Abstract
Our understanding of the origins of new metabolic functions is based upon anecdotal genetic and biochemical evidence. Some auxotrophies can be suppressed by overexpressing substrate-ambiguous enzymes (i.e., those that catalyze the same chemical transformation on different substrates). Other enzymes exhibit weak but detectable catalytic promiscuity in vitro (i.e., they catalyze different transformations on similar substrates). Cells adapt to novel environments through the evolution of these secondary activities, but neither their chemical natures nor their frequencies of occurrence have been characterized en bloc. Here, we systematically identified multifunctional genes within the Escherichia coli genome. We screened 104 single-gene knockout strains and discovered that many (20%) of these auxotrophs were rescued by the overexpression of at least one noncognate E. coli gene. The deleted gene and its suppressor were generally unrelated, suggesting that promiscuity is a product of contingency. This genome-wide survey demonstrates that multifunctional genes are common and illustrates the mechanistic diversity by which their products enhance metabolic robustness and evolvability.
Keywords: catalytic promiscuity, directed evolution, multicopy suppression, substrate ambiguity
Introduction
Our goal is to understand the origins of novel catalytic functions. This process underlies the evolution of new metabolic pathways and the adaptive speciation of microorganisms (Hegeman and Rosenberg 1970). New genes are primarily produced by duplication (Ohno 1970; Force et al. 1999); however, the relationship between gene duplication and the emergence of new biochemical functions remains incompletely understood (Roth et al. 2007). Questions about origins underlie those about evolution, but they cannot be addressed in the same way. Protein sequence comparisons can reveal phylogenetic relationships and suggest the occurrence of adaptive evolution. Recent advances in covariation analysis can highlight specific relationships between protein sequences, structures, and functions (Suel et al. 2003; Merlo et al. 2007). Biochemical techniques, including site-directed mutagenesis and reconstructive gene synthesis, can also be applied to test hypotheses about adaptive evolution experimentally (Gaucher et al. 2003; Thomson et al. 2005; Bridgham et al. 2006).
Comparisons of extant proteins, however, generally reveal little about the emergence of new catalytic functions. Many extant activities are ancient and their origins apparently precede the last universal ancestor. The predominance of functionally neutral mutations (Kimura 1983), the complicated nature of “fitness” in situ, and the complexity of protein structure/function relationships all tend to obscure the earliest stages of an enzyme’s evolutionary history. Nevertheless, novel catalytic functions continue to emerge in the biosphere, such as when bacteria are challenged with a novel nutrient or a toxic molecule. An appealing hypothesis posits that the evolutionary starting point for these functions is the broad specificity and/or secondary activities of existing protein scaffolds (Jensen 1976; D’Ari and Casadesús 1998; Khersonsky et al. 2006). Indeed, it has long been recognized that many proteins retain the ability to catalyze identical chemical transformations on numerous substrates (substrate ambiguity). Some enzymes also display catalytic promiscuity in vitro; that is, they possess the ability to catalyze different transformations on one or more substrates (O’Brien and Herschlag 1999). These properties could serve as the molecular basis of contingency, which describes instances when “a feature evolved long ago for a different use has fortuitously permitted survival during a sudden and unpredictable change in rules” (Gould 1989).
We speculated that ambiguity and promiscuity are commonplace, even within the small proteome of a well-characterized organism such as Escherichia coli. Further, we hypothesized that adaptation routinely begins with the overexpression of ambiguous and/or promiscuous enzymes. Anecdotal evidence in support of this hypothesis includes early experiments showing that the enzymes of fucose metabolism could be recruited for the catabolism of l-1,2-propanediol (Cocks et al. 1974) and d-arabinose (St Martin and Mortlock 1976). Campbell, Hall, and Hartl also demonstrated that a repressor mutation enabling constitutive expression of a cryptic β-galactosidase, Ebg, was critical for its ability to suppress E. coli lacZ mutants (Hall 2003). Berg et al. (1998) demonstrated that avtA on a high copy number plasmid was able to suppress ilvE mutations but that a single chromosomal copy could not. More recently, Miller and Raines screened a library of E. coli genomic fragments and identified 4 glycokinases that were able to complement a glucokinase auxotroph, provided they were overexpressed using a strong T7 promoter (Miller and Raines 2004, 2005). If the evolutionary mechanisms that create new catalytic functions are similar to those that recreate old ones, auxotrophy suppression experiments emulate the responses of asexual bacterial populations to xenobiotic nutrients or toxins.
In this study, we have conducted a systematic survey to demonstrate the prevalence of ambiguity and promiscuity within the E. coli proteome, using 2 recently constructed functional genomics tools. The first is the Keio collection, which comprises 3,985 single-gene knockout strains (Baba et al. 2006). The second is the A Complete Set of E. coli K-12 Open Reading Frame Archive (ASKA) library, which contains every E. coli open reading frame (ORF), each individually cloned into the expression vector pCA24N (Kitagawa et al. 2005). By transforming each conditional Keio auxotroph with every clone from the ASKA library, we provide experimental evidence that the E. coli genome contains a large and diverse reservoir of multifunctional suppressors, which can be readily accessed via regulatory mutations or gene amplification events.
