic pre-miR-122a and buffer at 37°C for 2 hours. Activity was determined by quantitative RT-PCR of mature miR-122a levels relative to the recombinant dicer and is presented as Δ CT values. Constitutively expressed miRs were determined by miRNA microarray analyses and are presented as the percentage of miRs constitutively expressed in each cell line.
Figure 2. The HDACi, TSA, downregulates Dicer protein but not message levels in human JAR and JEG-3 trophoblasts and in a mouse B16 melanoma cell line.
JAR (50nM), JEG-3 (250nM), and B16 (100nM) cell lines were treated in vitro with TSA for 24hrs; total RNA was recovered and whole cell lysates were prepared. Western blots for Dicer protein expression and real time RT-PCR analyses (reported as CT values) of Dicer mRNA levels are shown here for treated and untreated samples.
Figure 3. HDACi effects on Dicer activity and expression.
A. Cytoplasmic extract from the JAR cell line or recombinant Dicer enzyme was assayed with 50nM TSA, a synthetic pre-miR-122a, and buffer at 37°C for 2 hours. Activity was determined by quantitative RT-PCR of mature miR-122a levels relative to the untreated samples and is presented as Δ CT values. B. JAR cells were treated with valproic acid (250μM for 24hrs) and TSA (50nM for 24hrs). Proteins were collected and analyzed by western blotting for Dicer and βactin levels.
Figure 4. Multiple stress pathways downregulate Dicer protein.
A. JAR trophoblast cells were treated with 500μM H2O2 for 4hrs, washed, and then cultured for an additional 20 hours. Proteins were collected and analyzed by western blotting for Dicer and βactin levels. B. IMR-90 human fibroblast cells were transduced with either an empty retroviral vector or an activated Ras construct (18). Proteins were collected and analyzed by western blotting for Dicer and βactin levels. C. JAR cells were treated with PMA/IM (500ng/mL PMA for 4hrs followed by the addition of 10mM IM for 24hrs). Proteins were collected and analyzed by western blotting for Dicer and βactin levels.
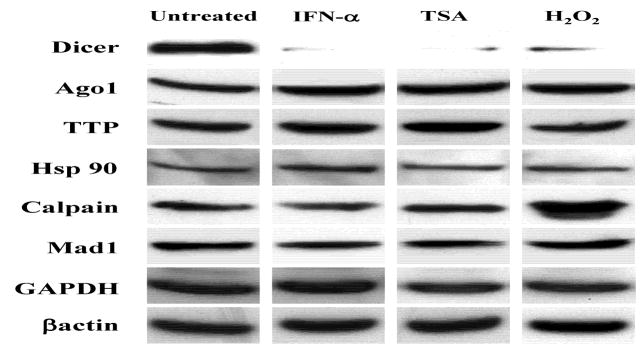
Figure 6. Dicer protein levels are selectively regulated by cellular stresses in JAR trophoblast cells.
Individual JAR whole cell extracts from the designated treatments (1,000U/mL IFN-α for 72hrs, 50nM TSA for 24hrs, and 250μM H2O2 for 15min) were analyzed by western blotting for Dicer, Argonaute 1, TTP, Hsp90, Calpain, Mad 1, GAPDH, and βactin. The results are representative of three independent experiments.
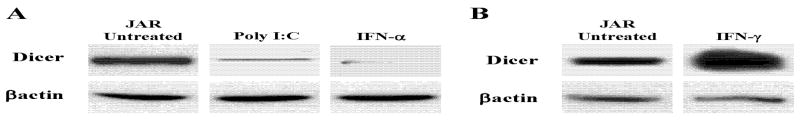
Toll-like receptors (TLRs) are sensors of various environmental stresses, including the dsRNA pathway, that activate Type I interferon, a major mediator of the TLR3 response (1, 14). As shown in Figure 5, dsRNA (poly IC) as well as IFN-α inhibited Dicer protein expression. Additionally, recombinant IFN-α2A and IFN-αrepressed Dicer in JAR cells (data not shown). However, IFN-γ at 500U/ml substantially enhanced its expression in JAR and HeLa cells (Fig. 5B). In B16 mouse cells 100U/ml of IFN-γ enhances Dicer protein levels (data not shown). It seems therefore, that IFN-α/β and several different types of stresses share the common property of repressing Dicer protein expression and that this is largely post-transcriptional, possibly at the level of translation which is known to be a major focus of IFN-α/β regulation (30, 34, 38). MiRs have antiviral activity and Dicer knockdown is reported to enhance the susceptibility of mice to viral infection (28). Since IFN-α is a primary host defense mechanism it is difficult to appreciate how inhibition of Dicer by IFN-α would be a component of antiviral defense. However, as pointed out recently, Dicer contributes to both the production of host antiviral miRs, as well as viral responses which may target Dicer (26).
