Abstract
K-Ras mutations are frequently found in various cancers, and are associated with resistance to treatment or poor prognosis. Similarly, poor outcomes have recently been observed in cancer patients with overexpression of protein kinase C iota (PKCι), an atypical protein kinase C that is activated by oncogenic Ras protein and is required for K-Ras-induced transformation and colonic carcinogenesis in vivo. Thus far, there is no effective agent for treatment of cancers with K-Ras mutations or PKCι overexpression. By synthetic lethality screening, we identified a small compound (designated oncrasin-1) that effectively kills various human lung cancer cells with K-Ras mutations at low or submicromolar concentrations. The cytotoxic effects correlated with apoptosis induction as was evidenced by increase of apoptotic cells and activation of caspase-3 and caspase-8 upon the treatment of oncrasin-1 in sensitive cells. Treatment with oncrasin-1 also led to abnormal aggregation of PKCι in nucleus of sensitive cells but not in resistant cells. Furthermore, oncrasin-1 induced apoptosis was blocked by siRNA of K-Ras or PKCι suggesting that oncrasin-1 is targeted to a novel K-Ras/PKCι pathway. The in vivo administration of oncrasin-1 suppressed the growth of K-ras mutant human lung tumor xenografts by >70% and prolonged the survival of nude mice bearing these tumors, without causing detectable toxicity. Our results indicate that oncrasin-1 or its active analogues could be a novel class of anticancer agents which effectively kill K-Ras mutant cancer cells.
Keywords: Synthetic Lethality, Cancer, K-Ras, PKCiota
Introduction
Activating mutations of three oncogenic Ras genes—H-Ras, K-Ras, and N-Ras—play important roles in tumorigenesis and the maintenance of malignant phenotypes (1). Among them, K-Ras mutations are the most frequently found in tumors, especially adenocarcinomas of the pancreas, colon, and lung (1,2). K-Ras mutations are also associated with resistance to chemotherapy and radiotherapy, and thus a poor prognosis (3). Therefore, mutant Ras proteins are important targets for anticancer therapy.
As a subfamily of small guanine nucleotide-binding proteins, Ras proteins cycle between an active GTP-bound form and an inactive GDP-bound form (4). Binding of Ras with GTP is facilitated by guanine nucleotide exchange factors through catalyzing the release of GDP and is required for the interaction of Ras with target proteins (5). Ras mutations that diminish the GTPase activity or decrease the GDP binding capacity render Ras constitutively active and GTP bound. Ras protein can also be activated by other mechanisms. In the absence of a Ras mutation, increased Ras activity is frequently detected in human cancer because of gene amplification (6), overexpression, and an increase in upstream signals from tyrosine-kinase growth-factor receptors such as Her2 (7).
Because Ras proteins must be translocated to the inner leaflet of the plasma membrane to be activated, agents were developed that interrupt the posttranslational modifications required for Ras trafficking to the plasma membrane as a means of suppressing Ras function. One group of such agents is the farnesyltransferase inhibitors (FTIs), which have now been intensively investigated in preclinical and clinical trials as a cancer therapy (8). This approach, however, may be effective in preventing the membrane translocation of H-Ras, but not K-Ras and N-Ras, because in the presence of FTIs, N-Ras and K-Ras proteins are geranylgeranylated and transferred to the membrane (9,10). Several phase II and phase III clinical trials also showed that FTIs did not have significant single-agent activity in lung, pancreatic, colorectal, bladder, and prostate cancers (8). Thus, compounds with novel mechanisms of action and high specificity for cancer cells with hyperactive Ras will be valuable for anticancer therapy.
Synthetic lethality screening has recently merged as a new approach to identify cytotoxic agents targeted to cancer cells with mutations in a particular gene (11). Using genetically defined cell lines to screen compounds that kill cells with a particular mutant gene only but not the normal counterparts will allow us to identify compounds that target to a protein whose functional change is synthetic lethal to an inactivated tumor suppressor gene or an activated oncogene. Using this approach, we identified a small molecule that can effectively kill K-Ras mutant cancer cells, but not normal isogenic cells or H-Ras or N-Ras mutant cancer cells. The mechanistic studies revealed that apoptosis induction by this compound is blocked by knockdown of K-Ras or PKCι, suggesting that oncrasin-1 is synthetic lethal to active K-Ras and PKCι.
Materials and Methods
Cell lines
The human non-small cell lung carcinoma H1299, H322, H460, H358, H2887, H2087, and A549 cell lines were routinely propagated in a monolayer culture in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 100 units/ml penicillin, and 100 mg/ml streptomycin. The establishing of T29, T29Kt1, and T29Ht1 cells and in vitro and in vivo biological characterization of those cell lines have been reported previously (12). In vitro cell growth and/or cell cycle progression of those three cell lines were similar. The cells were maintained in Dulbecco’s modified Eagle’s medium with the same supplements. All cells were maintained in the presence of 5% CO2 at 37°C.
Chemicals and antibodies
A chemical library with 10,000 compounds, including oncrasin-1, was obtained from Chembridge Corporation (San Diego, CA). The chemicals in the library are provided at a concentration of 5 mg/ml in dimethyl sulfoxide (DMSO). Antibodies to the following proteins were used for the Western blot analysis: PKCι, PKCζ, Raf-1 and caspase-3 (Santa Cruz Biotechnology, Santa Cruz, CA); caspase-8 (PharMingen, San Diego, CA); and β-actin (Sigma, St. Louis, MO).