Materials and Methods
Escherichia coli Strains
The Keio knockout and green fluorescent protein (GFP)-tagged ASKA collections were purchased from Hirotada Mori (Nara Institute of Science and Technology, Nara, Japan). These resources were constructed as part of the National BioResource Project (National Institute of Genetics, Shizuoka, Japan) and have been described elsewhere (Kitagawa et al. 2005; Baba et al. 2006). The ASKA library includes duplicates of many recently reannotated ORFs; in total, it contains 5,272 clones.
Identification of Conditionally Essential Genes
Two recent reports have assessed the growth of every Keio strain in glucose-supplemented 3-cCmorpholinopropa-nesulfonic acid medium (Baba et al. 2006) and in glycerol-supplemented M9 medium (Joyce et al. 2006). By combining the results of these studies, we derived a list of 155 strains that we predicted would be unable to form colonies on M9-glucose. Each strain was cultured in Luria-Bertani (LB) medium (0.4 ml) supplemented with kanamycin (Kan; 30 μg ml-1). After overnight growth at 37 °C, each saturated culture was washed and resuspended to a final OD600 ≈ 0.1 with sterile M9 salts (1× concentration; Difco, Sparks, MD). A 3-μl aliquot of the resuspended cells was streaked on M9-glucose medium, which comprised M9 salts (1×), agar (1.5%, w/v), MgSO4 (2 mM), CaCl2 (0.1 mM), glucose (0.4%, w/v), and Kan. Plates were incubated at 37 °C for up to 7 days, and colony formation was monitored.
Selection of Rescuing Activities
Aliquots (25 μl) of the 5,272 clones in the ASKA library were mixed, and plasmid DNA (pCA24N-[ASKA]) was isolated from the resulting pool. Deletion strains that failed to grow on M9-glucose were made electrocompetent and transformed with pCA24N-[ASKA] DNA. Transformed cells were washed and resuspended in M9 salts (1×), then spread on M9-glucose-isopropyl-β-d-thiogalactoside (IPTG) medium. The latter was M9-glucose as described above, further supplemented with IPTG (50 μM) and chloramphenicol (Cam; 34 μg ml-1). An average (median) of 30,500 transformed cells (lower quartile = 21,400 and upper quartile = 50,500) were plated in each experiment, thereby ensuring that >95% of the ASKA library was represented (Firth and Patrick 2005). Plates were placed in airtight containers with moistened paper towels (to prevent overdrying) and incubated at 37 °C for 4-5 weeks.
Identification of Rescuing Clones
Colonies that appeared on the M9-glucose-IPTG selection plates were used to inoculate LB-Cam-Kan medium. After overnight growth at 37 °C, a 0.5-μl aliquot of each saturated culture was used as the source of template DNA for PCR, using the pCA24N-specific primers pCA24N.for (5′-GAT AAC AAT TTC ACA CAG AAT TCA TTA AAG AG) and pCA24N.rev (5′-CCC ATT AAC ATC ACC ATC TAA TTC AAC). Each rescuing clone was identified by sequencing the amplified product using primer pCA24N.for.
Retransformation Test
The rescuing activity of each selected clone was verified by retransformation. Plasmid DNA was isolated from the corresponding member of the ASKA library using EconoSpin plasmid mini-prep columns (Epoch Biolabs, Houston, TX). The purified plasmid was used to transform a fresh aliquot of the appropriate deletion strain by electroporation. Approximately 100-1,000 transformed cells were washed and resuspended in M9 salts (1×), then spread on M9-glucose-IPTG medium. A negative control plasmid (pCA24N-lacZ) was used to transform the same strain, and growth at 37 °C was monitored.
Pairwise Structural Alignments
The MAMMOTH algorithm (Ortiz et al. 2002) and the DBAli database (http://salilab.org/DBAli/; Pieper et al. 2006) were used to superimpose the structure of each deleted protein upon that of its non-cognate suppressor. When an experimentally determined structure was not available, a homology model from ModBase (http://modbase.compbio.ucsf.edu/modbase-cgi/index.cgi; Pieper et al. 2006) was used for the structural alignment.
Results and Discussion
High-Throughput Selection of Multicopy Suppressors
The objective of this study was to identify enzymes that evince biologically relevant substrate ambiguity or catalytic promiscuity. The first step was to develop a high-throughput screen for multifunctional genes derived from the E. coli genome. To begin, we identified 107 strains from the Keio knockout collection (each with a single-gene deletion) that were unable to form colonies when streaked on M9-glucose. Three strains (ΔguaA, Δlpd, and ΔthyA) were subsequently excluded because they could not be transformed efficiently by plasmid DNA. Most of the remaining 104 strains contained deletions in genes of central metabolism, including 62 in amino acid metabolism (3 of which encode regulatory proteins), 20 in cofactor biosynthesis, and 17 in nucleotide biosynthesis (see supplementary tables 1 and 2, Supplementary Material online). The failure of these conditional auxotrophs to grow on M9-glucose implies that each is suffering from a severe metabolic imbalance.
The ASKA ORF library was constructed by individually cloning every protein-coding E. coli gene; each ORF is overexpressed as a fusion protein with N-terminal (His)6 and C-terminal GFP tags (Kitagawa et al. 2005). We propagated each of the 5,272 clones to saturation in 96-well microplates, pooled equal volumes of each culture and isolated plasmid DNA (presumably containing genes that could rescue the 104 auxotrophs) from the pool. We reasoned that the deletion of an essential gene from the chromosome of a Keio strain would lead to the accumulation of an “orphaned” substrate for the corresponding enzyme, perhaps to concentrations in excess of 1 mM within the cytoplasm (∼2 fL total volume). Concomitant overexpression of a native protein from the ASKA library (concentration >1 μM) would then facilitate the discovery of weak but physiologically relevant catalytic activities.