Figure 5. Toll-like receptor and interferon mediated effects on Dicer expression.
A. JAR cells were treated with poly I:C (2mg/mL for 24hrs), a TLR3 ligand, and IFN-α (1000U/mL for 72hrs), produced by multiple TLR stimuli. Proteins were collected and analyzed by western blotting for Dicer and βactin levels. B. JAR cells were also treated with 500U/mL IFN-γ for 24hrs to compare with IFN-α results. Proteins were collected and analyzed by western blotting for Dicer and βactin levels. Similar results to those shown above were seen in HeLa cells (data not shown).
A fundamental event in the response of organisms to stress and apoptosis is a global shutdown of protein synthesis as a cellular conservation measure, while the expression of a group of ‘survival’ proteins, principally those that have internal ribosome entry sites (IRES), continue to be produced (35). In this regard, multiple types of stresses and apoptotic stimuli globally inhibit translation (13, 41). However, as illustrated in Figure 6, when levels of the cellular proteins are compared there is no significant change in expression. We cannot exclude changes in protein levels that are not seen by the relatively insensitive western analysis.
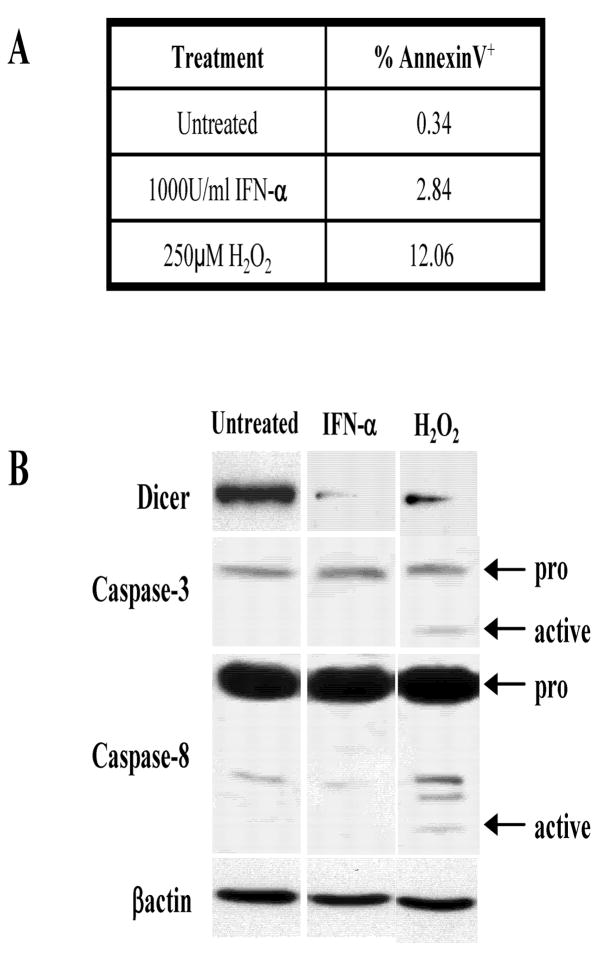
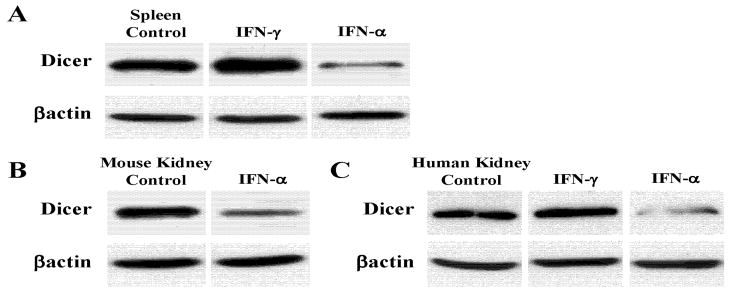
The stresses employed here, depending on the concentration and timing can induce apoptosis. A key issue therefore, is whether the Dicer inhibition reported here is related to apoptotic caspase activity. It is known that caspases cleave multiple proteins (23). However, in JAR cells we found that IFN-α(1000U/ml) treatment that represses Dicer does not induce apoptosis as measured by Annexin V staining (Fig. 7A) and Caspase 3 and 8 activation (Fig. 7B). Moreover the pan-caspase inhibitor ZVAD-FMK does not alter Dicer inhibition by IFN-α (data not shown). Thus, although Dicer levels may be altered in apoptotic cells, the above results indicate that IFN-α can inhibit Dicer in non-apoptotic cells. Additionally, Dicer repression by IFN-α occurs in normal mouse spleen (Fig. 8A) and kidney (Fig. 8B) and Dicer in fresh human kidney tissues is inhibited by IFN-α(Fig. 8C).
