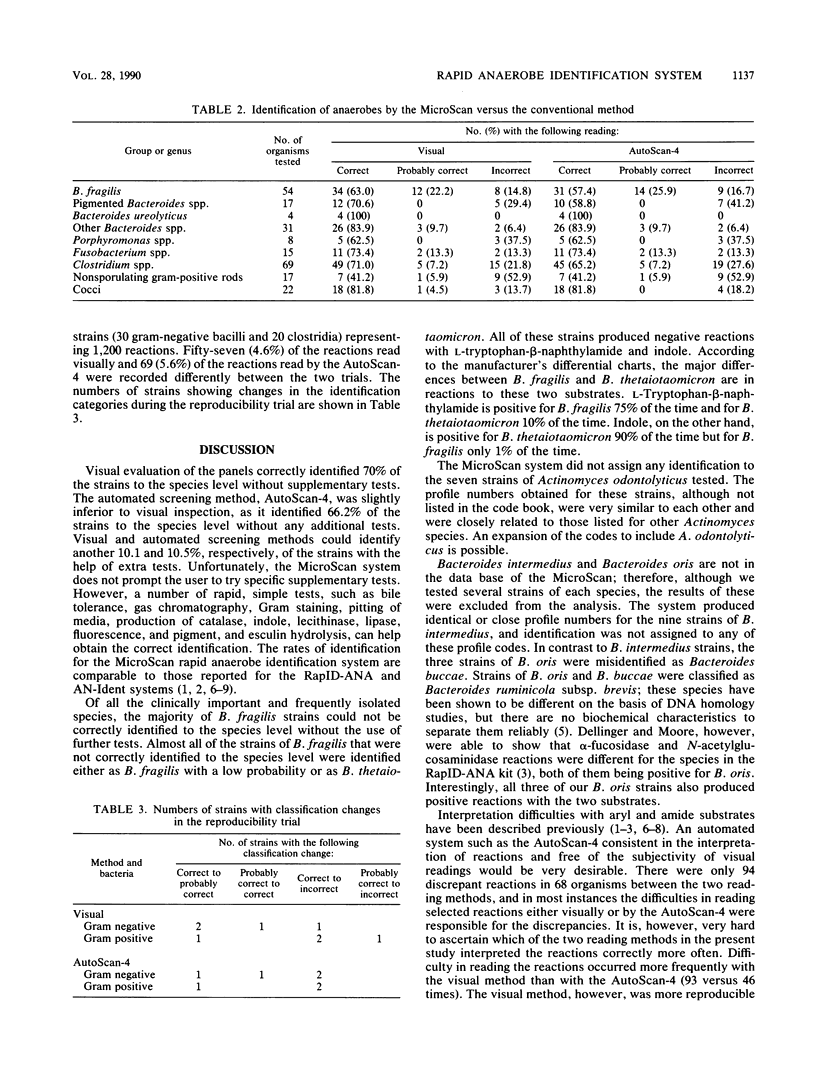
Abstract
Rapid anaerobe identification (MicroScan) panels (4 h) were evaluated both visually and by the AutoScan-4, a computer-controlled microplate reader. The results of both reading methods were compared with identifications obtained by the conventional (Virginia Polytechnic Institute) method. In total, 237 anaerobes were tested. Correct identifications were obtained for 166 strains (70%) by visual reading and 157 strains (66.2%) by the AutoScan-4. Supplementary tests resulted in 80.1 and 76.7% total correct identifications, respectively. Comparison of the two reading methods revealed complete agreement for 169 strains. Differences between the two reading methods were due to difficulties in reading specific reactions. This was especially true with the clostridial species. The performance of the MicroScan system in the identification of anaerobic bacteria appears comparable to that of other 4-h identification systems for anaerobes, but this system shows significant variance from the conventional system. Improvements in the trays and data base are required before the system can be recommended for routine use.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appelbaum P. C., Kaufmann C. S., Depenbusch J. W. Accuracy and reproducibility of a four-hour method for anaerobe identification. J Clin Microbiol. 1985 Jun;21(6):894–898. doi: 10.1128/jcm.21.6.894-898.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burlage R. S., Ellner P. D. Comparison of the PRAS II, AN-Ident, and RapID-ANA systems for identification of anaerobic bacteria. J Clin Microbiol. 1985 Jul;22(1):32–35. doi: 10.1128/jcm.22.1.32-35.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellinger C. A., Moore L. V. Use of the RapID-ANA System to screen for enzyme activities that differ among species of bile-inhibited Bacteroides. J Clin Microbiol. 1986 Feb;23(2):289–293. doi: 10.1128/jcm.23.2.289-293.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain Z., Lannigan R., Schieven B. C., Stoakes L., Kelly T., Groves D. Comparison of RapID-ANA and Minitek with a conventional method for biochemical identification of anaerobes. Diagn Microbiol Infect Dis. 1987 May;7(1):69–72. doi: 10.1016/0732-8893(87)90073-3. [DOI] [PubMed] [Google Scholar]
- Karachewski N. O., Busch E. L., Wells C. L. Comparison of PRAS II, RapID ANA, and API 20A systems for identification of anaerobic bacteria. J Clin Microbiol. 1985 Jan;21(1):122–126. doi: 10.1128/jcm.21.1.122-126.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray P. R., Weber C. J., Niles A. C. Comparative evaluation of three identification systems for anaerobes. J Clin Microbiol. 1985 Jul;22(1):52–55. doi: 10.1128/jcm.22.1.52-55.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summanen P., Jousimies-Somer H. Comparative evaluation of RapID ANA and API 20 A for identification of anaerobic bacteria. Eur J Clin Microbiol Infect Dis. 1988 Dec;7(6):771–775. doi: 10.1007/BF01975045. [DOI] [PubMed] [Google Scholar]