Each of the 104 Keio strains was transformed with the pooled plasmids of the ASKA library, plated on M9-glucose (supplemented with IPTG to induce the overexpression of each ASKA-encoded gene), and incubated at 37 °C. Any colony that appeared in 4-5 weeks was analyzed by PCR and DNA sequencing to identify the ASKA clone responsible for suppression. In 92 selections (88%), the plasmid-borne copy of the chromosomally deleted gene could effect colony formation. This is consistent with a previous study, in which 61% of the clones in the ASKA library were soluble when overexpressed and purified (Arifuzzaman et al. 2006). The implication is that only a minority of ASKA clones are inactivated by fusion to an N-terminal (His)6 tag and/or a C-terminal GFP tag.
As expected, the observed “self-complementation” was usually rapid, with colonies appearing in 2-5 days. Slower growing self-complementers included expression from pCA24N-pdxH in the ΔpdxH strain (9 days to form colonies); bioF/ΔbioF, cysD/ΔcysD, and cysN/ΔcysN (each 11 days to form colonies); metA/ΔmetA (16 days); and glnA/ΔglnA (29 days). In the latter strain, colonies expressing AsnB (asparagine synthetase) actually appeared more rapidly (20 days) than those expressing the self-complementer, suggesting that the tagged version of GlnA (glutamine synthetase) possesses substantially reduced catalytic activity. However, the primary focus of this study was the numerous noncognate suppressors that were also discovered (see below).
Retransformation Tests
To confirm the rescue phenotypes, plasmid DNA corresponding to each suppressor was purified from the (un-pooled) ASKA collection. Naïve cells of the rescued Keio strain were transformed with the clonal plasmid preparations, and the growth of transformants on M9-glucose-IPTG plates was monitored. These retests were designed to focus this study on monogenic, cell-autonomous suppressors. Complex (multigene and/or multicell) suppression mechanisms are important but were excluded from this initial survey. For example, proY, which is annotated as a cryptic proline transporter (Keseler et al. 2005), was selected in experiments with the ΔhisB, ΔhisH, and ΔhisI strains. However, it did not suppress these strains when retested in the absence of neighboring cells expressing other ASKA-encoded proteins. ProY apparently catalyzes histidine transport in E. coli (rather than proline), but ProY-expressing cells need an extracellular source of histidine (e.g., from the ΔhisB strain harboring pCA24N-hisB) to enable cell survival.
The retransformation tests also eliminated instances of chromosomal suppression from our analysis. The minimal medium used for retesting contained Cam, which ensured that only cells containing the plasmid-encoded multicopy suppressor were viable. A small population of transformed cells (typically 100-1,000; median = 640) was plated, to ensure that low-frequency chromosomal mutation events were unlikely to be observed. In general, 100% of the plated transformants formed colonies. In a few exceptional cases, smaller numbers of colonies arose at uniform growth rates across the plate. Each strain was transformed with pCA24N-lacZ, and a similar number of transformants were spread on a separate M9-glucose-IPTG plate; de novo chromosomal suppressors were never observed under these selection conditions. Overall, 55% of the putative suppressors passed this stringent retesting process.
Diverse Mechanisms of Multicopy Suppression
Remarkably, the retransformation tests confirmed that 21 of the 104 strains (20%) could be rescued by ASKA plasmids encoding a noncognate gene. Each of these strains was rescued by 1-4 ASKA clones; in total, 41 examples of suppression were identified (table 1). Most suppressors displayed specificity in their action: isogenic strains, each with deletions in the same metabolic pathway, were rescued by a common suppressor in only 3 cases (overexpression of avtA suppressed the ΔilvD and ΔilvE lesions, menF suppressed ΔpabA and ΔpabB, and yneH suppressed ΔserA and ΔserC). Growth rates were highly variable, with colonies taking 2-28 days to form (median = 6 days; supplementary table 2, Supplementary Material online). In 6 cases, growth did not proceed beyond pinprick-sized colonies, but all rescued strains grew significantly faster than the corresponding negative control (i.e., the same strain expressing β-galactosidase from pCA24N-lacZ).
Table 1. Multicopy Suppressors Rescue 21 Conditional Auxotrophs, via 8 Proposed Mechanisms.