Cell viability assay
The inhibitory effects of oncrasin-1 and other agents on cell growth were determined by using the sulforhodamine B (SRB) assay, as described previously (13). Each experiment was performed in quadruplicate and repeated at least three times. The IC50 value, a dose that causes a 50% reduction in surviving cells as compared with the number of control cells, was determined by using the CurveExpert Version 1.3 program.
Flow cytometry assay
Cells were seeded at a density of 1 × 106 cells per dish in 100-mm dishes and allowed to grow overnight. After the treatment, cells were harvested with trypsin, washed twice with phosphate-buffered saline (PBS). Cells were fixed with 75% ethanol at the cell density of 1 × 106 cells/ml overnight. Next, the cell pellets were harvested and re-suspended with propidium iodide (PharMingen) for 15 minutes in the dark at room temperature. The cells were analyzed using an EPICS Profile II flow cytometer (Coulter Corp., Hialeah, FL) with the Multicycle Phoenix Flow Systems program (Phoenix Flow Systems, San Diego, CA). All experiments were repeated three times.
Western blot and Immunofluorescent staining
Western blot analysis was performed as we have previous described (14). For immunofluorescent staining, cells were seeded at a density of 1X105 cells per well in six well plates containing a 1% gelatin-treated cover slide. Cells were allowed to grow overnight. Cells were treated with different compounds or radiation as indicated. After the treatment, cells were washed with PBS twice, then fixed with 2% paraformaldehyde 20 min, permeablized with 0.1% Triton-100 20 min, blocked with 5% normal goat serum 1 h. The slides were incubated with primary antibodies followed by FITC or Rhodamine-linked secondary antibodies. After PBS wash three times, the slides were taken out and mounted with Prolong Gold antifade reagent (Molecular Probes, Carsbad, CA). The slides were read under Olympus fluorescence microscope (Olympus, Melville, NY).
Animal experiments
Animal experiments were carried out in accordance with Guidelines for the Care and Use of Laboratory Animals (NIH publication number 85–23) and the institutional guidelines of M. D. Anderson Cancer Center. Subcutaneous tumors were established in 4- to 6-week-old female nude mice (Charles River Laboratories Inc., Wilmington, MD) by inoculation of 1.5 × 106 H460 cells into the dorsal flank of each mouse. After the tumors grew to 2–3 mm in diameter, the mice were treated with intraperitoneal administration of oncrasin-1 at a dose of 100 mg/kg/day (dissolved in 0.5 ml solvent containing 10% DMSO, 10% Cremophore EL, and 10% ethanol) or solvent alone. Tumor volumes were calculated by using the formula a × b2 × 0.5, where a and b represented the larger and smaller diameters, respectively. Mice were killed when the tumors grew to 15 mm in diameter. To evaluate the toxicity of treatment, blood samples were collected from the tail vein before treatment and 2 days after the last treatment, and serum alanine transaminase, aspartate transaminase, blood urea nitrogen, and creatinine levels were determined as described elsewhere (15).
Statistical analysis
Differences between the treatment groups were assessed by analysis of variance (ANOVA) using statistical software (StatSoft, Tulsa, OK). Differences between the results of the in vivo tumor growth experiment were assessed using ANOVA with a repeated measurement module. P values of < 0.05 were regarded as significant.
Results
Library screening for oncogenic Ras-targeted compounds
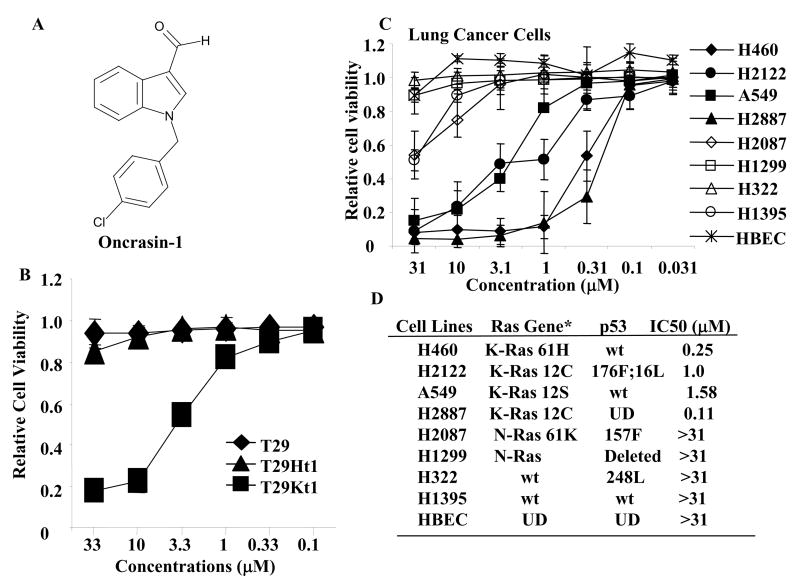
We used human ovarian surface epithelial cells immortalized with the catalytic subunit of human telomerase reverse transcriptase and the SV40 early genomic region (designated T29), and its tumorigenic derivatives transformed with either mutant H-Ras (T29Ht1) or mutant K-Ras (T29Kt1) [12], to screen a chemical library from Chembridge Corporation (San Diego, CA) for compounds that showed an ability to selectively kill tumor cells. Cells seeded in a 96-well plate were treated with each compound at a final concentration of about 5 μg/ml (about 20–30 μM). Cells treated with solvent (dimethyl sulfoxide [DMSO]) were used as controls. A lethal effect was determined in a sulforhodamine B (SRB) assay 4 days after treatment [13]. Compounds found to kill more than 50% of cells were then subjected to two additional dose-response tests to confirm the SRB assay finding. Among 10,000 compounds screened, we identified a compound, 1-[(4-chlorophenyl)methyl]-1H-indole-3-carboxaldehyde, designated oncrasin-1 (oncogenic Ras tumor-inhibiting compound 1), that was highly selective for T29Kt1 cells at a wide range of doses (Fig. 1AB). Oncrasin-1 induced dose-dependent cytotoxicity in T29Kt1 cells, with a 50% inhibitory concentration (IC50) of 4.81 μM. In contrast, no cytotoxicity was detectable in T29 or T29Ht1 cells with concentrations of 33 μM, the highest we tested.