Figure 7. Apoptosis is not responsible for downregulation of Dicer with IFN-α treatment.
JAR cells were treated with 1000U/mL IFN-αfor 72hrs. For a positive control, JAR cells were treated with 250μM H2O2 for 15minutes. A. The presence of apoptotic cells was analyzed by Annexin V surface staining of treated cells. Treated JAR cultures were stained with Annexin V-FITC (see Materials and Methods). After single cell gating, a marker was set based on the untreated cells and the percentage of cells stained with Annexin V was determined. B. Activation of the apoptotic signaling cascade was analyzed by SDS-PAGE and western blotting for Dicer, Caspase-3, Caspase-8, and βactin.
Figure 8. Dicer protein levels in fresh tissues of mice and humans.
A. Splenocytes from C57BL/6 mice were treated in culture with 100U/mL IFN-γ for 24hrs or 1000U/mL IFN-αfor 72hrs. Protein was analyzed by SDS-PAGE and western blotting for Dicer and βactin. B. Fresh C57BL/6 kidneys were dissociated and cultured for 72hrs. Cultures were then treated with 1000U/mL IFN-α for 72hrs prior to whole cell lysate preparation and western analysis. C. Normal human kidney cells (obtained with IRB approval as excess tissue post-Pathology examination) were cultured and treated with 500U/mL IFN-γ for 24hrs or 10000U/mL IFN-α for 72hrs. Protein was analyzed by SDS-PAGE and western blotting.
Several issues pertinent to the mechanisms by which Dicer protein levels are controlled have not, as yet, been fully evaluated. As mentioned above, characterization of the 5′UTR of Dicer has defined three non-coding exon1 variants, as well as several alternatively spliced 5′ leader exons, and each exon1 variant uniquely affects translational efficiency and could be responsible, at least in part, for changes in Dicer protein levels in different cells in response to stresses (15, 33). Cell specific regulation of Dicer could also occur via mechanisms involving the Dicer 3′UTR and the binding of miRs. In this regard, recent computational studies have suggested that certain components of the miR pathway, including Dicer, are high probability targets of multiple miRs (2). For example, the miRNA, miR-122a, predicted to target Dicer is known to respond to various cellular stresses (4, 22) and is therefore a candidate repressor of Dicer during stress. Importantly, it is uncertain whether some or all the stresses examined inhibit Dicer by a common mechanism, perhaps involving the induction of Type I interferons, or whether there are pathways specific for each type of stress. Also noteworthy is the variation in protein expression levels between different cells derived from the same tissue, or even the same cells derived from a different freeze down. This type of unexplained experimental variation has been repeatedly observed especially in the response to IFN, especially IFN-α (38).
Type I interferon signals are transduced by multiple complex pathways, including the JAK/Stat, MAPK and PI3K/TOR (37). For example, the dsRNA activated kinase PKR mediates the early activation of a robust antiviral response and the later translational inhibition, which may be accompanied by apoptotic cell death (11). Thus certain stresses may sequentially integrate complex opposing translational responses which, in the above studies, could lead to changes in Dicer and potentially miR expression levels. Type I interferons could, via their effects on Dicer, allow de-repression of a set of survival stress genes. In view of the key role of interferons in immunity, and their systemic and local use in clinical treatment protocols, further exploration focusing on the underlying complex pathways is warranted. It will be important in future studies to identify the specific miRNAs and the genes regulated by Type I and II interferons as cells attempt to adapt to various stresses.