| Strain | Protein Deficiency | MS | Rescuing Gene Product |
|---|---|---|---|
| Isozyme overexpression | |||
| ΔglyA | Glycine hydroxymethyltransferase | ltaE | l-allo-threonine aldolase |
| ΔilvA | Threonine dehydratase (biosynthetic) | tdcB | Threonine dehydratase (catabolic) |
| ΔilvE | Branched-chain amino acid transferase | avtA | Valine-pyruvate AT |
| ΔmetC | Cystathionine β-lyase/cysteine desulfhydrase | malY | Cystathionine β-lyase/maltose regulon repressor |
| Substrate ambiguity | |||
| ΔglnA | Glutamine synthetase | asnB | Asparagine synthetase |
| ΔpdxB | Erythronate-4-phosphate dehydrogenase | tdh | Threonine dehydrogenase |
| ΔserB | Phosphoserine phosphatase | gph | Phosphoglycolate phosphatase |
| ΔserB | Phosphoserine phosphatase | hisB | IGP dehydratase/histidinol phosphate phosphatase |
| ΔserB | Phosphoserine phosphatase | ytjC | Predicted bisphosphatase; uncharacterized |
| Ambiguity in metabolite transport | |||
| Δfes | Enterochelin esterase | setB | Predicted lactose/IPTG efflux transporter |
| ΔilvA | Threonine dehydratase (biosynthetic) | emrD | Multidrug efflux transporter |
| Δppc | Phosphoenolpyruvate carboxylase | ecfM | Predicted extracytoplasmic factor; uncharacterized |
| Δppc | Phosphoenolpyruvate carboxylase | yccT | Predicted outer membrane protein; uncharacterized |
| ΔptsI | Carbohydrate PTS system, enzyme I | fucP | Fucose transporter |
| ΔptsI | Carbohydrate PTS system, enzyme I | xylE | Xylose transporter |
| Catalytic promiscuity | |||
| Δfes | Enterochelin esterase | thiL | Thiamine monophosphate kinase |
| ΔmetC | Cystathionine β-lyase/cysteine desulfhydrase | alr | Alanine racemase |
| ΔpabA | Aminodeoxychorismate synthase, small subunit | menF | Isochorismate synthase |
| ΔpabB | Aminodeoxychorismate synthase, large subunit | menF | Isochorismate synthase |
| ΔpdxB | Erythronate-4-phosphate dehydrogenase | purF | Glutamine PRPP amidotransferase |
| ΔpurK | Carboxyaminoimidazole ribonucleotide synthetase | anmK | Anhydro-N-acetylmuramic acid kinase |
| Maintaining metabolic flux | |||
| ΔilvD | Dihydroxy-acid dehydratase | avtA | Valine-pyruvate AT |
| ΔpurK | Carboxyaminoimidazole ribonucleotide synthetase | purE | Carboxyaminoimidazole ribonucleotide mutase |
| Metabolic pathway bypass and/or alternate source of downstream substrate | |||
| ΔcarA | Carbamoyl phosphate synthase, small subunit | carB | Carbamoyl phosphate synthase, large subunit |
| ΔglyA | Glycine hydroxymethyltransferase | tdh | Threonine dehydrogenase |
| ΔglyA | Glycine hydroxymethyltransferase | yneH | Predicted glutaminase; uncharacterized |
| ΔhisH | IGP synthase; small subunit | hisF | IGP synthase; large subunit |
| ΔpabA | Aminodeoxychorismate synthase, small subunit | pabB | Aminodeoxychorismate synthase, large subunit |
| ΔptsI | Carbohydrate PTS system, enzyme I | galE | UDP-glucose 4-epimerase |
| ΔserA | Phosphoglycerate dehydrogenase | yneH | Predicted glutaminase; uncharacterized |
| ΔserC | Phosphoserine AT/phosphohydroxythreonine AT | yneH | Predicted glutaminase; uncharacterized |
| Regulatory effects | |||
| ΔcarA | Carbamoyl phosphate synthase, small subunit | ygiT | Predicted transcriptional regulator; uncharacterized |
| ΔglyA | Glycine hydroxymethyltransferase | rsd | Anti-sigma factor |
| ΔmetC | Cystathionine β-lyase/cysteine desulfhydrase | fimE | Recombinase for fimbrial phase switching |
| ΔmetR | Transcriptional activator of methionine biosynthesis | metE | Homocysteine transmethylase |
| ΔpurK | Carboxyaminoimidazole ribonucleotide synthetase | cpdA | Cyclic adenosine monophosphate phosphodiesterase |
| Unknown | |||
| ΔcarA | Carbamoyl phosphate synthase, small subunit | cho | Stress (SOS)-induced endonuclease for DNA repair |
| ΔcarA | Carbamoyl phosphate synthase, small subunit | yncK | Predicted transposase; uncharacterized |
| ΔpurF | Glutamine PRPP amidotransferase | chbA | Carbohydrate phosphotransferase system, enzyme IIChb |
| ΔyhhK | Putative acetyltransferase | murI | Glutamate racemase |
| ΔyhhK | Putative acetyltransferase | purF | Glutamine PRPP amidotransferase |
Note.-MS, multicopy suppressor; AT, aminotransferase; IGP, imidazole glycerol phosphate; PRPP, phosphoribosylpyrophosphate; UDP, uridine diphosphate.
The suppressors in table 1 are grouped according to their proposed mechanisms of rescue, which were assigned using the EcoCyc database of E. coli genes and metabolic pathways (Keseler et al. 2005). To our knowledge, only 8 of the 41 cases had been described previously. These included suppression of the ΔglyA, ΔilvA, ΔilvE, and ΔmetC auxotrophies by expression of the known isozymes LtaE (Liu et al. 1998), TdcB (Datta et al. 1987), AvtA (Berg et al. 1988), and MalY (Awano et al. 2005), respectively. The ΔglyA mutation was also suppressed by tdh, consistent with a previous observation that glyA mutants can be rescued in an epistatic, 2-step pathway catalyzed by the tdh and kbl gene products (Aronson et al. 1989). MetR normally activates transcription of metE (Maxon et al. 1989), so it was reassuring that IPTG-induced expression of MetE rescued ΔmetR. Finally, PurK deficiency could be complemented by overexpression of PurE, which follows PurK in the purine biosynthetic pathway. Depletion of the PurK reaction product (due to PurE-catalyzed turnover) perturbs the equilibrium in vivo and favors its spontaneous, nonenzymatic formation (Firestine et al. 1994). Overall, these rediscoveries confirmed that the ASKA and Keio clones behave like regular expression vectors and deletion mutants, respectively, and that our less intuitive results cannot be ascribed to any systematic peculiarity in plasmid or strain construction.