Figure 1.
Library screening. (A) Chemical structures of oncrasin-1. (B) Dose effect of oncrasin-1 in T29, T29Ht1, and T29Kt1 cells. The cells were treated with various concentrations (ranging from 0.1 μM to 33 μM) of oncrasin-1. Cell viability was determined 3 days after treatment by SRB assays. Control cells were treated with solvent (DMSO), and their value was set as 1. (C) Dose-Response in human lung cancer cell lines. Lung cancer cells with various oncogenic Ras gene status were treated with oncrasin-1 at various concentrations. Cell viability was determined as in B. The values shown are the means + SD of two assays done in quadruplicate. Control cells were treated with solvent (DMSO), whose value was set as 1. (D) The status of Ras gene and p53 gene mutations, and IC50 of oncrasin-1 in lung cancer cell lines tested in C. *Based on published data and the Cancer Genome Project Database (http://www.sanger.ac.uk/genetics/CGP/). Wt, wild type; UN, unknown; HBEC, human bronchial epithelial cells.
Testing of oncrasin-1 on native human cancer cell lines showed that it effectively killed K-Ras–mutant H460, H2122, H2887, and A549 cells with an IC50 of ≤3 μM (Figure 1C). However, oncrasin-1 had minimal effects on normal human bronchial epithelial cells. It also had minimal effects on H322 and H1395 lung cancer cells, which have wild-type Ras genes, and on H1299 and H2087 lung cancer cells, which harbor mutant N-Ras genes. Those results suggested that oncrasin-1 is effective against various lung cancer cells with K-Ras mutations.
Induction of apoptosis by oncrasin-1
Chemically, this compound has the same core structure as indole-3-carbinol, a naturally occurring constituent of many plant foods that has been tested for the prevention and treatment of cancer (16). It has also structural similarity to lonidamine (17), which have been evaluated preclinically and clinically for treatment of cancers. However, both indole-3-carbinol and lonidamine did not have any cytotoxic effects in T29, T29Kt1, T29Ht1, and H460 cells at any of the concentrations tested (up to 100 μM; data not shown), suggesting that oncrasin-1 may have different anticancer mechanisms from those two agents.
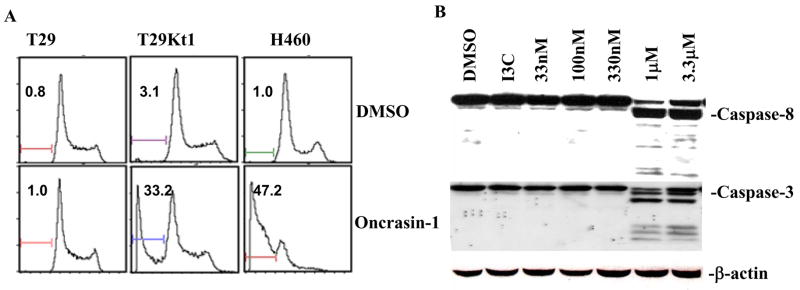
To determine whether the antitumor activity of oncrasin-1 is due to the suppression of cell proliferation or to cell killing, we performed flow cytometric analysis after treatment with oncrasin-1 at 10 μM (for T29 or T29Kt1 cells) or 1 μM (for H460 cells), the doses of about IC80 for T29Kt1 and H460, respectively. At 12 h after the treatment with oncrasin-1, apoptotic cells account for 47.2% in H460 and 33.2% in T29Kt1 cells (Fig. 2A). In contrast, apoptotic cells in H460 and T29Kt1 cells, which were treated with dimethyl sulfoxide (DMSO), and in the T29 cells treated with oncrasin-1 were <3.1%. This result indicated that oncrasin-1 can effectively induce cell killing in T29Kt1 and H460 cells and that apoptosis induction is a major mechanism of oncrasin-1-induced cell killing. Western blot analysis showed further that the treatment of H460 cells with 1 μM oncrasin-1 effectively activated caspases 3 and 8 (Fig. 2B), whereas treatment with DMSO or indole-3-carbinol at 30 μM did not induce detectable cleavages of the caspases, indicating that its cytotoxic effect in cancer cells is due to its induction of apoptosis.