Acknowledgments
We thank the members of the Tomasi laboratory for technical assistance and advice and William J. Magner for reviewing this manuscript. This work was supported by a National Institutes of Health grant HD 17013 and utilized core facilities of Roswell Park Cancer Institute’s NCI Cancer Center Support Grant CA16056.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Akira S, Takeda K. Toll-like receptor signaling. Nature Rev Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 2.Asirvatham AJ, Gregorie CJ, Magner WJ, Hu Z, Tomasi TB. MicroRNA Targets in Immune Genes and the Dicer/Argonaute and ARE Machinery Components. Mol Immunol. 2007;45:1995–2006. doi: 10.1016/j.molimm.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Bhattacharyya SN. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 5.Bihani T, Chicas A, Lo CP-K, Lin AW. Dissecting the Senescence-like Program in Tumor Cells Activated by Ras Signaling. J Biol Chem. 2007;282:2666–2675. doi: 10.1074/jbc.M608127200. [DOI] [PubMed] [Google Scholar]
- 6.Billy E, Brondani V, Zhang H, Müller U, Filipowicz W. Specific interference with gene expression induced by long, double-stranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci (USA) 2001;98:14428–13333. doi: 10.1073/pnas.261562698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Caillaud A, Prakash A, Smith E, Masumi A, Hovanessian AG, et al. Acetylation of interferon regulatory factor-7 by p300/CREB-binding protein (CBP)-associated factor (PCAF) impairs its DNA binding. J Biol Chem. 2002;277:49417–49421. doi: 10.1074/jbc.M207484200. [DOI] [PubMed] [Google Scholar]
- 8.Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, et al. Overexpression of Dicer in Precursor Lesions of Lung Adenocarcinoma. Cancer Res. 2007;67:2345–2350. doi: 10.1158/0008-5472.CAN-06-3533. [DOI] [PubMed] [Google Scholar]
- 9.Chiou TJ. The role of microRNAs in sensing nutrient stress. Plant Cell Environ. 2007;30:323–32. doi: 10.1111/j.1365-3040.2007.01643.x. [DOI] [PubMed] [Google Scholar]
- 10.Chou S-D, Khan ANH, Magner WJ, Tomasi TB. Histone Acetylation Regulates the Cell Type Specific CIITA Promoters, MHC Class II Expression and Antigen Presentation in Tumor Cells. Internatl Immunol. 2005;17:1483–1494. doi: 10.1093/intimm/dxh326. [DOI] [PubMed] [Google Scholar]
- 11.Donzé O, Deng J, Curran J, Sladek R, Picard D, et al. The protein kinase PKR: a molecular clock that sequentially activates survival and death programs. EMBO J. 2004;23:564–571. doi: 10.1038/sj.emboj.7600078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gartel AL, Kandel ES. miRNAs: Little known mediators of oncogenesis. Sem Cancer Biol. 2008;18:103–110. doi: 10.1016/j.semcancer.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 13.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nature Rev Mol Cell Biol. 2005;6:318– 327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 14.Honda K, Yanai H, Takaoka A, Taniguchi T. Regulation of the type I IFN induction: a current view. Internatl Immunol. 2005;17:1367–1378. doi: 10.1093/intimm/dxh318. [DOI] [PubMed] [Google Scholar]
- 15.Irvin-Wilson CV, Chaudhuri G. Alternative initiation and splicing in dicer gene expression in human breast cells. Breast Cancer Res. 2005;7:R563–569. doi: 10.1186/bcr1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karube Y, Tanai H, Osada H, Tomida S, Tatematsu Y, et al. Reduced expression of Dicer associated with poor prognosis in lung cancer patients. Cancer Sci. 2005;96:111–115. doi: 10.1111/j.1349-7006.2005.00015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nature Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 18.Lin AW, Barradas M, Stone JC, van Aelst L, Serrano M, et al. Premature senescence involving p53 and p16 is activated in response to constitutive MEK/MAPK mitogenic signaling. Genes Dev. 1998;12:3008–3019. doi: 10.1101/gad.12.19.3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 21.Magner WJ, Kazim AL, Stewart C, Romano MA, Catalano G, et al. Activation of MHC class I, II, and CD40 gene expression by histone deacetylase inhibitors. J Immunol. 2000;165:701–724. doi: 10.4049/jimmunol.165.12.7017. [DOI] [PubMed] [Google Scholar]
- 22.Marsit CJ, Eddy K, Kelsey KT. MicroRNA responses to cellular stress. Cancer Res. 2006;66:10843–10848. doi: 10.1158/0008-5472.CAN-06-1894. [DOI] [PubMed] [Google Scholar]
- 23.Matskevich AA, Moelling K. Stimuli-dependent cleavage of Dicer during apoptosis. Biochem J. 2008;412:527–534. doi: 10.1042/BJ20071461. [DOI] [PubMed] [Google Scholar]