In 3 further experiments, overexpression of the large subunit of a protein complex (CarB, HisF, and PabB) rescued strains in which the small subunit was deleted (ΔcarA, ΔhisH, and ΔpabA). In each case, the large subunit is a synthase, whereas the small subunit possesses glutaminase activity. It has been reported previously that each large subunit is able to function (albeit inefficiently) using exogenous ammonia, rather than the ammonia generated by small subunit-catalyzed glutamine hydrolysis (Rubino et al. 1987; Ye et al. 1990; Klem and Davisson 1993). These results therefore reflected the ready availability of free ammonia from the M9 salts of the selection medium.
Suppression Does Not Imply Homology
It is becoming clear that the traditional “one enzyme, one substrate” view of enzyme specificity does not reflect biological reality (D’Ari and Casadesús 1998; O’Brien and Herschlag 1999; Khersonsky et al. 2006). Substrate ambiguity and catalytic promiscuity can provide starting points for the evolution of new catalytic functions, both in nature and in the laboratory. Studies of adaptive molecular evolution are often guided by structural similarities and differences among enzyme family and superfamily members (Orengo and Thornton 2005; Glasner et al. 2007). Therefore, we assessed structure-based superimpositions of each deleted protein with its multifunctional suppressor (input chains for each alignment are listed in supplementary table 3, Supplementary Material online). Significant structural similarity was only detected for 6 of the 41 protein pairs (table 2), 3 of which were pairs of isozymes. In the remainder of cases, the deleted protein and its suppressor were not structural homologues.
Table 2. All Instances of Structural Homology between Deleted Proteins and Their Suppressors (out of 41 cases; table 1).
| Deleted Protein | Rescuing Protein | P Value* |
|---|---|---|
| IlvA | TdcB | 31.3 |
| GlyA | LtaE | 30.7 |
| PabB | MenF | 28.7 |
| MetC | MalY | 22.9 |
| SerB | Gph | 15.1 |
| SerB | HisB | 9.1 |
A P value > 5.2 from the alignment was considered evidence of homology (Ortiz et al. 2002).
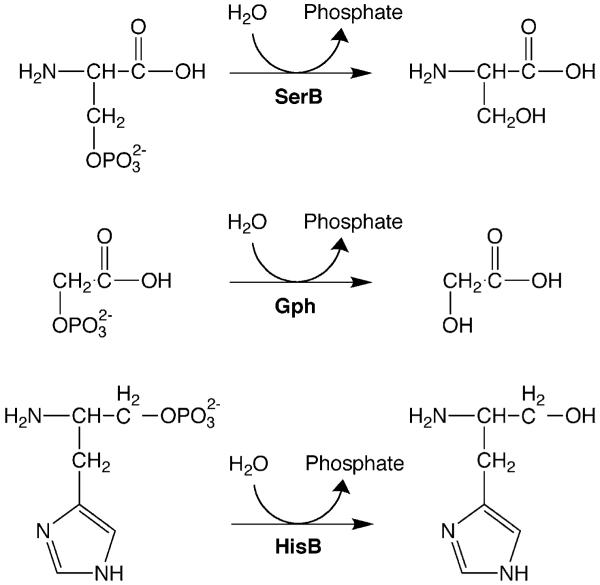
The phosphoserine phosphatase, SerB, provides an illustrative example. The ΔserB strain was rescued by overexpression of Gph (phosphoglycolate phosphatase), HisB (histidinol phosphate phosphatase), and YtjC. In vitro, the homologous haloacid dehalogenase (HAD)-like phosphatases SerB and Gph possess overlapping specificities (Kuznetsova et al. 2006), and in vivo, their metabolic substrates are similar (fig. 1). We might therefore have predicted this result. The ΔserB mutation was also suppressed by a third HAD-like phosphatase, HisB, even though this enzyme showed no activity in vitro (Kuznetsova et al. 2006) and acts on a considerably bulkier substrate than phosphoserine in histidine biosynthesis (fig. 1). This result validated our application of highly sensitive genetic selections, rather than in vitro assays, to evaluate promiscuous activities. Most unpredictably of all, the ΔserB strain was rescued by YtjC, which is not a HAD-like phosphatase, but which instead possesses limited sequence similarity to fructose-2,6-bisphosphatases. Moreover, cells expressing YtjC formed colonies more rapidly (3 days) than those expressing the most “logical” suppressor, Gph (5 days).
Fig. 1.
Phosphatase substrate ambiguity. SerB is the phosphoserine phosphatase that catalyzes the final step in the biosynthesis of serine from 3-phosphoglycerate (top). Deletion of serB can be rescued by expression of 2 other HAD-like phosphatases: Gph, which hydrolyzes the phosphate monoester 2-phosphoglycolate (middle), and HisB, which ordinarily catalyzes the phosphorolysis of histidinol phosphate (bottom).