Figure 2.
Apoptosis induction by oncrasin-1. (A) T29, T29Kt1, and H460 cells were treated with 10 μM (for T29 or T29Kt1) or 1 μM (for H460) oncrasin-1 and then harvested 12 hours later. Apoptosis was detected by flow cytometiric analysis. The number in each panel indicates percentage of apoptotic cells. (B) Western blot analysis. H460 cells were treated with various concentrations of oncrasin-1 as indicated or with 30 μM indole-3-carbinol (I3C), an inactive analogue, for 24 hours. Activation of caspases 3 and 8 was detected by Western blot analysis. β-actin was used as the loading control.
K-Ras is required for effective apoptosis induction by oncrasin-1
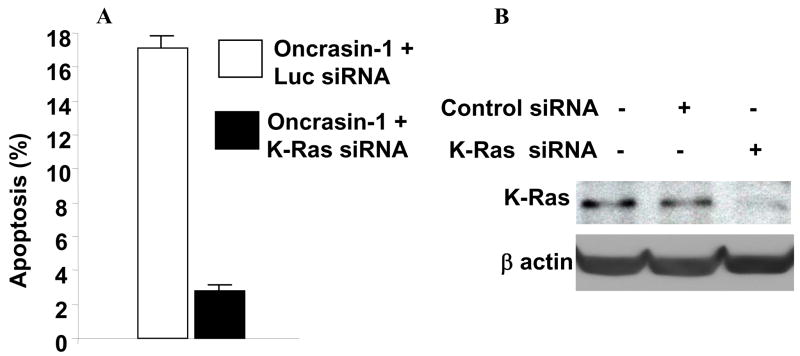
Because oncrasin-1 was identified by synthetic lethality screening of K-Ras-mutant cells, we investigated the role of the K-Ras gene in oncrasin-induced cell death. We treated H460 cells with 200 pM K-Ras-specific siRNA or control siRNA for 24 h. The cells were then treated with DMSO or 1 μM oncrasin-1. After another 12 h, the cells were harvested for apoptosis detection using fluorescence-activated cell-sorting (FACS) analysis. The cell lysate was also used to detect K-Ras gene expression. The results showed that treatment with K-Ras-specific siRNA but not with control siRNA suppressed K-Ras expression in H460 cells. Cells transfected with either K-Ras siRNA or control siRNA alone had a background apoptosis level of about 9–11%. Nevertheless, in cells treated with control siRNA, oncrasin-1 resulted in an increase of about 17% of apoptotic cells, compared with that of DMSO + siRNA treated cells. In contrast, in K-Ras siRNA-treated cells, treatment with oncrasin-1 yielded only about 3% increase in apoptotic cells when compared with DMSO + siRNA treated cells (Fig. 3). This result demonstrated that K-Ras activity is required for oncrasin-1 to induce apoptosis in H460 cells.
Figure 3.
K-Ras knockdown inhibited oncrasin-mediated apoptosis. (A) H460 human lung cancer cells were treated with either control (luciferase [Luc]) siRNA or K-Ras siRNA and then treated with DMSO or 1 μM oncrasin-1 for 12 hours. The values represent differences of percentage of apoptotic cells between oncrasin-1 + siRNA and DMSO + siRNA groups. The values shown are the means + SD of two experiments. (B) Western blot analysis of siRNA-mediated K-Ras knockdown in H460 cells. cells treated with phosphate-buffered saline, K-Ras siRNA, or control siRNA were harvested for testing K-Ras proteins by Western blot analysis. β-Actin was used as the loading control.
Oncrasin-1 induced abnormal aggregation of PKCiota in nucleus
To investigate the molecular mechanisms of apoptosis induction by oncrasin-1, we determined the levels and/or phosphorylation status of several proteins that are involved in apoptosis and/or Ras signaling pathways, including Bax, Bik, Bcl2, and Bcl-XL in apoptosis pathway, and Raf-1, B-Raf, Akt, Mst1, and the atypical protein kinases C (aPKCs), PKCζ and PKCι in Ras signaling pathway. Western blot analysis of those molecules produced little elucidative information about mechanisms of oncrasin-1 induced apoptosis (Data not shown).