- 24.Mayo MW, Denlinger CE, Broad RM, Yeung F, Reilly ET, et al. Ineffectiveness of Histone Deacetylase Inhibitors to Induce Apoptosis Involves the Transcriptional Activation of NF-κB through the Akt Pathway. J Biol Chem. 2003;278:18980–18989. doi: 10.1074/jbc.M211695200. [DOI] [PubMed] [Google Scholar]
- 25.Meister G. miRNAs Get an Early Start on Translational Silencing. Cell. 2007;131:25– 28. doi: 10.1016/j.cell.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 26.Müller S, Imler J-L. Dicing with Viruses: MicroRNAs as Antiviral Factors. Immunity. 2007;27:1–3. doi: 10.1016/j.immuni.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 27.Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a Binds the 5′UTR of Ribosomal Protein mRNAs and Enhances Their Translation. Mol Cell. 2008;30:400–471. doi: 10.1016/j.molcel.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 28.Otsuka QJ, Georgel P, New L, Chen J, Mols J, Kang YJ, et al. Hypersusceptibility to Vesicular Stomatitis Virus Infection in Dicer1-Deficient Mice Is Due to Impaired miR24 and miR93 Expression. Immunity. 2007;27:123–134. doi: 10.1016/j.immuni.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 29.Ozaki K, Minoda A, Kishikawa F, Kohno M. Blockade of the ERK pathway markedly sensitizes tumor cells to HDAC inhibitor-induced cell death. Biochem Biophys Res Comm. 2006;339:1171–1177. doi: 10.1016/j.bbrc.2005.11.131. [DOI] [PubMed] [Google Scholar]
- 30.Platanias LC. Mechanisms of type-I and type-II interferon-mediated signaling. Nature Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 31.Rosato RR, Almenara JA, Grant S. The Histone Deacetylase Inhibitor MS-275 Promotes Differentiation or Apoptosis in Human Leukemia Cells through a Process Regulated by Generation of Reactive Oxygen Species and Induction of p21CIP1/WAF1 1. Cancer Res. 2003;63:3637–3645. [PubMed] [Google Scholar]
- 32.Ruefli AA, Ausserlechner MJ, Bernhard D, Sutton VR, Tainton KM, et al. The histone deacetylase inhibitor and chemotherapeutic agent suberoylanilide hydroxamic acid (SAHA) induces a cell-death pathway characterized by cleavage of Bid and production of reactive oxygen species. Proc Natl Acad Sci (USA) 2001;98:10833–10838. doi: 10.1073/pnas.191208598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Singh S, Bevan SC, Patil K, Newton DC, Marsden PA. Extensive Variation in the 5′-UTR of Dicer mRNAs Influences Translational Efficiency. Biochem Biophys Res Comm. 2005;335:643–650. doi: 10.1016/j.bbrc.2005.07.138. [DOI] [PubMed] [Google Scholar]
- 34.Stetson DB, Medzhitov R. Type I Interferons in Host Defense. Immunity. 2006;25:373–382. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 35.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 36.Thomson JM, Newman M, Parker JS, Morin-Kensicki EM, Wright T, et al. Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev. 2006;20:2202–2207. doi: 10.1101/gad.1444406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thyrell L, Hjortsberg L, Arulampalam V, Panaretakis T, Uhles S, et al. Interferon α-induced Apoptosis in Tumor Cells Is Mediated through the Phosphoinositide 3-Kinase/Mammalian Target of Rapamycin Signaling Pathway. J Biol Chem. 2004;279:24152–24162. doi: 10.1074/jbc.M312219200. [DOI] [PubMed] [Google Scholar]
- 38.van Boxel-Dezaire AHH, Rani MRS, Stark GR. Complex Modulation of Cell Type-Specific Signaling in Response to Type I Interferons. Immunity. 2006;25:361–372. doi: 10.1016/j.immuni.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 39.van Rooji E, Sutherland LB, Qi X, Richardson JA, Hill J, et al. Control of Stress-Dependent cardiac growth and gene expression by a micro-RNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 40.Vasudevan S, Tong Y, Steitz JA. Switching from Repression to Activation: MicroRNAs Can Up-Regulate Translation. Science. 2007;318:1931–1934. doi: 10.1126/science.1149460. [DOI] [PubMed] [Google Scholar]
- 41.Yamasaki S, Anderson P. Reprogramming mRNA translation during stress. Curr Opin Cell Biol. 2008;20:222–226. doi: 10.1016/j.ceb.2008.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu C, Friday BB, Lai J-P, McCollum A, Atadja P, et al. Abrogation of MAPK and Akt Signaling by AEE788 Synergistically Potentiates Histone Deacetylase Inhibitor-Induced Apoptosis through Reactive Oxygen Species Generation. Clin Cancer Res. 2007;13:1140–1148. doi: 10.1158/1078-0432.CCR-06-1751. [DOI] [PubMed] [Google Scholar]