In addition to enzymes, we also discovered examples in which an overexpressed transport protein displayed substrate ambiguity (table 1). In rescuing the ΔptsI strain, for example, it seems likely that the fucose and xylose transporters, FucP and XylE, are importing glucose instead and thereby enabling glucokinase-mediated cell survival in the absence of a functioning carbohydrate phosphotransferase system. The substrate ambiguity of transporter proteins is of practical utility as it offers a means to introduce novel feedstocks or pharmaceutical precursors into the cytoplasmic compartment.
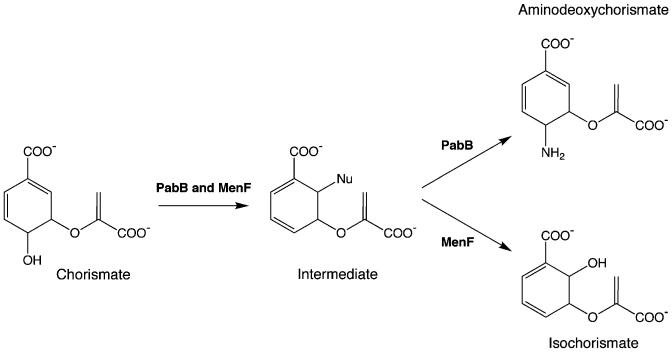
Catalytic promiscuity might be expected to occur less frequently than substrate ambiguity as it is predicated upon a finer degree of molecular recognition—between the enzyme and 2 or more potentially unrelated transition states. Nevertheless, 6 strains were rescued by catalytically promiscuous enzymes (table 1). These examples run the gamut from recently diverged enzymes that catalyze shared part reactions (PabB and MenF; fig. 2) to nonhomologous enzymes that both utilize the cofactor pyridoxal 5′-phosphate (MetC and Alr) and to enzymes in which the promiscuous activity appears to be completely serendipitous (PdxB and PurF). None of these results were particularly intuitive. Even in the case of promiscuous homologues, PabB (aminodeoxychorismate synthase) appears mechanistically more similar to TrpE (anthranilate synthase) than to the isochorismate synthase, MenF (He et al. 2004). Moreover, entC, encoding a second E. coli isochorismate synthase, was not identified in the ΔpabB selection experiment. These results demonstrate that even homologous isozymes (such as MenF and EntC) can show important differences in their promiscuous activities. Previous workers have demonstrated that mutating enzymes to increase their catalytic promiscuity can rescue otherwise auxotrophic E. coli (Jürgens et al. 2000; Schmidt et al. 2003). The promiscuous activity of a purified wild-type enzyme is usually orders of magnitude less than its primary activity (O’Brien and Herschlag 1999). Here, we show than these weak activities are nevertheless sufficient to overcome a number of chemically distinct metabolic imbalances in vivo.
Fig. 2.
Catalytic promiscuity among chorismate-utilizing enzymes. PabB catalyzes the formation of 4-amino-4-deoxychorismate; the ΔpabB strain can be rescued by expression of its homologue, MenF (isochorismate synthase). The 2 enzymes act via a common intermediate, generated in an SN2′ mechanism that leads to addition of a nucleophile at C2, with concomitant elimination of the C4 hydroxyl group (He et al. 2004). In PabB, the nucleophile is the ∊-amino group of an active site lysine; the result is a covalent intermediate. In MenF, the nucleophile is assumed to be water, although its isozyme, EntC, can also utilize ammonia. Our results suggest that product release by MenF is sufficiently slow that ammonia can enter the active site and effect a second SN2′ displacement, yielding aminodeoxychorismate in quantities that enable cell survival in the absence of PabB.
Implications for Directed Evolution
Our results demonstrate that the ambiguous and promiscuous activities of proteins are often products of contingency. Homology-based site-directed mutagenesis and directed evolution can be used to test evolutionary hypotheses, but the functional interconversion of paralogs remains challenging (Jürgens et al. 2000; Hedstrom 2002; Schmidt et al. 2003). In contrast, examples such as the suppression of ΔserB by ytjC suggest that functional convergence of otherwise unrelated protein scaffolds can be readily achieved. The ASKA library encodes a vast assortment of protein folds and offers an unbiased starting point for directed evolution. ORF library-based evolution experiments are also intrinsically less contrived than those based on semirationally chosen single genes and should provide greater insight into natural adaptive processes.
Conclusion
We have conducted the first quantitative survey of enzymatic ambiguity and promiscuity. Even under stringent selection conditions, many genetic lesions could be suppressed by the overexpression of preexisting genes. We have used the EcoCyc database to propose possible mechanisms by which the suppressors act (table 1); future studies will explore specific examples in greater biochemical and genetic detail. In spite of the comprehensive annotation of the E. coli genome, the majority of these secondary activities were previously undiscovered. It has been postulated that, compared with extant enzymes, primitive catalysts possessed very broad specificity and that gene duplication afforded the “luxury of increased specialization and improved metabolic efficiency” (Jensen 1976). Here we have demonstrated that contemporary enzymes remain surprisingly broad in reaction specificity. This general property of proteins enhances metabolic robustness and generates the seeds of evolutionary innovation, as it has since primordial times.