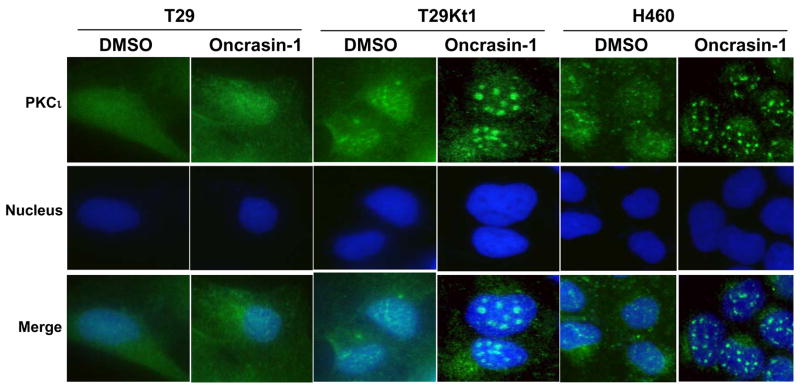
Because Ras proteins execute their biological functions mainly in cell membranes, through interacting with a diversity of membrane receptors and modulating signal transduction of a variety signaling pathways that govern cell growth, proliferation, differentiation and death, we then tested the subcellular localization of several molecules involved in the Ras signaling pathway. Treatment with oncrasin-1 induced no obvious changes in the subcellular localization of Raf-1, B-Raf, Akt, PKCzeta;, PKCdelta, and p53. However, a substantial change in the subcellular localization of PKCι was detected after oncrasin-1 treatment (Figure 4). In both H460 and T29Kt1 cells, both PKCι and PKCζ were diffusely distributed, with high concentrations or tiny dots on some area, especially in the nucleus and on the cell membrane, findings that are consistent with previous ones that aPKCs contain both nuclear localization signals and nuclear export signals and can shuttle between the cytoplasm and the nucleus (18). Whereas treatment with oncrasin-1 resulted in no obvious changes in the subcellular localization of PKCζ, the same treatment resulted in formation of large foci of PKCι in nuclei of both T29Kt1 and H460 cells (Figure 4) but not in oncrasin-1-resistant T29 and H1299 cells. We then tested whether aggregation of PKCι into large nuclear foci is inducible by other chemotherapeutic agents, such as 5-fluorouracil and paclitaxel. The results showed that treatment of T29Kt1 cells with the chemotherapeutic agents 5-fluorouracil, paclitaxel or with radiation at doses that resulted in cell killing similar to that caused by oncrasin-1 did not induce aggregation of PKCι in the nuclei, suggesting that such aggregation is not a general phenomenon occurring in dying cells (data not shown).
Figure 4.
Oncrasin-induced aggregation of PKCι T29, T29Kt1, and H460 cells were treated with DMSO or 10 μM (for T29 or T29Kt1) or 1 μM (for H460) oncrasin-1 for 12 hours, and then immunofluorescent staining was performed to test for intracellular localization of PKCι. Nuclear PKCι aggregated into large foci after oncrasin-1 treatment in sensitive T29Kt1 and H460 cells but not in the resistant T29 cells.
Roles of PKCiota in oncrasin-induced apoptosis
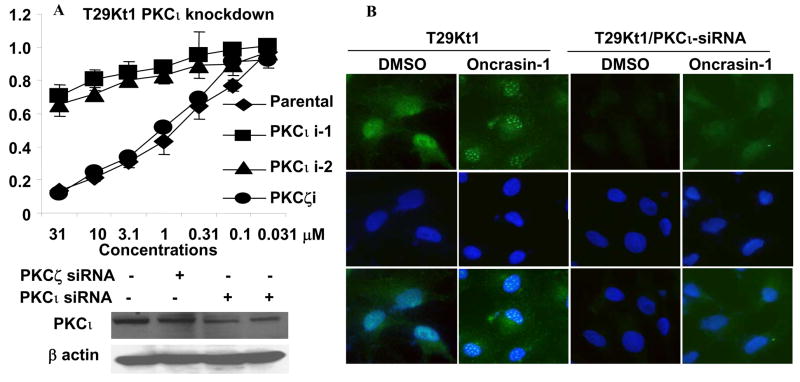
To investigate the possible role of PKCι in oncrasin-1-mediated cytotoxicity, we knocked down PKCι in T29Kt1 cells by using plasmids encoding shRNA for PKCι or PKCζ provided by the Origene Corporation (Rockville, MD). The plasmids were transfected to T29Kt1 cells by using FuGENE 6 (Roche Diagnostics Corp., Indianapolis, IN) as instructed by the manufacturer. Cells were pooled together after a brief selection with puromycin and tested for levels of PKCι expression. Cell viability analysis showed that stable knockdown of PKCι about 50% in T29Kt1 cells by siRNA constructs resulted in almost complete resistance to oncrasin-1 (Figure 5A). The IC50 in PKCι-knockdown T29Kt1 cells was 100 times higher than it was in parental T29Kt1 cells. This value was comparable to that of oncrasin-resistant T29 cells. In contrast, transfection with PKCζ shRNA encoded by the same vector system did not change the susceptibility of T29Kt1 cells to oncrasin-1. Immunofluorescent analysis showed that knockdown of PKCι in T29Kt1 cells abolished the abnormal aggregation of nuclear PKCι (Fig. 5B), consisting with the results observed in T29 cells. Together, those results indicated that oncrasin-1 has synthetic lethality for PKCι.
Figure 5.
Effects of PKCι on oncrasin-1-induced cytotoxicity. (A) Susceptibility to oncrasin-1 after PKCι Knockdown. Six plasmids encoding PKCι specific shRNA were divided into two groups (PKCι i-1 and PKCι i-2) and were used in the experiment. PKCζ shRNA plasmids (a mixture of 4) were used as a control. Cell viability was determined as described in Figure 1. Western blot analysis of PKCι in T29Kt1 cells after plasmid transfection and a brief selection was also shown. (B) Knockdown of PKCι abolished abnormal nuclear PKCι aggregation. The experiment was performed as described in Figure 4.
Antitumor activity in vivo
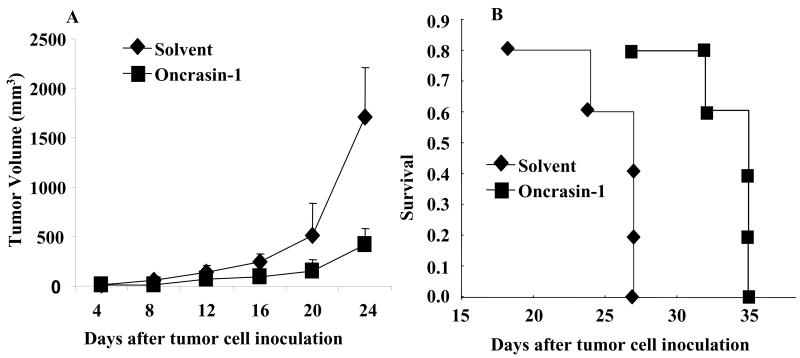
We also investigated the in vivo antitumor activity of oncrasin-1. For this purpose, 1.5 × 106 H460 cells were inoculated into the dorsal flank of each nude mouse. After the tumors grew to 2–3 mm in diameter, the mice were treated with intraperitoneal injections of oncrasin-1 for 10 days at a dose of 100 mg/kg/injection daily, or they were given intraperitoneal injections of solvent alone. The tumor volumes were monitored every other day. Blood samples were collected from the tail vein before treatment and 2 days after the last treatment for testing of serum liver enzymes. The results showed that, in comparison with solvent, oncrasin-1 suppressed tumor growth by 75.4% (Fig. 6A). Treatment with oncrasin-1 also prolonged survival (Fig. 6B): the mean survival times for mice treated with solvent and oncrasin-1 were 24 and 32, respectively. No differences were observed in the body weights of mice treated with solvent or oncrasin-1. In addition, the serum ALT, AST, and creatinine levels were within the normal ranges in all mice tested, regardless of the treatment received, suggesting that in vivo antitumor activity can be achieved without observable toxicity. Those data demonstrated that oncrasin-1 might be useful for the treatment of cancers with K-Ras mutations.
Figure 6.
Antitumor activity in vivo. (A) Tumor growth. Mice with subcutaneous tumors derived from H460 cells were treated with oncrasin-1 or solvent. Tumor volumes were monitored over time after the treatments. The values are the means ± SD of data from 5 mice per group. The mean tumor volume in the mice treated with oncrasin-1 differed significantly from that of the solvent-treated mice (p <.05). (B) Survival curve. The mean survival times in mice treated with solvent and oncrasin-1 were 24 and 32 days, respectively.
Discussion
Synthetic lethality was originally referred to as a lethal phenotype caused by mutations of 2 genes (19); i.e., mutations of the 2 genes are viable if they occur separately, but if they occur together, the combination is lethal. A synthetic lethal phenotype often indicates that the 2 genes or 2 related pathways affect a common essential biologic function. Assume that the biologic event E, which is critical for the cell to survive, is affected by signals A and B. Signal A is transduced via proteins A1, A2, and A3, whereas signal B is transduced via B1, B2, and B3 and then to E. An abnormality occurring in either signal pathway is insufficient to cause dysfunction of E; therefore, cell is viable. However, abnormalities in both signal pathways lead to the disorder of E and thus to cell death. In this case, A1 is synthetic lethal with B1, B2, or B3, but not with A2 or A3, and vise verse. Nevertheless, our current knowledge of molecular networks in normal or cancer cells is not adequate for us to predict what genes are synthetic lethal to an oncogene. The advantage of synthetic lethality screening is that a compound targeting to a protein that is synthetic lethal to an oncogenic protein can be identified, without prior knowledge of such a protein target. The disadvantage of this approach is that identifying targets of such a compound remains challenging. On the basis of our results, we hypothesize that oncrasin-1 affect some cellular targets that share an essential biologic function with the K-Ras/PKCiota pathway. Oncrasin-1 may not function by inhibition of K-Ras or PKC⌈ activities but require K-Ras or PKCι activity for its apoptosis-induction in cancer cells, because it did not inhibit PKCiota activity in an in vitro enzyme assay with recombinant PKCiota protein nor inhibit S35-GTPγ binding to K-Ras (data not shown), and because the siRNAs of K-Ras and PKCiota induced an antagonistic but not an additive effect with oncrasin-1. Although the direct target or targets of the oncrasin-1 remain to be characterized, our results led us to hypothesize that it affects some cellular targets whose reduced or enhanced activity is lethal for cancer cells with increased K-Ras and/or PKCiota activity. In fact, oncogenic Ras can induce either cell transformation or apoptosis, depending on cell types and contexts (20,21). Expression of oncogenic Ras in primary human or rodent cells often results in apoptosis or senescence, whereas expression of oncogenic Ras in immortal cells or cells with inactivation of p53, p16, or the transcriptional activator interferon regulatory factor 1 leads to transformation and tumorigenesis (20,21). It is possible that, in comparison with cancer cells, the cellular targets of oncrasin-1 in primary cells are at hypo- or hyper-status that is not viably compatible with increased K-Ras/PKCiota activity.