Supplementary Material
Acknowledgments
We thank P. Mars for technical assistance and J. Reed for helpful discussions. We also thank J. Bull, J. Gallivan, M. Gerth, M. Hecht, T. Schlenke, and D. Tawfik for their critiques of the manuscript. This study was supported by grants from the National Institutes of Health (R01 GM074264) and the National Science Foundation (CHE-0404677). W.M.P. and I.M. conceived the project and wrote the paper. W.M.P. and E.M.Q. designed the protocols for screening and selection, conducted the selection experiments, and analyzed the data. D.B.S. screened for usable Keio strains and W.M.P. conducted the retransformation tests. W.M.P. performed the pairwise structural alignments.
Footnotes
Laura Katz, Associate Editor
Literature Cited
- Arifuzzaman M, Maeda M, Itoh A, et al. Large-scale identification of protein-protein interaction of Escherichia coli K-12. Genome Res. 2006;16:686–691. doi: 10.1101/gr.4527806. 23 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson BD, Somerville RL, Epperly BR, Dekker EE. The primary structure of Escherichia colil-threonine dehydrogenase. J Biol Chem. 1989;264:5226–5232. [PubMed] [Google Scholar]
- Awano N, Wada M, Mori H, Nakamori S, Takagi H. Identification and functional analysis of Escherichia coli cysteine desulfhydrases. Appl Environ Microbiol. 2005;71:4149–4152. doi: 10.1128/AEM.71.7.4149-4152.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg CM, Wang MD, Vartak NB, Liu L. Acquisition of new metabolic capabilities: multicopy suppression by cloned transaminase genes in Escherichia coli K-12. Gene. 1988;65:195–202. doi: 10.1016/0378-1119(88)90456-8. [DOI] [PubMed] [Google Scholar]
- Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312:97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- Cocks GT, Aguilar T, Lin EC. Evolution of l-1,2-propanediol catabolism in Escherichia coli by recruitment of enzymes for l-fucose and l-lactate metabolism. J Bacteriol. 1974;118:83–88. doi: 10.1128/jb.118.1.83-88.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Ari R, Casadesús J. Underground metabolism. Bioessays. 1998;20:181–186. doi: 10.1002/(SICI)1521-1878(199802)20:2<181::AID-BIES10>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Datta P, Goss TJ, Omnaas JR, Patil RV. Covalent structure of biodegradative threonine dehydratase of Escherichia coli: homology with other dehydratases. Proc Natl Acad Sci USA. 1987;84:393–397. doi: 10.1073/pnas.84.2.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firestine SM, Poon SW, Mueller EJ, Stubbe J, Davisson VJ. Reactions catalyzed by 5-aminoimidazole ribonucleotide carboxylases from Escherichia coli and Gallus gallus: a case for divergent catalytic mechanisms. Biochemistry. 1994;33:11927–11934. doi: 10.1021/bi00205a031. [DOI] [PubMed] [Google Scholar]
- Firth AE, Patrick WM. Statistics of protein library construction. Bioinformatics. 2005;21:3314–3315. doi: 10.1093/bioinformatics/bti516. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaucher EA, Thomson JM, Burgan MF, Benner SA. Inferring the palaeoenvironment of ancient bacteria on the basis of resurrected proteins. Nature. 2003;425:285–288. doi: 10.1038/nature01977. [DOI] [PubMed] [Google Scholar]
- Glasner ME, Gerlt JA, Babbitt PC. Mechanisms of protein evolution and their application to protein engineering. Adv Enzymol Relat Areas Mol Biol. 2007;75:193–239. doi: 10.1002/9780471224464.ch3. [DOI] [PubMed] [Google Scholar]
- Gould SJ. Wonderful life: the burgess shale and the nature of history. W W Norton and Company; New York: 1989. [Google Scholar]
- Hall BG. The EBG system of E. coli: origin and evolution of a novel β-galactosidase for the metabolism of lactose. Genetica. 2003;118:143–156. [PubMed] [Google Scholar]
- He Z, Stigers Lavoie KD, Bartlett PA, Toney MD. Conservation of mechanism in three chorismate-utilizing enzymes. J Am Chem Soc. 2004;126:2378–2385. doi: 10.1021/ja0389927. [DOI] [PubMed] [Google Scholar]
- Hedstrom L. Serine protease mechanism and specificity. Chem Rev. 2002;102:4501–4524. doi: 10.1021/cr000033x. [DOI] [PubMed] [Google Scholar]
- Hegeman GD, Rosenberg SL. The evolution of bacterial enzyme systems. Annu Rev Microbiol. 1970;24:429–462. doi: 10.1146/annurev.mi.24.100170.002241. [DOI] [PubMed] [Google Scholar]
- Jensen RA. Enzyme recruitment in evolution of new function. Annu Rev Microbiol. 1976;30:409–425. doi: 10.1146/annurev.mi.30.100176.002205. [DOI] [PubMed] [Google Scholar]