K-Ras mutations are frequently found in various types of human cancer and often associated with resistance to radiotherapy and chemotherapy, including novel anticancer agents such as cetuximab and erlotinib (22). As a result, the outcome for cancer patients with K-Ras mutations is frequently poor (23). Similarly, poor outcomes have recently been observed in cancer patients with overexpression of PKCiota (24,25). Unlike other protein kinase C, PKCζ and PKCι are insensitive to regulation by diacylglycerol, Ca2+, or phorbol esters but are activated by phosphoinositide 3-kinase (PI3K) and its lipid product phosphotidylinositol-3,4,5-triphosphate (PIP3) (26). Phosphorylation of Thr410 (PKCζ)/Thr403 (PKCι) in aPKCs by 3-phosphoinositide-dependent protein kinase-1 (PDK1) is PI3K dependent and serves as a direct on–off switch for the aPKCs (27). PDK1 is a constitutively active kinase, although its access to substrates is regulated by phosphoinositides (28). Ras directly interacts in a GTP-dependent manner with the catalytic subunit of the PI3Ks and activates them (29,30), leading to the generation of a short-lived second-messenger product such as phosphatidylinositol 3,4,5-phosphate (PIP3) and to the activation of many PI3K/PDK1-dependent kinases, including aPKCs (27) and Akt (31). Moreover, Ras proteins can directly interact with aPKCs in vitro and in vivo, regulating the activities of the aPKCs (32). Thus, the aPKCs might function as Ras downstream effectors, mediating signal transduction in certain important Ras signaling pathways (33). A recent study showed that PKCι is required for K-Ras-induced transformation and colonic carcinogenesis in vivo (34). Nevertheless, how K-Ras and PKCι interact and what signal transduction they mediate remain to be characterized. Our study showed that treatment with oncrasin-1 led to aggregation of nuclear PKCι into large foci. Whether this abnormal PKCι aggregation is related to oncrasin-1 induced apoptosis is not yet clear, although this phonotype is only observed in sensitive cells. Because of the critical roles of increased K-Ras/PKCι activity in oncogenesis and in the poor outcome of cancer patients (23,25), oncrasin-1 or its analogues that can induce synthetic lethality with oncogenic K-Ras and PKCι can be useful agents for cancer treatment or useful tools for characterize biological functions of oncogenic K-Ras and PKCι, even though the direct targets of oncrasin-1 are not yet clear.
Acknowledgments
We thank Dr. Jack A. Roth for helpful suggestions, Karen F. Philips for editorial review on the manuscript, Li Wang, Henry Peng and Jennifer Wang for technical assistance. This work was supported by National Cancer Institute grants R01 CA 092487 (B. Fang), R01 CA 098582 (B. Fang), lung SPORE (CA 70907) developmental award, and Cancer Center Support Grant CA 16672.
References
- 1.Bos JL, Fearon ER, Hamilton SR, et al. Prevalence of ras gene mutations in human colorectal cancers. Nature. 1987;327:293–7. doi: 10.1038/327293a0. [DOI] [PubMed] [Google Scholar]
- 2.Vogelstein B, Fearon ER, Hamilton SR, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319:525–32. doi: 10.1056/NEJM198809013190901. [DOI] [PubMed] [Google Scholar]
- 3.Bernhard EJ, Stanbridge EJ, Gupta S, et al. Direct evidence for the contribution of activated N-ras and K-ras oncogenes to increased intrinsic radiation resistance in human tumor cell lines. Cancer Res. 2000;60:6597–6600. [PubMed] [Google Scholar]
- 4.Bar-Sagi D, Hall A. Ras and Rho GTPases: a family reunion. Cell. 2000;103:227–38. doi: 10.1016/s0092-8674(00)00115-x. [DOI] [PubMed] [Google Scholar]
- 5.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nature Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 6.Hoa M, Davis SL, Ames SJ, Spanjaard RA. Amplification of wild-type K-ras promotes growth of head and neck squamous cell carcinoma. Cancer Res. 2002;62:7154–6. [PubMed] [Google Scholar]
- 7.Ehrhardt A, David MD, Ehrhardt GR, Schrader JW. Distinct mechanisms determine the patterns of differential activation of H-Ras, N-Ras, K-Ras 4B, and M-Ras by receptors for growth factors or antigen. Mol Cell Biol. 2004;24:6311–23. doi: 10.1128/MCB.24.14.6311-6323.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sebti SM, Adjei AA. Farnesyltransferase inhibitors. Sem Oncol. 2004;31:28–39. doi: 10.1053/j.seminoncol.2003.12.012. [DOI] [PubMed] [Google Scholar]
- 9.Whyte DB, Kirschmeier P, Hockenberry TN, et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–64. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- 10.James G, Goldstein JL, Brown MS. Resistance of K-RasBV12 proteins to farnesyltransferase inhibitors in Rat1 cells. Proc Natl Acad Sci USA. 1996;93:4454–8. doi: 10.1073/pnas.93.9.4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nature Rev Cancer. 2005;5:689–98. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 12.Liu J, Yang G, Thompson-Lanza JA, et al. A genetically defined model for human ovarian cancer. Cancer Res. 2004;64:1655–63. doi: 10.1158/0008-5472.can-03-3380. [DOI] [PubMed] [Google Scholar]
- 13.Rubinstein LV, Shoemaker RH, Paull KD, et al. Comparison of in vitro anticancer-drug-screening data generated with a tetrazolium assay versus a protein assay against a diverse panel of human tumor cell lines. J Natl Cancer Inst. 1990;82:1113–8. doi: 10.1093/jnci/82.13.1113. [DOI] [PubMed] [Google Scholar]
- 14.Teraishi F, Wu S, Sasaki J, et al. JNK1-dependent antimitotic activity of thiazolidin compounds in human non-small-cell lung and colon cancer cells. Cell Mol Life Sci. 2005;62:2382–89. doi: 10.1007/s00018-005-5365-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gu J, Kagawa S, Takakura M, et al. Tumor-specific transgene expression from the human telomerase reverse transcriptase promoter enables targeting of the therapeutic effects of the Bax gene to cancers. Cancer Res. 2000;60:5359–64. [PubMed] [Google Scholar]
- 16.Chinni SR, Li Y, Upadhyay S, Koppolu PK, Sarkar FH. Indole-3-carbinol (I3C) induced cell growth inhibition, G1 cell cycle arrest and apoptosis in prostate cancer cells. Oncogene. 2001;20:2927–36. doi: 10.1038/sj.onc.1204365. [DOI] [PubMed] [Google Scholar]
- 17.Ravagnan L, Marzo I, Costantini P, et al. Lonidamine triggers apoptosis via a direct, Bcl-2-inhibited effect on the mitochondrial permeability transition pore. Oncogene. 1999;18:2537–46. doi: 10.1038/sj.onc.1202625. [DOI] [PubMed] [Google Scholar]
- 18.White WO, Seibenhener ML, Wooten MW. Phosphorylation of tyrosine 256 facilitates nuclear import of atypical protein kinase C. J Cell Biochem. 2002;85:42–53. [PubMed] [Google Scholar]
- 19.Dobzhansky TH. Genetics of natural populations. XIII Recombination and variability and populations of drosophila pseudoobscura. Genetics. 1946;31:269–90. doi: 10.1093/genetics/31.3.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tanaka N, Ishihara M, Kitagawa M, et al. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell. 1994;77:829–39. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 21.Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- 22.Lievre A, Bachet JB, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–5. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 23.Keohavong P, DeMichele MA, Melacrinos AC, Landreneau RJ, Weyant RJ, Siegfried JM. Detection of K-ras mutations in lung carcinomas: relationship to prognosis. Clin Cancer Res. 1996;2:411–8. [PubMed] [Google Scholar]
- 24.Regala RP, Weems C, Jamieson L, et al. Atypical protein kinase C iota is an oncogene in human non-small cell lung cancer. Cancer Res. 2005;65:8905–11. doi: 10.1158/0008-5472.CAN-05-2372. [DOI] [PubMed] [Google Scholar]
- 25.Eder AM, Sui X, Rosen DG, et al. Atypical PKCiota contributes to poor prognosis through loss of apical-basal polarity and cyclin E overexpression in ovarian cancer. Proc Natl Acad Sci USA. 2005;102:12519–24. doi: 10.1073/pnas.0505641102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakanishi H, Brewer KA, Exton JH. Activation of the zeta isozyme of protein kinase C by phosphatidylinositol 3,4,5-trisphosphate. J Biol Chem. 1993;268:13–6. [PubMed] [Google Scholar]
- 27.Le Good JA, Ziegler WH, Parekh DB, Alessi DR, Cohen P, Parker PJ. Protein kinase C isotypes controlled by phosphoinositide 3-kinase through the protein kinase PDK1. Science. 1998;281:2042–5. doi: 10.1126/science.281.5385.2042. [DOI] [PubMed] [Google Scholar]
- 28.Pullen N, Dennis PB, Andjelkovic M, et al. Phosphorylation and activation of p70s6k by PDK1. Science. 1998;279:707–10. doi: 10.1126/science.279.5351.707. [DOI] [PubMed] [Google Scholar]
- 29.Rodriguez-Viciana P, Warne PH, Dhand R, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–32. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 30.Pacold ME, Suire S, Perisic O, et al. Crystal structure and functional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–43. doi: 10.1016/s0092-8674(00)00196-3. [DOI] [PubMed] [Google Scholar]
- 31.Stephens L, Anderson K, Stokoe D, et al. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science. 1998;279:710–4. doi: 10.1126/science.279.5351.710. [DOI] [PubMed] [Google Scholar]
- 32.Diaz-Meco MT, Lozano J, Municio MM, et al. Evidence for the in vitro and in vivo interaction of Ras with protein kinase C zeta. J Biol Chem. 1994;269:31706–10. [PubMed] [Google Scholar]
- 33.Berra E, Diaz-Meco MT, Dominguez I, et al. Protein kinase C zeta isoform is critical for mitogenic signal transduction. Cell. 1993;74:555–63. doi: 10.1016/0092-8674(93)80056-k. [DOI] [PubMed] [Google Scholar]
- 34.Murray NR, Jamieson L, Yu W, et al. Protein kinase Ciota is required for Ras transformation and colon carcinogenesis in vivo. J Cell Biol. 2004;164:797–802. doi: 10.1083/jcb.200311011. [DOI] [PMC free article] [PubMed] [Google Scholar]