- Joyce AR, Reed JL, White A, Edwards R, Osterman A, Baba T, Mori H, Lesely SA, Palsson BO, Agarwalla S. Experimental and computational assessment of conditionally essential genes in Escherichia coli. J Bacteriol. 2006;188:8259–8271. doi: 10.1128/JB.00740-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jürgens C, Strom A, Wegener D, Hettwer S, Wilmanns M, Sterner R. Directed evolution of a (βα)8-barrel enzyme to catalyze related reactions in two different metabolic pathways. Proc Natl Acad Sci USA. 2000;97:9925–9930. doi: 10.1073/pnas.160255397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keseler IM, Collado-Vides J, Gama-Castro S, Ingraham J, Paley S, Paulsen IT, Peralta-Gil M, Karp PD. EcoCyc: a comprehensive database resource for Escherichia coli. Nucleic Acids Res. 2005;33:D334–D337. doi: 10.1093/nar/gki108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr Opin Chem Biol. 2006;10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge University Press; Cambridge: 1983. [Google Scholar]
- Kitagawa M, Ara T, Arifuzzaman M, Ioka-Nakamichi T, Inamoto E, Toyonaga H, Mori H. Complete set of ORF clones of Escherichia coli ASKA library (a complete set of E. coli K-12 ORF archive): unique resources for biological research. DNA Res. 2005;12:291–299. doi: 10.1093/dnares/dsi012. [DOI] [PubMed] [Google Scholar]
- Klem TJ, Davisson VJ. Imidazole glycerol phosphate synthase: the glutamine amidotransferase in histidine biosynthesis. Biochemistry. 1993;32:5177–5186. doi: 10.1021/bi00070a029. [DOI] [PubMed] [Google Scholar]
- Kuznetsova E, Proudfoot M, Gonzalez CF, et al. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem. 2006;281:36149–36161. doi: 10.1074/jbc.M605449200. 14 co-authors. [DOI] [PubMed] [Google Scholar]
- Liu JQ, Dairi T, Itoh N, Kataoka M, Shimizu S, Yamada H. Gene cloning, biochemical characterization and physiological role of a thermostable low-specificity l-threonine aldolase from Escherichia coli. Eur J Biochem. 1998;255:220–226. doi: 10.1046/j.1432-1327.1998.2550220.x. [DOI] [PubMed] [Google Scholar]
- Maxon ME, Redfield B, Cai XY, Shoeman R, Fujita K, Fisher W, Stauffer G, Weissbach H, Brot N. Regulation of methionine synthesis in Escherichia coli: effect of the MetR protein on the expression of the metE and metR genes. Proc Natl Acad Sci USA. 1989;86:85–89. doi: 10.1073/pnas.86.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlo LM, Lunzer M, Dean AM. An empirical test of the concomitantly variable codon hypothesis. Proc Natl Acad Sci USA. 2007;104:10938–10943. doi: 10.1073/pnas.0701900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller BG, Raines RT. Identifying latent enzyme activities: substrate ambiguity within modern bacterial sugar kinases. Biochemistry. 2004;43:6387–6392. doi: 10.1021/bi049424m. [DOI] [PubMed] [Google Scholar]
- Miller BG, Raines RT. Reconstitution of a defunct glycolytic pathway via recruitment of ambiguous sugar kinases. Biochemistry. 2005;44:10776–10783. doi: 10.1021/bi0506268. [DOI] [PubMed] [Google Scholar]
- O’Brien PJ, Herschlag D. Catalytic promiscuity and the evolution of new enzymatic activities. Chem Biol. 1999;6:R91–R105. doi: 10.1016/S1074-5521(99)80033-7. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Springer-Verlag; New York: 1970. [Google Scholar]
- Orengo CA, Thornton JM. Protein families and their evolution—a structural perspective. Annu Rev Biochem. 2005;74:867–900. doi: 10.1146/annurev.biochem.74.082803.133029. [DOI] [PubMed] [Google Scholar]
- Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieper U, Eswar N, Davis FP, et al. MODBASE: a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 2006;34:D291–D295. doi: 10.1093/nar/gkj059. 14 co-authors. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth C, Rastogi S, Arvestad L, Dittmar K, Light S, Ekman D, Liberles DA. Evolution after gene duplication: models, mechanisms, sequences, systems, and organisms. J Exp Zoolog B Mol Dev Evol. 2007;308B:58–73. doi: 10.1002/jez.b.21124. [DOI] [PubMed] [Google Scholar]
- Rubino SD, Nyunoya H, Lusty CJ. In vivo synthesis of carbamyl phosphate from NH3 by the large subunit of Escherichia coli carbamyl phosphate synthetase. J Biol Chem. 1987;262:4382–4386. [PubMed] [Google Scholar]
- Schmidt DM, Mundorff EC, Dojka M, Bermudez E, Ness JE, Govindarajan S, Babbitt PC, Minshull J, Gerlt JA. Evolutionary potential of (β/α)8-barrels: functional promiscuity produced by single substitutions in the enolase superfamily. Biochemistry. 2003;42:8387–8393. doi: 10.1021/bi034769a. [DOI] [PubMed] [Google Scholar]
- St Martin EJ, Mortlock RP. Natural and altered induction of the l-fucose catabolic enzymes in Klebsiella aerogenes. J Bacteriol. 1976;127:91–97. doi: 10.1128/jb.127.1.91-97.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. doi: 10.1038/nsb881. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37:630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye QZ, Liu J, Walsh CT. p-Aminobenzoate synthesis in Escherichia coli: purification and characterization of PabB as aminodeoxychorismate synthase and enzyme X as aminodeoxychorismate lyase. Proc Natl Acad Sci USA. 1990;87:9391–9395. doi: 10.1073/pnas.87.23.9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.